Abstract
Cytotoxic necrotizing factor 1 (CNF1), the paradigm of Rho GTPase activating bacterial toxins has been shown to promote E. coli invasion of human brain microvascular endothelial cells (HBMEC), which constitute the blood-brain barrier, but its synthesis and secretion is unclear. In this study, we performed mini Tn5 mutagenesis screen to identify genetic requirements for CNF1 production and secretion. Transposon mutagenesis screen of meningitis-causing E. coli K1 strain RS218 revealed that CNF1 production was markedly decreased in a transposon mutant (NBC-28G9) where transposon insertion occurred in the 5′ end of gidA gene. In contrast, total deletion of gidA gene has less drastic effect on the production of CNF1. The N-terminus truncated GidA exhibited dominant negative effect on the production of CNF1. The inhibition of CNF1 production by N-terminus truncated GidA was shown to occur at the translational level. This was supported by our demonstrations that cnf1 mRNA transcription levels did not differ between strains RS218 and NBC-28G9; and the production of recombinant CNF1 under the control of artificial promoter was also repressed by truncated GidA. Progressive deletion of DNA regions in cnf1 gene identified two putative regions that were responsible for translational inhibition mediated by truncated GidA.
Keywords: CNF1, GidA, mRNA, E. coli, gene expression
1. Introduction
CNF1 is a classical A-B toxin, in which a catalytic A domain is located in the C-terminus, and a receptor binding B domain is located in the N-terminus. CNF1 catalyzes the constitutive activation of Rho GTPases by deamidation (Flatau et al., 1997; Schmidt et al., 1997). This activation is, however, attenuated by ubiquitin-mediated proteasomal degradation of activated Rho protein (Doye et al., 2002; Lerm et al., 2002). Rho proteins are essential regulatory molecules, controlling the host cell actin cytoskeleton organization and dynamic, which affect cell polarity, movement, differentiation or phagocytosis (Lemonier et al., 2007; Munro et al., 2005). Both Rho protein activation and deactivation confer phagocytic properties on epithelial and endothelial cells (Doye et al., 2002; Lemonier et al., 2007; Munro et al., 2005).
CNF1 has been shown to be an important virulence factor for meningitis-causing E. coli K1 by contributing to E. coli invasion of human brain microvascular endothelial cells (HBMEC), which constitute the blood-brain barrier, and penetration into the brain (Khan et al., 2002; Kim, 2003; Kim, 2008). This was shown by the demonstration that the CNF1 deletion mutant exhibited significantly decreased HBMEC invasion as well as penetration into the brain compared to the parent E. coli strain despite having similar high levels of bacteremia (Khan et al., 2002).
We have also shown that CNF1 contributes to E. coli invasion of HBMEC and penetration into the brain via its interaction with the receptor, 37 laminin receptor precursor (37LRP)/67 laminin receptor (67LR) (Chung et al., 2003; Kim et al., 2005). CNF1, however, is the cytoplasmic protein, and to exhibit its biological activity in host cells, it has to be produced and secreted. There is little information on the CNF1 production as well as secretion pathway. No typical signal peptide is found in the CNF1 sequence. Recent studies have shown that CNF1 is transported to the culture supernatant in a complex with outer membrane vesicles (OMVs) in uropathogenic E. coli (UPEC) strains J96 and CP9 (Davis et al., 2006; Kouokam et al., 2006). It has been documented that a cis-acting mRNA element corresponding to the first 48 codons of cnf1 is involved in the dual translational regulation of CNF1 synthesis (Fabbri et al., 1999). We have previously demonstrated that SelB is involved in translational regulation of CNF1 expression without incorporation of selenocysteine in CNF1 protein (Yu and Kim, 2011), but the underlying molecular mechanisms of CNF1 expression remain unclear.
In order to study the genetic requirements for regulation of CNF1 expression and secretion in meningitis-causing E. coli, we designed a Tn5 mutational screening strategy by applying TEM β-lactamase as the reporter to monitor CNF1 secretion. From this screen, we identified a mutant that had transposon insertion at gidA gene and exhibited drastically decreased production of CNF1. GidA is known to be a FAD-binding tRNA modification enzyme (Brégenon et al., 2001; Moukadiri et al., 2009). In this study, we investigated the involvement of GidA in production of CNF1 in E. coli.
2. Materials and methods
2.1 Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids are shown in Table 1. E. coli K1 strain RS218 (O18:K1:H7) is the cerebrospinal fluid isolate from a neonate with meningitis (Khan et al., 2002). E. coli K-12 strain DH5α was used as the host for plasmids, and EC100D pir116+ (Epicentre Biotechnologies, Madison, WI) as the host for R6kγ origin plasmid. E. coli strains were routinely grown at 37°C in Luria Broth (LB). Where appropriate, the medium was supplemented with ampicillin (100 μg/ml), spectinomycin (100 μg/ml), tetracycline (10 μg/ml), or chloramphenicol (20 μg/ml).
Table 1.
