Abstract
Variegated expression of variable NK cell receptors for polymorphic MHC class I broadens the range of an individual’s NK cell response, and the capacity for populations and species to survive disease epidemics and population bottlenecks. On evolutionary time-scales this component of immunity is exceptionally dynamic, unstable and short-lived, being dependent upon co-evolution of ligands and receptors subject to varying, competing selection pressures. Consequently these systems of variable NK cell receptors are largely species-specific and have recruited different classes of glycoprotein, even within the primate order of mammals. Such disparity helps explain substantial differences in NK cell biology between humans and animal models, for which the population genetics is largely ignored. KIR3DL1/S1, that recognizes the Bw4 epitope of HLA-A and –B and is the most extensively studied of the variable NK cell receptors, exemplifies how variation in all possible parameters of function is recruited to diversify the human NK cell response.
Keywords: Natural Killer Cells, MHC, Comparative Immunology/Evolution, Antigens/Peptides/Epitopes
Investigation of natural killer (NK) cell function began in the 1970s, with its emphasis on anti-tumor immunity (1, 2). During the following decade, the capacity of NK cells to kill cellular targets was inversely correlated with the amount of MHC class I on the target cell surface (3). This seminal observation led to the missing-self hypothesis and the search for NK cell receptors that recognize MHC class I (4). The 1990s first saw cellular, then molecular, definition of receptors (5, 6), and characterization of the genomic regions that encode them: the Natural Killer Complex (NKC) (7) and the Leukocyte Receptor Complex (LRC) (8, 9). Whereas the binding domains of NKC receptors resemble C-type lectins (10), their LRC counterparts are Ig-like (11, 12), a qualitative difference revealing that NK cell receptors for MHC class I evolved independently in the two genetic complexes.
Species-specific evolution of variable NK cell receptors
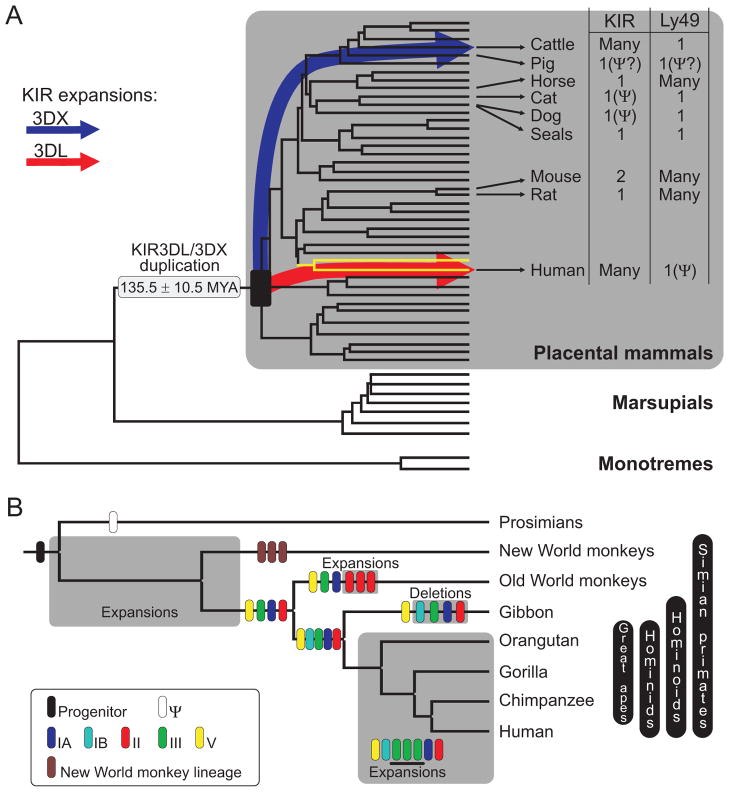
That some NK cell receptors for MHC class I evolve rapidly, became apparent from species comparison (13) (Fig. 1). The rodent NKC encodes variable Ly49 receptors that diversify NK cell function in rats (14) and mice (15), while the LRC family of killer cell immunoglobulin-like receptors (KIR) provides a comparable system for humans and other simian primates (16). Mammalian species having only single-copy Ly49 and KIR genes can survive and flourish (17), but no species has yet been found to have both variable Ly49 and KIR (Fig. 1A). In the context of variable KIR, human Ly49 is a single-copy pseudogene (18); in the context of variable Ly49, the mouse KIR locus left the LRC for the X chromosome (19), where it comprises two genes: one expressed by NK cells and T cells (20), the other by brain cells (21). Such contrasting situations, point to past crises in mammalian evolution when species-specific expansion of a new type of NK cell receptor accompanied extinction of an older form.
Figure 1.