Strains and plasmids used in the current study
| Strains or Plasmid | Relevant characteristic(s) | Reference or Source |
|---|---|---|
| Strains | ||
| E. coli RS218 | E. coli RS218 (O18:K1:H7), the cerebrospinal fluid isolate from a neonate with meningitis | (Khan et al., 2002) |
| E. coli EC100D | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL nupG pir+(DHFR). | Epicentre Biotechnologies |
| E. coli DH5α | F′ Phi80dlacZ DeltaM15 Delta(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK-mK+)phoA supE44 lambda- thi-1 | Lab stock |
| NBC | β-lactamase reporter gene was translationally fused to the C- terminal of cnf1 gene in the chromosome of strain RS218 | (Yu and Kim, 2010) |
| YK3 | E. coli RS218 (O18:K1:H7) gidA deletion mutant | This study |
| YK4 | E. coli RS218 (O18:K1:H7) gidA deletion mutant compelemented with gidA under the control of its native promoter. Complementation was achieved by Tn7 site- specific insertion into a second benign site in the chromosome. | This study |
| YK05 | E. coli RS218 (O18:K1:H7) gidA deletion mutant, compelemented with N-terminal truncated GidA under the control of arabinose promoter. Complementation was achieved by Tn7 site-specific insertion into a second benign site in the chromosome. | This study |
| YK06 | E. coli RS218 (O18:K1:H7) with additional copy of N- terminal truncated GidA under the control of arabinose promoter. Complementation was achieved by Tn7 site- specific insertion into a second benign site in the chromosome. | This study |
| Plasmids | ||
| pCXN | CNF1 coding region was cloned into the KpnI site of pCX340, tetracycline resistance. | (Yu and Kim, 2010) |
| pKD3 | Containing chromphenicol resistance gene, R6kγ replication origin | (Datsenko and Wanner, 2000) |
| pKD47 | a derivative of pKD46 (Datsenko & Wanner, 2000), with the only modification that blaM in pKD46 was replaced by spectinomycin resistance gene. | (Yu and Kim, 2010) |
| pGRG36 | Tn7 insertion vector, ampicillin resistance, temperature sensitive | (Mckenzie and Craig, 2006) |
| pGAP | Multiple cloning site of pGRG36 was ligated into PvuII site of pBC-KS, resulting in pGRGM. Then AraC and arabinose promoter (pBAD) was cloned into AvrII and XhoI site of pGRGM. | (Yu and Kim, 2010) |
| pCX340 | PBR322 derivative, cloning vector used to fuse CNF1 to the mature form of TEM1 β-lactamase, tetracycline resistance. | (Charpentier and Oswald, 2004) |
| pSR | Tn5 vector, spectinomycin resistance, R6kγ replication origin | This study |
| pGAP- gidAΔN | The coding region of N-terminal truncated GidA was cloned into NdeI and NotI site of pGAP | This study |
| pG-gidAΔN | DNA fragment containing truncated gidA gene that obtained from pGAP-gidAΔN by digestion with AvrII and PacI, and ligated into same sites of pGRG36. | This study |
| pG-gidA | DNA fragment containing gidA gene that obtained by PCR amplification from genomic DNA, and ligated into same sites of pGRG36. | This study |
2.2 β-lactamase (Bla) activity assay
Bla activity assay was done as described previously (Yu and Kim, 2010). Nitrocefin was added to bacterial culture supernatant at the final concentration of 0.1 mM, which was then incubated at 37° C for 20 hrs. Bla hydrolyzes its substrate, nitrocefin (yellow) into a red product, and Bla activity was read as positive if the color change to red occurred. Spectrophotometric assays for Bla activity using nitrocefin were also carried out by measuring absorbance at 486 nm.
2.3 Genomic DNA isolation and sequencing
Genomic DNA was isolated from individual Tn5 mutants as described previously (Yu and Kim, 2010). Quantity of chromosomal DNA was measured with the Quant-iT™ dsDNA BR assay kit (Invitrogen, Carlsbad, CA). 12 μl genomic DNA (0.5 μg/μl) and 12 μl sequencing primer SR-Seq (8 μM) were sent to DNA Synthesis and Sequencing Facility (Johns Hopkins University School of Medicine) for sequencing.
2.4 Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed as previously described (Yu et al., 2007). Bacterial total RNA was extracted using the RNeasy minikit (Qiagen, Valencia, VA) with a modification of applying bead mill to disrupt bacteria cells. Residual genomic DNA was removed by in-column DNaseI digestion. The amount of RNA was measured by Quant-iT RNA assay kit (Invitrogen). Primers cnf1-RTP and rplU-RTP were used for reverse transcription of mRNA of cnf1 and rplU, respectively. 1 μg of total RNA was reverse transcribed by the SuperScript first-strand synthesis system (Invitrogen) in 25 μl reaction volume. After reverse transcription, total RNA was removed by RNaseA treatment, and then 0.5 μl cDNA was applied for each regular PCR with JumpStart Red Taq premix (Sigma, St. Louis, MO). Primers (cnf1-RTs & cnf1-RTa, and rplU-RTs & rplU-RTa) were used for amplification of cnf1 and rplU, respectively. The amplifcation cycles were determined by independent experiments so that the amplifcation was still under the exponential stage. In this experiment, 28 cycles were used for both cnf1 and rplU gene.