Evolution and variability of KIR and Ly49 NK cell receptors in mammalian species. A. On the right is shown the number of KIR and Ly49 genes in modern species. Emerging by gene duplication in an ancestral placental mammal, KIR3DL expanded in primates (red arrow) and KIR3DX expanded in cattle (blue arrow). The tree is adapted from that of Murphy et al (111). Ψ, pseudogene and the primate branches are yellow. MYA, million years ago. B. KIR diversification in primates. The primate KIR3DL progenitor (black) became a pseudogene in prosimians (empty box) but flourished in simian primates to form five hominoid lineages: IA (dark blue), IB (light blue), II (red), III (green) and V (yellow), and a unique New World monkey lineage (brown). Modern KIR haplotypes evolved through species-specific gene duplications (lineage II in Old World monkeys and lineage III in hominids) and deletions (lineages I–III in gibbons).
Further evidence for independent evolution of MHC class I receptors is seen within the LRC. Flanking the KIR locus is the gene family encoding the leukocyte immunoglobulin-like receptors (LILR) (22). Of these, LILRB1 is an NK cell receptor that binds to the more conserved Ig-like domains (α3 and β2-m) of MHC class I (12), whereas the variable α1 and α2 domains of MHC class I are the target for KIR (11). Embedded within the LILR locus is a KIR pseudogene (23), KIR3DX, which diverged from genes of the functional KIR locus ~120 million years ago, before the radiation of placental mammals (24). Cattle, even toed ungulates, are the only non-primates known to have variable KIR (20), but in cattle it was KIR3DX that became the variable gene family, while KIR3DL, the ancestral founder of the variable primate KIR, remained a single-copy gene(24) (Figure 1A).
Common to mice (25) and humans (26) are NKC-encoded MHC class I receptors comprising heterodimers of CD94 and an NKG2 family member. These CD94:NKG2 receptors recognize complexes of a conserved non-classical class I molecule (mouse Qa1 or human HLA-E) and peptides derived from the leader sequences of other MHC class I molecules (10). Like KIR (11), the CD94:NKG2 receptors interact with the upward face of the α1 and α2 domains and are sensitive to residues of the bound peptide (27, 28). Although CD94, NKG2 and HLA-E are conserved in humans (29), analysis of the grey mouse lemur, showed the potential of CD94:NKG2 to be a variable NK cell receptor. This prosimian primate has single Ly49 and KIR genes, but three CD94 genes and eight (five expressed, three pseudogenes) NKG2 genes. Equally distinctive are the lemur’s MHC class I genes; while four class I pseudogenes remain part of the MHC, the cluster of six functional class Igenes, left the MHCfor a different chromosome (30).
Co-evolution of MHC class I and KIR in simian primates
That prosimians and most non-primates have just one KIR gene shows that the diverse family of human KIR genes originated during simian primate evolution, following their separation from pro-simians ~58–69 mya (31) (Fig. 1B). Simian primates comprise New World monkeys, Old World monkeys, lesser apes (gibbons), and hominids (great apes, and humans). Distinctive lineages of human KIR recognize epitopes carried by different HLA class I molecules: notably, lineage II KIR recognize some HLA-A and B allotypes, and lineage III KIR recognize HLA-C and some HLA-B allotypes (32–34). These functional interactions are the result of the co-evolution of ligands with receptors during simian primate diversification (35).
Lacking counterparts to HLA-A, B and C, New World monkeys have distinctive MHC class I and KIR, showing they took a different evolutionary tack from that followed by other simian primates (36, 37). The abundance of Old World monkey MHC class I genes resembling either HLA-A or HLA-B (38, 39) correlates with increased numbers of lineage II KIR genes (40–42). Associated with the emergence of MHC-C in hominids is a multiplicity of lineage III KIR genes (34). While KIR evolution in Old World monkeys and hominids is marked by gene expansions, the lesser apes took a different road (43). Although having orthologs of most human class I genes and pseudogenes, gibbon MHC haplotypes lack an ortholog of HLA-G that provides the ligand for KIR2DL4 (44). Correlating with this absence, KIR2DL4 has been either deleted from gibbon KIR haplotypes or disabled. Gibbon MHC haplotypes also lack an HLA-C ortholog, and correspondingly gibbon KIR haplotypes lack the multiplicity of lineage III KIR genes characterizing species with MHC-C (43). Spared from the deletions and mutations that diminished and disordered the gibbon KIR locus were lineage II KIR3DL1, predicted to recognize MHC-A and/or MHC-B, and KIR3DL3 (lineage V) for which neither ligand nor function is known (45, 46).