2.5 gidA gene deletion and complementation
To delete gidA gene, a chloramphenicol resistance cassette was amplified from pKD3 using primers gidA-KOF and gidA-KOR (Table 2). The PCR product was inserted into the chromosome by Lambda Red-mediated allele replacement, as described previously (Datsenko and Wanner, 2000). The correct insertion was verified by PCR with primers gidA-ckf and gidA-ckr (Table 2).
Table 2.
Primers used in the study
| Primer | Sequence (5′-3′)a |
|---|---|
| gidA-KOF | TCTTGATGCGTTGCCTGGTAAGCGGGTGCTTACCAGGCATTTTTAATGCGGTGTAGGCTGGAGCTGCTTC |
| gidA-KOR | CGGCCCGGGCTTCAATCCATTTTCATACCGCTTTATGCGAGGCAATCACCCATATGAATATCCTCCTTAG |
| gidA-ckf | AGTAAGGAGAGTTTGTTGAGCA |
| gidA-ckr | AAGCCGACAAAGTTGAGTAGA |
| gidA-a3 | CGCGTTAATTAACAGTAAGGAGAGTTTGTTGAGCA |
| gidA-s | GCGCCTAGGCCTTTTGTGGGGCTATCG |
| gidA-s2 | CGCTGGCATATGAGCTGCAACC |
| Spc-SeqR | GCCTTGCTGTTCTTCTACGG |
| Tn7-ckf | ACGGTCGGGAACTGGAAC |
| Tn7-ckr | TGACCAGCCGCGTAACCT |
| CNF1-RTP | CCTTCTTTTCGGGCAAC |
| CNF1-RTs | TTTGAAGCCGCTAATGCTGATG |
| CNF1-RTa | CATTCGCCCCACGAGCAG |
| RplU-RTP | GCCCTGCTGCTTACGATA |
| RplU-RTs | CTTACGATAGTGTTTACGACGAC |
| RplU-RTa | CCTGGAAAAGCTGGACATC |
| cnf1-s3 | GCGCGGTACCATGGGTAACCAATGGCAA |
| cnf1-a | GGATCCGGTACCAAATTTTTTTGAAATACCTTCA |
| cnf1-s2 | GGCGCCATATGGGTAACCAATGGCAA |
| cnf1-A | GCCGGATCCAAATTTTTTTGAAATACCTTCA |
| cnf1-s8 | CGCGTTTCATATGGTTAATCATTGGGCAAT |
| cnf1-a8 | GCCGAATTCCCTTTATTGCTAAGTGTCTTATTGG |
| cnf1-s11 | TGCGCACATATGGATCCTGATAATGCTTATTTCAT |
| cnf1-s12 | TGCGCACATATGACAGAAGTACTGACACTCACTCA |
| cnf1-s13 | TGCGCACATATGAAAAATGGAGAGCTTGATGA |
| cnf1-S10 | CTGCGGTACCCAAGATGGAGTTTCCTATGC |
| cnf1-a13 | GCCGAATTCCCATAAAGATACAAAGCAGAAGAAAT |
| cnf1-s19 | TGCGCACATATGGGATATTACGCATCTGATATT |
| cnf1-s21 | TGCGCACATATGAACGCCTTAAATCGCA |
| cnf1-a7 | GCCGAATTCCCCCTGTCAACCACAGCCAG |
| cnf1-s9 | CGCGGCATATGACCATACGTAAATTGTTATCTCTA |
| cnf1-s14 | TGCGCACATATGAGCGGAAATCTAAGTGGTTGT |
| cnf1-a14 | GCGGAATTCGTCGTACAACCACTTAGATTTC |
| cnf1-s17 | TGCGCACATATGACAAAAGAACCAATACCTCGC |
Restriction sites for cloning are underlined.
For gene complementation, Tn7 site-specific insertion of gene into the second benign site in the chromosome of mutant was carried out as described previously (Yu and Kim, 2010; Mckenzie and Craig, 2006). Briefly, gidA gene together with its native promoter was amplified from the genomic DNA of strain RS218 by primers gidA-s and gidA-a3 (Table 2), and then ligated into AvrII and PacI sites of pGRG36, and the resulting plasmid was designated as pG-GidA. N-terminal truncated GidA was obtained by amplifying from the genomic DNA of strain RS218 by primers gidA-s2 and gidA-a3 (Table 2), and ligated into NdeI and NotI site of pGAP (Yu and Kim, 2010), yielding plasmid pGAP-GidAΔN. The DNA fragment containing araC, arabinose promoter and gidA gene was obtained by digesting pGAP-GidAΔN with restriction enzymes AvrII and PacI, and was subsequently ligated into the same sites of pGRG36 (McKenzie and Craig, 2006). The resulting plasmid was designated pG-GidAΔN. pG-GidA and pG-GidAΔN were transposed into the gidA mutant or RS218, and successful Tn7 transposition was verified by PCR with primers Tn7-ckf and Tn7-ckr (Table 2).