Although first studied in the context of tumor immunity (1, 2), NK cells are now firmly placed in the response to infection (47) where they cooperate with dendritic cells (48). NK cells also play a seminal role in reproduction through cooperation with extravillous trophoblast to enlarge maternal blood vessels that supply the placenta and nourish the fetus (49). All such cellular interactions of NK cells can be influenced by KIR engagement of MHC class I. Whereas most cell types express HLA-A, B and C, only HLA-C is expressed by extravillous trophoblast (50). It is also the only normal tissue to express HLA-G, which binds avidly to LILRB1 (51) and interacts with lineage I KIR2DL4 in endosomes (44).
The tissue distributions of HLA-C and G, and the fate of the gibbon KIR locus in their absence (43) suggest that selective pressures from reproduction induced MHC-C to evolve away from its MHC-B-like ancestor: to be expressed on trophoblast and recognized by the lineage III KIR preferentially expressed on uterine NK cells (52). In this model, HLA-C interactions with lineage III KIR are subject to selection pressures from both immunity and reproduction, whereas the interactions of lineage II KIR with HLA-A and HLA-B evolve principally under selection by infection. As a consequence HLA-A and HLA-B evolved to be exceptionally variable, as has lineage II KIR3DL1/S1 that recognizes a broad range of HLA-A and B allotypes. In contrast, KIR3DL2, which recognizes a narrow range of HLA-A allotypes, has variability that does not stand out from the mass of human genes (Fig. 2). Because of these features, the genetic and functional properties of KIR3DL1/S1 have been most extensively studied, making it an exemplary variable NK cell receptor.
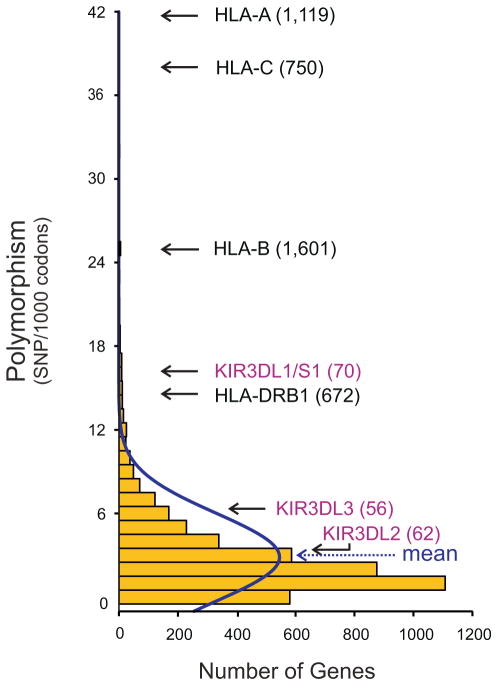
Figure 2. KIR3DL1/S1, like HLA-A, B, C and HLA-DRB1, is one of the most highly polymorphic human genes.
Shows comparison of coding-sequence diversity in the genomes of two Asian individuals. For the four alleles of each gene the number of single nucleotide polymorphisms (SNP) normalized to the number of codons in the gene. These values are presented in a histogram (yellow bars) and a continuous distribution (blue line). The genes form a normal distribution, with KIR3DL1/S1, HLA-A, B, and C, and HLA-DRB1 being outliers. For the named genes, the number of allotypes described worldwide is in parentheses (112, 53). Although KIR3DL2 and KIR3DL3 have many alleles, they differ by one or a few substitutions. Per-gene summary statistics were from Wang et al. 2008 (113) and Kim et al. 2009 (114), and analyzed using ‘Statistica software version 8’.
KIR3DL1 recognizes the Bw4 epitope of HLA-A and HLA-B
Sequences for >1,500 HLA-B allotypes are now known (53), but when first described in the 1960s, HLA-B was a simple serological dimorphism comprising the 4a and 4b antigens (54), later renamed the Bw4 and Bw6 epitopes, respectively (55). Every HLA-B allotype carries either Bw4 or Bw6, while some HLA-A allotypes also carry Bw4. Correlating with the Bw4/Bw6 difference are polymorphic sequence motifs at residues 77–83 in the helix of the HLA class I α1 domain (56), and the capacity for Bw4+ HLA-A and -B to be ligands for the inhibitory KIR3DL1 NK cell receptor(57, 58), formerly known as NKB1 (59).
Of the five residues that distinguish Bw4 and Bw6 motifs (77, 80, 81, 82, and 83) only arginine 83 is essential for binding KIR3DL1 (60). This contrasts with the position 80 dimorphism specifying the C1 and C2 epitopes recognized by lineage III KIR (61). As a further important structural difference, lineage III KIR interaction with HLA-C is accomplished with two Ig-like domains (D1 and D2), whereas an additional domain (D0) is necessary for 3DL1 to bind Bw4 (62, 63). Although a crystallographic structure for KIR3D has yet to be achieved, the combination of mutagenesis and modeling, based on the three-dimensional structures of KIR2D bound to HLA-C (11), predicts that the D0, D1, and D2 domains contribute equally to the HLA-binding surface in which a central pocket grasps arginine 83 (64). That all lineage III KIR genes contain an exon encoding D0, but which is no longer used, points to the more recent evolution of the interaction between HLA-C and lineage III KIR (65).
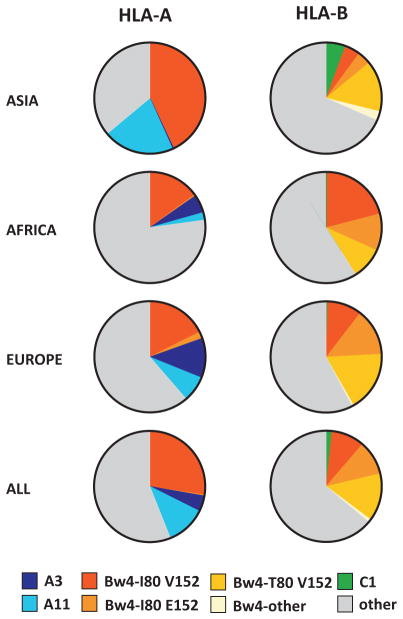
Some 25–42% of HLA-B and 15–43% of HLA-A allotypes carry the Bw4 epitope (Fig. 3). The Bw4 and Bw6 sequence motifs are frequent targets for short interallelic conversion events. Thus > 51 pairs of HLA-B allotypes differ only in presence or absence of Bw4. Gene conversion similarly introduced Bw4 into HLA-A, where it spread by interallelic conversion to the HLA-A*23, A*24, A*25, A*26, and A*32 allotypes. Although individual allotype frequencies vary between populations, the Bw4 and Bw6 frequencies remain remarkably constant (66), with around 50% of HLA haplotypes providing the Bw4 epitope. This even balance points to complementary functions for Bw4− allotypes more focused on T cell immunity and Bw4+ allotypes contributing to both NK cell and T cell immunity.
Figure 3. Distribution of HLA-A and B epitopes recognized by KIR in human populations.
Each pie represents the HLA-A or HLA-B allotypes in a population. Subdivisions within each pie are colored according to the epitope recognized by KIR, or shaded grey if they do not engage KIR. The A3/11 epitope is subdivided into A3 and A11, the Bw4 epitope is subdivided according to polymorphisms at positions 80, within the Bw4 sequence, and 152 that influence the avidity of Bw4 for KIR (115). HLA frequencies for representative Asian [n=24], African [n=13] and European [n=10] populations were obtained from http://www.allelefrequencies.net (116)
KIR3DL1 and KIR3DS1 segregate as functionally divergent alleles of the KIR3DL1/S1 gene
KIR3DL1 and KIR3DS1 are, respectively, inhibitory and activating receptors that diverge in the domains mediating signal transduction, but have very similar ligand-binding Ig-like domains. On the basis of their opposing signaling functions, 3DL1 and 3DS1 were initially considered to be the products of different genes (67), but with segregation studies, their allelic relationship was recognized(68) and signified by naming the gene KIR3DL1/S1 and numbering the 3DS1and 3DL1 variants as a single series of alleles (69). Complicating the situation, unequal crossing over, has produced several KIR haplotypes that either lack 3DL1/S1 (70), have both 3DS1 and 3DL1 (66, 71, 72), or have a fusion of 3DL1/S1 with 3DL2 (73).
Functional difference between 3DL1 and 3DS1 is not restricted to signaling. Whereas KIR3DL1 recognition of Bw4 can be readily detected in assays of binding and NK cell function (57, 58, 64), 3DS1 has no demonstrable interaction with Bw4, or any other HLA class I epitope (74–76). Despite this apparent lack of function, 3DS1 is present at significant frequency in every human population (66, 77). Another difference is in the variation: 3DL1 being highly polymorphic and 3DS1 conserved. All human populations have a balance between several 3DL1 alleles, even genetically less variable populations such as Japanese (five alleles) (78) and Yucpa Amerindians (3 alleles) (79), whereas 3DS1*013 dominates all populations, except Sub-Saharan Africans, and is the most abundant 3DL1/S1 allele worldwide (66). When the six residues distinguishing the extracellular domains of 3DS1 from 3DL1 were individually introduced into 3DL1, three abrogated interaction with Bw4, while having only minor affects on conformation and cell-surface expression, consistent with 3DS1 having been subject to strong positive selection for losing its avidity for Bw4 (64).