2.6 Plasmid construction
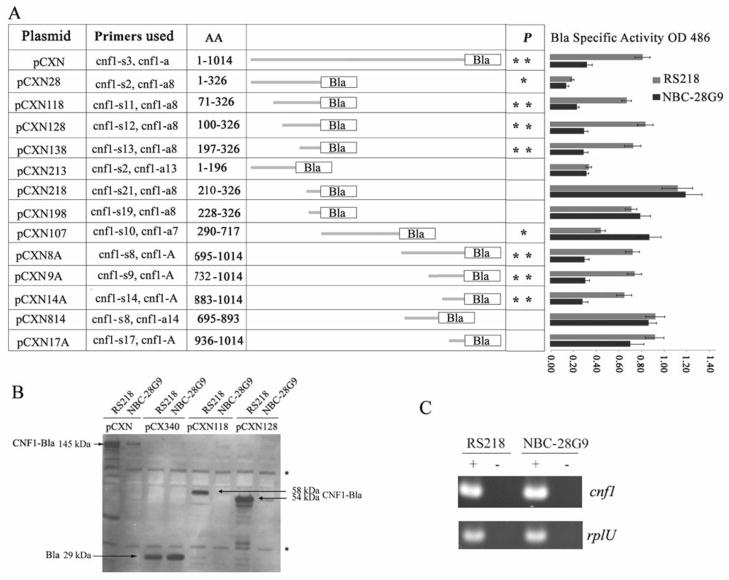
A number of translational fusions were constructed using the Bla reporter gene in pCX340 (Charpentier and Oswald, 2004). All the constructs were under the control of the trc promoter. Different regions of cnf1 were obtained by PCR amplification from RS218 genomic DNA as indicated in table 3. The PCR products were, then digested with appropriate restriction enzyme and ligated into the same sites of pCX340. Primers used for each constructs were listed in figure 3A (their sequences were listed in table 2).
Figure 3.
Inhibition of CNF1 expression by truncated GidA is mRNA context dependent.
A. Schematic representation of different regions of cnf1 gene fused with β-lactamase reporter gene. AA indicates the amino acid regions in each construct that were being fused to Bla. For constructing each plasmid, the primers used were listed; their detailed sequence information is shown in table 2. For each reaction, 2 μg bacterial lysates were incubated with 0.1 mM nitrocefin, after 30 minutes of incubation; 1 mM tazobactam sodium was added to stop the reaction. The numbers in column RS218 and NBC-28G9 represent the average and standard deviation of three measurements of OD486 (Bla activity, means ± standard deviations) of each construct in strain RS218 and NBC-28G9, respectively. In NBC-28G9, CNF1-Bla that was expressed from its genome has been determined by strain NBC-28G9 without plasmid constructs. It was extremely low in the diluted bacterial lysates when tested at the same condition (The average OD486 reading was 0.016). The statistical significance of the differences in Bla activity between two strains with the same plasmid construct was calculated with t test (*, p<0.05; **, p<0.01). The constructs without * label in p column indicate that the CNF1 fragment and Bla fusion proteins from those constructs were produced at similar level between RS218 and NBC-28G9.
B. Western blot analysis of the expression of CNF1-Bla fusion proteins from artificial promoter trc in plasmid in strains RS218 and NBC-28G9. 50 μg bacterial crude extracts were loaded in each lane and then separated with 4–12% Bis-tris gel (Invitrogen). β-lactamase polyclonal antibody (Millipore) was used as the primary antibody. Different constructs in different strains were indicated as labeled. “*” denotes non-specific band. The intensity of non-specific band on the film indicates that the same amount of protein was loaded in each well. The position and size of CNF1-Bla expressed from pCXN, pCXN118 and pCXN128 is indicated along with the arrow.
C. Reverse transcribed PCR analysis of cnf1 mRNA in RS218 and NBC-28G9. House-keeping gene rplU was used as the marker for the integrity of mRNA and the loading control. “−” denotes negative control without reverse transcriptase. “+” denotes reactions with reverse transcriptase.
2.7. Immunoblot assays
Protein samples were separated by SDS-PAGE, and transferred to nitrocellulose membrane. The blots were blocked with 5% skim milk in TBS (25 mM Tris, pH 7.4, 150 mM NaCl) for 60 min at 22°C. The membrane was then incubated for two hours at 22°C with primary antibody, which includes anti-CNF1 monoclonal antibody (DD1) (Meysick et al., 2001) and anti-Bla polyclonal antibody (Millipore). The membrane was washed with 0.5% tween-20 in TBS and subsequently incubated for 60 min at room temperature with horseradish peroxidase-linked secondary antibodies. The membrane was developed with an enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech, Piscataway, NJ).