Human KIR haplotypes form two groups, A and B, that differ in gene content, allele content, variability and disease association (80–82). Both haplotype groups are present in all human populations, often at even frequency, and are maintained by balancing selection (79). KIR3DL1 is characteristic of A haplotypes, which have mainly polymorphic genes encoding inhibitory receptors, whereas 3DS1 is a characteristic B haplotype gene. B haplotypes are enriched for genes encoding activating receptors with either reduced (2DS1) or undetectable (2DS2 and 3DS1) avidity for HLA class I, compared to their inhibitory counterparts (83, 84). Differential KIR and HLA associations with infectious and reproductive disease suggest that the balance between A and B haplotypes might derive from the former favoring resolution of infection, the latter successful reproduction (79, 85).
Balanced polymorphism between three lineages of KIR3DL1/S1 alleles
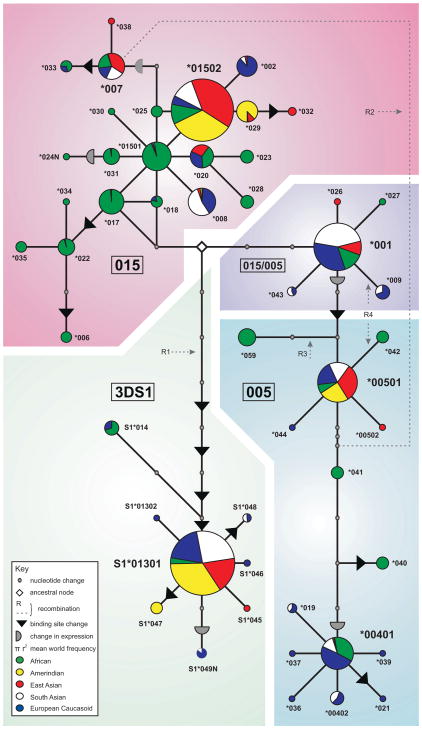
KIR3DL1/S1 alleles represent three phylogenetic lineages: 3DS1, 005, and 015, that have existed for >3 million years and are present in all populations. KIR3DS1*01301, 3DL1*005, and 3DL1*01502 are considered the prototypical alleles of the 3DS1, 005, and 015 lineages, respectively, because they are the only alleles present in all human populations (Fig. 4). Simulations point to their maintenance by balancing selection, indicating that each receptor lineage makes distinctive, complementary contributions to NK cell biology. The 015 lineage is uniquely diversified in African populations (Fig. 4, green shading) with commensurate reduction of the 005 and 3DS1 lineages, whereas the 3DS1 lineage is highly represented in Amerindians, and the 005 lineage in Caucasians (66).
Figure 4. Three divergent 3DL1/S1 allelic lineages are maintained in all human populations.
The minimum-spanning network shows the phylogenetic relationships and geographic distribution of 3DL1/S1 alleles. The distance between two nodes corresponds to one nucleotide change in the coding region. (
 ) denotes substitutions altering surface abundance, (▶) denotes substitutions in the ligand binding site (66). Nodes with colored circles are the alleles present in the modern human population, the area representing the frequency worldwide and the different colors the distribution between major population groups. Allelic lineages are denoted by the background shading: 015, magenta; 005, cyan; and 3DS1, green. 3DL1*001, a recombinant of the 015 and 005 lineages, has a purple background. Dashed lines indicate four other recombination events: R1, acquisition of activating signaling function to form 3DS1 from 3DL1 (The 22 unique substitutions in the 3DS1 signaling domain are not shown as nodes, because they were acquired en bloc.); R2, causing 3DL1*007 and 3DL1*004-like alleles to have the same cytoplasmic tail; R3, forming a chimera of 3DL1 and 3DL2; and R4, representing two independent events when 3DL1 acquired the D0 domain of 3DS1 to give the 3DL1*007 and 3DL1*042 alleles. The network was generated using the program TCS 1.21 (117) set to 99% confidence (by parsimony) that alleles formed by mutation not recombination.
) denotes substitutions altering surface abundance, (▶) denotes substitutions in the ligand binding site (66). Nodes with colored circles are the alleles present in the modern human population, the area representing the frequency worldwide and the different colors the distribution between major population groups. Allelic lineages are denoted by the background shading: 015, magenta; 005, cyan; and 3DS1, green. 3DL1*001, a recombinant of the 015 and 005 lineages, has a purple background. Dashed lines indicate four other recombination events: R1, acquisition of activating signaling function to form 3DS1 from 3DL1 (The 22 unique substitutions in the 3DS1 signaling domain are not shown as nodes, because they were acquired en bloc.); R2, causing 3DL1*007 and 3DL1*004-like alleles to have the same cytoplasmic tail; R3, forming a chimera of 3DL1 and 3DL2; and R4, representing two independent events when 3DL1 acquired the D0 domain of 3DS1 to give the 3DL1*007 and 3DL1*042 alleles. The network was generated using the program TCS 1.21 (117) set to 99% confidence (by parsimony) that alleles formed by mutation not recombination.
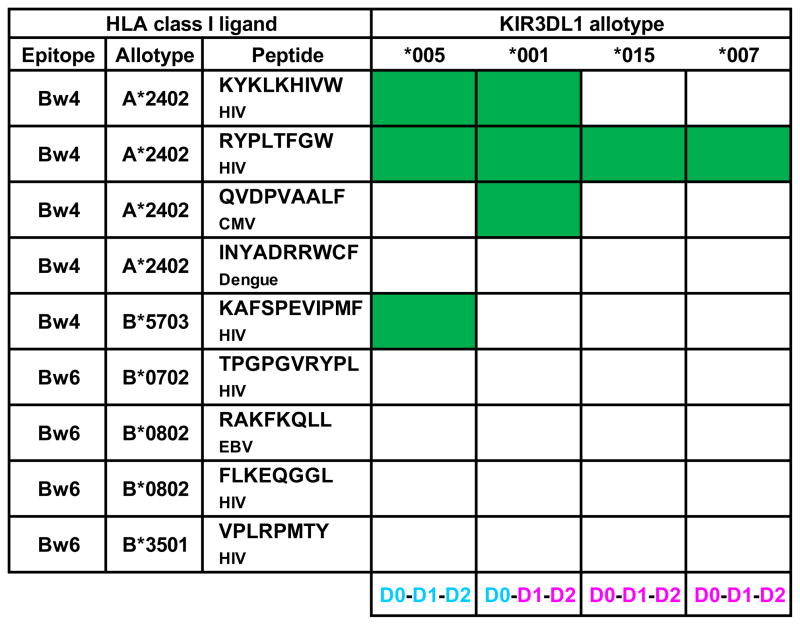
NK cell killing assays show that the interaction of KIR3DL1 with Bw4+ HLA class I is sensitive to polymorphisms in the Bw4 motif, notably position 80 (57), to polymorphism at positions away from the Bw4 motif that affect peptide binding (60) and to the sequence of the bound peptide (86–88). KIR3DL1 polymorphism also affects specificity for HLA class I, as seen in both cellular (78, 89) and direct-binding assays (90), as well as inferred by disease-associations (91). For example, measurement of binding for five complexes of defined viral peptide and Bw4+ HLA class I to four common KIR3DL1 allotypes gave three patterns of reaction and only eight of the 20 possible reactions (Table I) (90), a proportion identical to that seen in an earlier study using cytotoxicity assays (88). 3DL1*015 and 3DL1*007 have identical Ig-like domains and the same narrow reaction pattern, whereas 3DL1*005 has a broader specificity. 3DL1*001 combines the D0 domain of 3DL1*005, with the D1 and D2 domains of 3DL1*015 and also has a broad but distinctive specificity, illustrating the importance of the D0 domain in ligand-binding specificity.
Table I. KIR3DL1 polymorphism affects specificity for HLA class I.
A summary of the binding interactions of four KIR3DL1 allotypes with nine complexes of HLA class I and a viral peptide, as determined by Thananchai et al 2007 (90). Boxes shaded green denote significant binding. Under peptide the amino acid sequence of the peptide is given and the viral pathogen from which it derives: Human immunodeficiency virus (HIV), cytomegalovirus (CMV), Epstein-Barr virus (EBV). The relationship of the D0, D1, and D2 domains for each 3DL1 allotype is shown below; blue identical to 3DL1*005 and red identical to 3DL1*015
|
KIR polymorphism was originally observed through its influence on the proportion of NK cells expressing 3DL1 and the amount of 3DL1 on their surfaces (92). For example, of five common 3DL1 allotypes in Japanese, 3DL1*005 and 3DL1*007 are expressed at low level, 3DL1*001 at intermediate level, and 3DL1*020 and 3DL1*01502 at high level; a hierarchy reflected also in the proportion of NK cells expressing each allele (78). The relative level of 3DS1 expression remains uncertain because it is detected only by weak crossreactivity with anti-KIR3DL1 antibody (74, 75). Although KIR3DL1 allotypes differ in their capacity to educate NK cells and inhibit NK cell effector function, these differences do not correlate in a simple way with the level of cell-surface expression. Common in Caucasians, Africans and South Asians (Fig. 4), 3DL1*004 represents an extreme case with a very low level of cell-surface expression (93). Inefficient folding causes most of the protein to be retained within the cell, but the small amount reaching the surface can deliver inhibitory signals (94) and educate NK cells (95). Substitution at position 86 in the D0 domain is largely responsible for poor folding of 3DL1*004, with a minor contribution from position 182 in D1. Mutagenesis of 3DL1*015 at 40 sites of natural 3DL1/S1 variation showed that the great majority of substitutions had no affect or caused modest decrease in cell-surface abundance (as detected by antibodies), suggesting protein stability is not the only variable causing allele-specific differences in cell-surface expression(64) ; another likely source being transcriptional variation.