3. Results
3.1 Tn5 mutagenesis screen identified the gidA gene
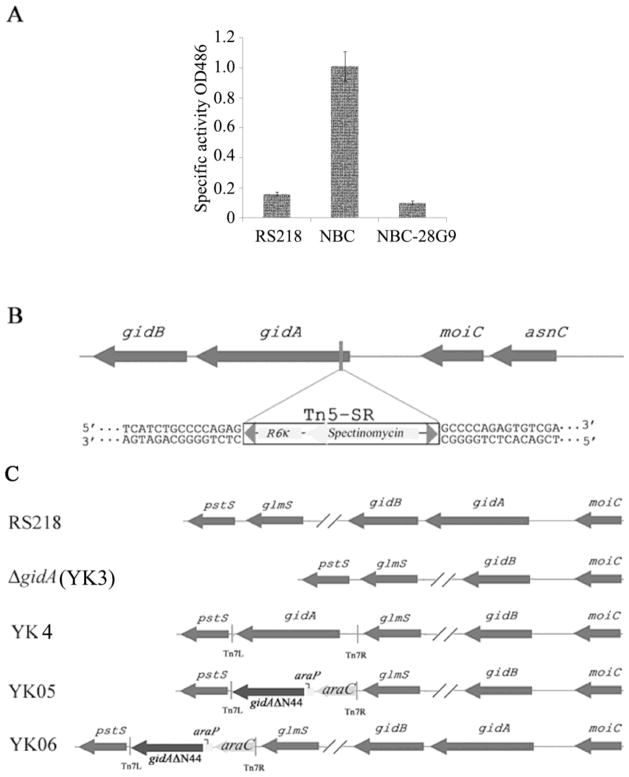
In order to investigate the genetic requirement for CNF1 secretion in RS218, we performed mini-Tn5 in vitro mutagenesis and constructed a mutant library of strain NBC, in which β-lactamase (Bla) was fused to the C-terminal of CNF1 in the chromosome of RS218 (Yu and Kim, 2010). In strain NBC, Bla secretion is entirely dependent on CNF1 secretion machinery and we measured the Bla activity in the culture supernatant to identify mutants with reduced CNF1-Bla fusion protein secretion. For Bla assays, the NBC strain was used as a positive control, while the wild-type strain RS218 was used as a negative control. We identified a mutant (NBC-28G9) that exhibited no Bla activity based on visual color change and the spectrometric reading (Fig. 1A). Next, we determined the location of the transposon insertion by direct DNA sequencing of the mutant’s genomic DNA. The insertion was shown to occur within the gidA gene (Fig. 1B).
Figure 1.
Identification of transposon mutant strain NBC-28G9.
A. The Bla activity in the culture supernatant of strain RS218, NBC and NBC-28G9. Specific Bla activity was measured as described in Materials and Methods. The represented Bla activity (means ± standard deviations) represents the results from three experiments in triplicate.
B. Schematic representation of the chromosomal structure of transposon insertion within gidA gene in the mutant strain NBC-28G9
C. Schematic representation of the chromosomal structure of gidA mutants constructed in this study. The chloramphenicol resistance gene in gidA deletion mutant (ΔgidA) was eliminated by using a helper plasmid expressing the FLP recombinase, which acts on the repeated FRT (FLP recognition target) sites flanking the resistance gene. YK4, YK05 and YK06 are complemented strains, in which gidA gene was inserted at a specific benign site in the chromosome by Tn7 complementation vector as described previously (Yu and Kim, 2010). In YK4, gidA gene with its native promoter was intergrated at the genome of ΔgidA; in YK05, truncated gidA gene under the control of arabinose promoter was intergrated at the genome of ΔgidA; in YK06, truncated gidA gene under the control of arabinose promoter was intergrated at the genome of RS218.
3.2 N-terminal truncated GidA has dominant negative effect on CNF1 synthesis
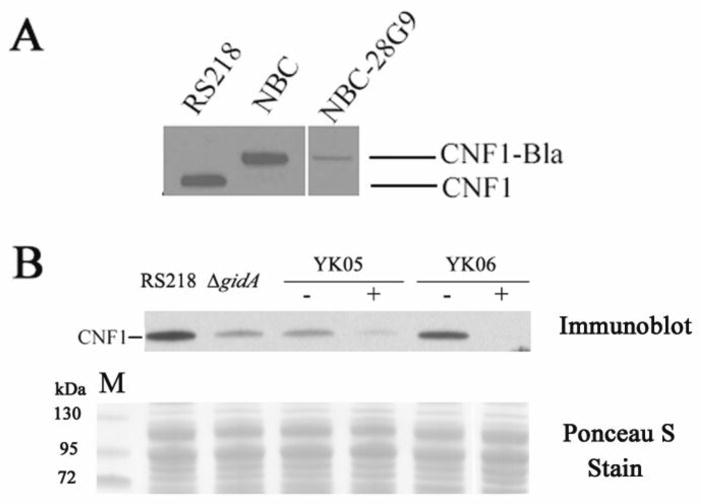
To examine whether or not the decreased CNF1-Bla secretion in NBC-28G9 was due to reduced production of CNF1-Bla, we examined and compared the amount of CNF1-Bla in NBC-28G9 mutant and NBC strain. As revealed by Western blot analysis, CNF1-Bla level was much lower in NBC-28G9 than in the parent strain NBC (Fig. 2A). To exclude a possibility that the low level of CNF1-Bla production in NBC-28G9 is not the result of a polar effect from transposon insertion, we constructed gidA deletion mutant and examined its CNF1 production. To our surprise, CNF1 production was less affected when gidA gene was totally removed from the genome (Fig. 2B). The decreased CNF1 expression in ΔgidA was restored in strain YK4 by complementation of gidA gene (data not shown). In mutant NBC-28G9, the transposon insertion occurred at its very end of N-terminal (44 amino acids was truncated from its N-terminal by transposon). Therefore, the markedly decreased CNF1-Bla production in NBC-28G9 could be due to polar effect induced by transposon insertion or by the presence of a partially truncated GidA protein. To examine such a possibility, we constructed strains named YK05 and YK06 (Fig. 1C). In strains YK05 and YK06, an N-terminus truncated gidA gene (GidAΔN44) was amplified by PCR and cloned after an arabinose inducible promoter, and was inserted at the benign Tn7 attachment site in the genome of the ΔgidA mutant and strain RS218, respectively. In both strains YK05 and YK06, CNF1 production was much lower when 0.2% arabinose was added to the culture medium, suggesting that reduced CNF1 production was caused by the presence of the N-terminus truncated GidA (Fig. 2B). Moreover, truncated GidA induced by arabinose arrested CNF1 production in wild type strain RS218 (Fig. 2B), suggesting that the N-terminus truncated GidA has dominant negative effect on CNF1 production in E. coli K1. It is interesting to note that residual truncated GidA has negative effect on the early stage of bacteria growth in YK05 and YK06 when no arabinose was added, however, switch on the expression of truncated GidA by adding arabinose did not alter the host cells’ growth curve (data was not shown). This finding differed from CNF1 synthesis, in which only induction of truncated GidA impacted CNF1 expression.