Organization and Variegated Expression of KIR genes
Transcription is controlled at the level of the entire KIR locus, which has an organizing framework comprising 3DL3 at the centromeric end, 2DL4 and the 3DP1 pseudogene in the center, and 3DL2 at the telomeric end. Regions of variable gene content lie between 3DL3 and 3DP1, and between 2DL4 and 3DL3. The intergenic regions containing the promoters are small (~2kb) and highly homologous, except the 13.4 kb region of unique sequence between 3DP1 and 2DL4(81).
In hematopoietic stem cells the KIR locus is inaccessible with transcription prevented by dense methylation, particularly of CpG islands in the promoter region (96, 97). The KIR locus opens up for transcription at a late stage in NK cell development, when it generates a repertoire of NK cells expressing diverse, combinations of KIR. The expressed KIR genes have hypomethylated promoters, whereas the promoters of the silenced genes are hypermethylated. The characteristic variegated expression of KIR by mature NK cells is thus determined by diverse patterns of promoter methylation(98, 99).
NK-cells express each KIR gene in one of three ways (100), exemplified by the three functional framework genes. All NK cells express 2DL4, a subset of NK cells expresses 3DL2, and very few NK cells express 3DL3. These differences correlate with promoter sequence variation that affects the binding of transcription factors, for which there are many potential sites (101). In studying the variegated expression of KIR genes, KIR3DL1/S1 has been the major subject for research (100, 102, 103).
The ~2kb intergenic region upstream of 3DL1/S1 contains two separate promoters. The proximal promoter (102), between nucleotides −1 and −255, has two non-overlapping sites, one promoting synthesis of sense mRNA, the other anti-sense mRNA(104, 105). The distal promoter (106), in the middle of the intergenic region >1kb from exon 1, promotes only sense mRNA. As transcription of a 3DL1/S1 allele begins, the distal promoter makes sense mRNA while the proximal promoter can favor either sense or antisense mRNA. If both promoters make sense mRNA the cell commits to long-term expression of the 3DL1/S1 allele. In contrast, if antisense mRNA is made from the proximal promoter it hybridizes to sense mRNA made from the distal promoter, which prevents transcription and leads to silencing of the 3DL1/S1 allele. The hybrid mRNAs give rise to a 28bp PINI-like RNA detectable only in the subset of 3DL1/S1− NK cells (107). In mature 3DL1/S1-expressing NK cells most transcripts arise from the proximal promoter but the distal promoter also contributes (108). Consistent with the distal promoter playing a decisive role in NK cell development is its activation by IL-15, a cytokine inducing NK cell differentiation (109).
That KIR3DL1/S1 alleles differ in their frequencies of expression in the NK cell population could arise from differences in the relative strengths of forward and reverse transcription at the proximal promoter. Stronger forward transcription favoring NK cell commitment to making the receptor, reverse transcription favoring commitment to not making the receptor. The 3DL1/S1 alleles expressed at high frequency by NK cells are those also expressed at high level on the cell surface (78), raising the intriguing possibility that competing dual activities of the bidirectional promoter also influence the amount of sense mRNA and protein made in mature NK cells.
Three substitutions in the 3DL1/S1 proximal promoter distinguish four different promoters: associated with 3DS1, the 015 lineage, 3DL1*005 and the combination of 3DL1*001 and 3DL1*004. In a cell-free system, measurement of the ratio of sense to antisense transcription distinguished the four promoters, but the values only partially correlated with the 3DL1/S1 phenotypes (105). Notably discordant was 3DS1, which gave the second lowest transcription ratio but is expressed by up to ~40% of NK cells in heterozygotes and ~80% in homozygotes (76, 110). Also unexplained by these promoter polymorphisms are the characteristic low cell-surface expression and cellular expression of 3DL1*007, which has the same promoter as high expressing 3DL1*015.
Conclusions
Variable NK cell receptors bind to the same complexes of peptide and MHC class I as the αβ TCR of CD8 T cells, and with sensitivity to the structural nuances of both peptide and MHC class I allotype. These interactions contribute to the education of NK cells during development and their effector functions when responding to cells compromised by infection or malignancy, or cells from somebody else, as occurs in pregnancy and transplantation. Within the individual, sets of inherited genes encoding polymorphic MHC class I and variable NK cell receptor cooperate to produce a diverse repertoire of functional NK cells, which gives versatility and specificity to the NK cell response. This individual variability is compounded at the population level where the number of possible KIR-HLA class I genotypes can exceed the size of the population.