Figure 2.
CNF1 expression was repressed in gidA mutants. Western blot was developed with anti-CNF1 monoclonal antibody DD1 (Meysick et al., 2001). 50 μg bacterial crude extracts was loaded in each lane and then separated with 4–12% Bis-tris gel (Invitrogen). The intensity of ponceau S stain of each lane revealed that the same amount of protein was loaded in each well.
A. Western blot analysis of bacterial crude extracts of strains RS218, NBC and NBC-28G9.
B. Western blot analysis of bacterial crude extracts of strains RS218, ΔgidA, YK05 and YK06. “−” denotes bacteria was grown without 0.2% arabinose, and “+” denotes bacteria was grown with 0.2% arabinose.
3.3 Decreased CNF1 production is not due to reduced mRNA level
To determine whether reduced CNF1 production was the result of reduced transcription of cnf1 gene, we analyzed mRNA level of cnf1 by semi-quantitative reverse transcribed PCR. In strain NBC-28G9, the level of cnf1 mRNA was comparable to that of wild type strain (Fig. 3C). The constitutively expressed house-keeping gene rplU was used as a control in this experiment. The similar levels of rplU bands from RS218 and NBC-28G9 samples indicate that an equal amount of RNA was present in both samples. Moreover, the production of CNF1 under the control of trc promoter from plasmid pCXN was also affected in strain NBC-28G9 (Fig. 3B). Trc promoter is a hybrid of the lac (-10 regions) and trp (-35 regions) promoters, and transcription initiated from trc promoter has been well characterized (Brosius et al., 1985; Egon et al., 1983). The expression of Bla from pCX340 was not affected in strain NBC-28G9 (Fig. 3B), indicating that activity of trc promoter was not affected by N-terminus truncated GidA. The decreased production of CNF1-Bla fusion protein (from pCXN) in NBC-28G9 is, therefore, not likely due to reduced transcription, which is consistent with the result of RT-PCR. Taken together, these findings demonstrate that GidA affects CNF1 production at translational level.
3.4 mRNA context dependent regulation of CNF1 translation by GidA
As the translation of CNF1-Bla fusions from pCXN was blocked in NBC-28G9, but not that of Bla from pCX340 (Fig. 3A&B), we hypothesize that specific regulation signal may be embedded in the mRNA region of cnf1. To address this hypothesis, we performed progressive deletions, and compared the Bla activity of various translational CNF1-Bla fusions between RS218 and NBC-28G9. We first divided cnf1 gene into three DNA fragments (corresponding to N-terminal domain, transmembrane domain and C-terminal domain, respectively), and cloned them into pCX340 to translationally fuse with Bla, resulting in plasmid pCXN28, pCXN107 and pCXN8A, respectively (Fig. 3A). After transforming each plasmid into RS218 and NBC-28G9, we found that the expression of CNF1-Bla hybrid from pCXN28 and pCXN8A was lower in NBC-28G9 than in RS218. The expression of CNF1-Bla in pCXN107 was not inhibited in NBC-28G9. Thereby, it is likely that specific signal is embedded in CNF1 regions that were cloned into pCXN28 and pCXN8A. To further identify those regions responsible for translational inhibition of CNF1 expression, we then progressively deleted DNA from pCXN28 and pCXN8A, and assayed Bla activity of each construct in RS218 and NBC-28G9, respectively.
Different constructs are listed in figure 3A and may have different production levels of CNF1-Bla in the same strain. For example, constructs pCX28 and pCXN213 have low expression levels in RS218, but pCXN118 has relatively high expression in RS218. This is consistent with earlier report that the mRNA corresponding to the first 48 codons of cnf1 has translational regulation role (Fabbri et al., 1999). We showed that the constructs devoid of DNA region corresponding to the first 48 codons in constructs increased CNF1-Bla production levels in RS218 but not in NBC-28G9 (pCXN118, pCXN128 and pCXN138), suggesting that the translational regulation mediated by GidA differs from the previously defined region corresponding to the first 48 codons (Fabbri et al., 1999).
The expression level of CNF1-Bla hybrid construct pCXN218 was similar between RS218 and NBC-28G9, while that from pCXN138 was repressed in NBC-28G9 (Fig. 3A). The only difference between pCXN138 and pCXN218 was that DNA region corresponding to codons 197 to 210 of cnf1 was present in pCXN138, but was absent in pCXN218, suggesting that cnf1 mRNA region corresponding to aa 197–210 is likely responsible for truncated-GidA-dependent CNF1 translational repression (Figure 3A).
In the C-terminal region of CNF1, when codons from aa 883–936 were deleted from pCXN14A, the expression of Bla fusion proteins in pCXN17A was no longer repressed in NBC-28G9, suggesting that cnf1 mRNA region corresponding to aa 883–936 is likely to represent another putative truncated-GidA-dependent CNF1 translational repression site (Figure 3A). On the other hand, CNF1-Bla hybrid encoded by constructs pCXN 213 and pCXN814 lacking those two putative mRNA regions exhibited similar expression levels between RS218 and NBC-28G9 (Fig. 3A). Collectively, our progressive deletion of CNF1 codons demonstrates that the two specific mRNA regions, aa 197–210 and aa 883–936, are responsible for translational inhibition of CNF1 translation mediated by truncated GidA, and support that such repression of CNF1 production is mRNA context dependent.
4. Discussion
In this study, we demonstrated that GidA, a tRNA modification enzyme is involved in regulation of CNF1 translation, and its regulation is mRNA context dependent.
Recent studies have shown that GidA is important for the virulence of many pathogenic bacteria, such as Pseudomonas syringae (Kinscherf and Willis, 2002), Aeromonas hydrophila (Sha et al., 2004), Shigella flexneri (Durand et al., 2000), Streptococcus suis (Li et al., 2010), Streptococcus pyogenes (Cho and Caparon, 2008) and Lactococcus garvieae (Menéndez et al., 2007), but little is known about their underlying mechanisms. GidA is one of the tRNA modification enzymes (Brégeon et al., 2001; Moukadiri et al., 2009) and found to be allelic with trmF (tRNA modification) (Brégeon et al., 2001). tRNA isolated from a trmF mutant carries 2-thiouridine instead of 5-methylaminomethyl-2-thiouridine modifications in the wobble position of the anticodon in certain tRNAs (Elseviers et al., 1984). GidA and MnmE can form a complex and act in the same tRNA modification pathway (Brégeon et al., 2001; Moukadiri et al., 2009). In the present study, GidA is shown to be involved in regulation of CNF1 translation, and two putative mRNA regions are likely to mediate such a regulation.
After transfer RNA precursor is transcribed, modifying enzymes introduce various chemical substitutions to its nucleotides, which are common in all living cells (Phizicky and Alfonzo, 2010). However, the biological functions of many of these alterations are relatively unknown (Phizicky and Alfonzo, 2010). CNF1 production was decreased when gidA was deleted, indicating that certain type of tRNA modification would enhance the translational efficiency of CNF1. This was probably achieved by stabilizing tRNA tertiary structure and/or by optimizing the mRNA decoding function (Phizicky and Alfonzo, 2010; Motorin and Helm, 2010).
Mutations in gidA and mnmE (by transposon insertion) have been found to be responsible for +2 translational frame shift in E. coli (Brégeon et al., 2001). It was suggested that this is due to the presence of GA repeats in mRNA and hypomodified uridine at the wobble position of tRNAGlu (Brégeon et al., 2001). The Tn10 mutations recovered in that study were shown to have insertions at positions of 707, 1025, 5, and 51 in the gidA gene (Brégeon et al., 2001). In our study, the transposon was inserted at the 5′ end of gidA gene, and it remains unknown how a truncated gidA gene can have promoter for its transcription and ribosome binding site for its translation. However, cloning the truncated gidA under the control of arabinose promoter, and then inducing the expressing of such a truncated gidA by arabinose clearly had negative effect on CNF1 expression, suggesting that the dominant negative effect on CNF1 translation was induced by N-terminus truncated GidA. Our hypothesis is that truncated GidA may have an unrestricted modification activity and tRNA modified by GidA could be further modified by truncated GidA, and generated hypomodified tRNA as suggested previously (Brégeon et al., 2001). The presence of both native GidA and N-terminus truncated GidA might be more toxic to CNF1 translation than the presence of only N-terminus truncated GidA (Fig. 2B). It remains, however, unclear whether or not the inhibited CNF1 translation is due to the ribosome slip in the presence of hypomodified tRNA.
We identified two putative mRNA regions that are responsible for inhibition of CNF1 translation in the presence of truncated GidA, indicating that such an inhibition is mRNA context dependent. The two regions probably contain codons that are decoded by modified tRNA, or contain the embedded signal of requiring modified tRNA for decoding certain codon(s). It would be of interest to find out why modified tRNA is selected to regulate the production of CNF1. Additional information on modified tRNA may help in elucidating the mechanisms involved in modulation of CNF1 translation
Acknowledgments
This work was supported by the NIH grants NS26310 and AI84984. We thank Dr. Alison O’Brien for providing the CNF1 monoclonal antibody, Dr. Nancy L. Craig for providing the pGRG36 plasmid, Dr. Fred Heffron for providing the plasmid pMini-Tn5 cycler, and Dr. Eric Oswald for providing plasmid pCX311 and pCX340. We also thank George Niemann for helpful suggestions.
Abbrevations
- CNF1
cytotoxic necrotizing factor 1
- HBMEC
human brain microvascular endothelial cell
- Bla
β-lactamase
- FAD
flavin adenine dinucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brégeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Erfle M, Storella J. Spacing of the -10 and -35 Regions in the tac Promoter. J Biol Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- Charpentier X, Oswald E. Identification of the secretion and translocation domain of the Enteropathogenic and Enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based reporter. J Bacteriol. 2004;186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KH, Caparon G. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun. 2008;76:3176–3186. doi: 10.1128/IAI.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JW, et al. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. J Biol Chem. 2003;278:16857–16862. [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia Coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Carvalho HM, Rasmussen SB, O’Brien AD. Cytotoxic necrotizing factor type 1 delivered by outer membrane vesicles of uropathogenic Escherichia coli attenuates polymorphonuclear leukocyte antimicrobial activity and chemotaxis. Infect Immun. 2006;74:4401–4408. doi: 10.1128/IAI.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye A, et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Durand JM, Dagberg B, Uhlin BE, Björk GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol Microbiol. 2000;35:924–935. doi: 10.1046/j.1365-2958.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- Egon A, Brosius J, Ptashne M. Vectors Bearing a Hybrid trp-lac Promoter Useful for Regulated Expression of Cloned Genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Elseviers D, Petrullo LA, Gallagher PJ. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri A, Gauthier M, Boquet P. The 5′ region of cnf1 harbours a translational regulatory mechanism for CNF1 synthesis and encodes the cell-binding domain of the toxin. Mol Microbiol. 1999;33:108–118. doi: 10.1046/j.1365-2958.1999.01453.x. [DOI] [PubMed] [Google Scholar]
- Flatau G, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–33. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Khan NA, et al. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J Biol Chem. 2002;277:15607–15612. doi: 10.1074/jbc.M112224200. [DOI] [PubMed] [Google Scholar]
- Kim KS. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol. 2008;6:625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J Biol Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Kim KS. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat Rev Neurosci. 2003;4:376–385. doi: 10.1038/nrn1103. [DOI] [PubMed] [Google Scholar]
- Kinscherf TG, Willis DK. Global regulation by gidA in Pseudomonas syringae. J Bacteriol. 2002;184:2281–2286. doi: 10.1128/JB.184.8.2281-2286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, Uhlin BE. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect Immun. 2006;74:2022–2030. doi: 10.1128/IAI.74.4.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonier M, Landraud L, Lemichez E. Rho GTPase-activating bacterial toxins: from bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol Rev. 2007;31:515–534. doi: 10.1111/j.1574-6976.2007.00078.x. [DOI] [PubMed] [Google Scholar]
- Lerm M, Pop M, Fritz G, Aktories K, Schmidt G. Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Immun. 2002;70:4053–4058. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu L, Qiu D, Chen H, Zhou R. Identification of Streptococcus suis serotype 2 genes preferentially expressed in the natural host. Int J Med Microbiol. 2010;300:482–488. doi: 10.1016/j.ijmm.2010.04.018. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 2006;6:39. doi: 10.1186/1471-2180-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez A, Fernández L, Reimundo P, Guijarro PA. Genes required for Lactococcus garvieae survival in a fish host. Microbiology. 2007;153:3286–3294. doi: 10.1099/mic.0.2007/007609-0. [DOI] [PubMed] [Google Scholar]
- Meysick KC, Mills M, O’Brien AD. Epitope mapping of monoclonal antibodies capable of neutralizing cytotoxic necrotizing factor type 1 of uropathogenic Esherichia coli. Infect Immun. 2001;69:2066–2704. doi: 10.1128/IAI.69.4.2066-2074.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- Moukadiri I, Prado S, Piera J, Velázquez-Campoy A, Björk GR, Armengod ME. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009;37:7177–7193. doi: 10.1093/nar/gkp762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro P, Lemichez E. Bacterial toxins activating Rho GTPases. Curr Top Microbiol Immunol. 2005;291:177–190. doi: 10.1007/3-540-27511-8_10. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2010;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Sha J, Kozlova EV, Fadl AA, Olano JP, Houston CW, Peterson JW, Chopra AK. Molecular characterization of a glucose-inhibited division gene, gidA that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect Immun. 2004;72:1084–1095. doi: 10.1128/IAI.72.2.1084-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kim KS. Ferredoxin is involved in secretion of cytotoxic necrotizing factor 1 across the cytoplasmic membrane in Escherichia coli K1. Infect Immun. 2010;74:2022–2030. doi: 10.1128/IAI.00674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kim KS. The involvement of SelB in the expression of cytotoxic necrotizing factor 1 in Escherichia coli. FEBS lett. 2011;585:1934–1940. doi: 10.1016/j.febslet.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Yu H, Yao Y, Liu Y, Jiao R, Jiang W, Zhao GP. A complex role of Amycolatopsis mediterranei GlnR in nitrogen metabolism and related antibiotics production. Arch Microbiol. 2007;188:89–96. doi: 10.1007/s00203-007-0228-7. [DOI] [PubMed] [Google Scholar]