Despite this diversity and versatility, comparative studies have uncovered an unprecedented degree of species specificity showing that individual receptors and entire systems of variable NK cell receptors have limited lifespans. Thus Ly49, KIR3DL, KIR3DX, and CD94:NKG2 are all seen as highly variable NK cell receptors, but in different species of placental mammals. Such evolutionary transience in NK cell receptors could arise from the competing demands of immunity and reproduction, botched compromise between the need for MHC class I to serve both NK-cell and T-cell receptors, or from obsolescence, being too specialized at fighting past infection and unable to adapt to current threats. The KIR system of variable antigen receptors is restricted to humans and other simian primates, species in which the co-evolution of KIR with MHC class I can be reconstructed. The effects of balancing selection are everywhere evident, particularly in humans with their distinctive A and B KIR haplotypes, and functionally disparate KIR3DL1 and KIR3DS1 alleles. Such striking qualitative differences are consistent with KIR-HLA class I interactions contributing to two essential functions in human biology: immune defense and reproduction.
Footnotes
Research in the authors’ laboratory was supported by grants from the National Institutes of Health.
References
- 1.Herberman RB, Holden HT. Natural cell-mediated immunity. Adv Cancer Res. 1978;27:305–377. doi: 10.1016/s0065-230x(08)60936-7. [DOI] [PubMed] [Google Scholar]
- 2.Kiessling R, Wigzell H. An analysis of the murine NK cell as to structure, function and biological relevance. Immunol Rev. 1979;44:165–208. doi: 10.1111/j.1600-065x.1979.tb00270.x. [DOI] [PubMed] [Google Scholar]
- 3.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 4.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari MC, Moretta L. Receptors for HLA class-I molecules in human natural killer cells. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama WM, Seaman WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- 8.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 9.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:129–139. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 11.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 12.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 13.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 14.Wilhelm BT, Mager DL. Rapid expansion of the Ly49 gene cluster in rat. Genomics. 2004;84:218–221. doi: 10.1016/j.ygeno.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol. 2009;182:3618–3627. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 19.Hoelsbrekken SE, Nylenna O, Saether PC, Slettedal IO, Ryan JC, Fossum S, Dissen E. Cutting Edge: Molecular Cloning of a Killer Cell Ig-Like Receptor in the Mouse and Rat. J Immunol. 2003;170:2259–2263. doi: 10.4049/jimmunol.170.5.2259. [DOI] [PubMed] [Google Scholar]
- 20.Dissen E, Fossum S, Hoelsbrekken SE, Saether PC. NK cell receptors in rodents and cattle. Semin Immunol. 2008;20:369–375. doi: 10.1016/j.smim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Bryceson YT, Foster JA, Kuppusamy SP, Herkenham M, Long EO. Expression of a killer cell receptor-like gene in plastic regions of the central nervous system. J Neuroimmunol. 2005;161:177–182. doi: 10.1016/j.jneuroim.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–225. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, Lifson JD, Carrington M, Beck S. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 25.Gunturi A, Berg RE, Forman J. The role of CD94/NKG2 in innate and adaptive immunity. Immunol Res. 2004;30:29–34. doi: 10.1385/IR:30:1:029. [DOI] [PubMed] [Google Scholar]
- 26.Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35:263–278. doi: 10.1385/IR:35:3:263. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J, Huyton T, Hopkins EJ, Reid HH, Wilce MC, Kabat J, Borrego F, Coligan JE, Rossjohn J, Brooks AG. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27:900–911. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci U S A. 2008;105:6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, Guethlein LA, Uhrberg M, Parham P. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 30.Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5:e1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee HJ, Ho SY, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK Cell Receptors of the Orangutan (Pongo pygmaeus): A Pivotal Species for Tracking the Coevolution of Killer Cell Ig-Like Receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 33.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 34.Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of Killer Cell Ig-Like Receptor (KIR) Genes: Definition of an Orangutan KIR Haplotype Reveals Expansion of Lineage III KIR Associated with the Emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 35.Parham P, Abi-Rached L, Matevosyan L, Moesta AK, Norman PJ, Older Aguilar AM, Guethlein LA. Primate-Specific Regulation of Natural Killer Cells. J Med Primatol. 2010;39:194–212. doi: 10.1111/j.1600-0684.2010.00432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 37.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 38.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol. 2004;21:2079–2091. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- 40.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O’Connor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. 2010;62:295–306. doi: 10.1007/s00251-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics. 2010;62:281–293. doi: 10.1007/s00251-010-0433-4. [DOI] [PubMed] [Google Scholar]
- 43.Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, Parham P, Walter L. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones DC, Hiby SE, Moffett A, Trowsdale J, Young NT. Nature of allelic sequence polymorphism at the KIR3DL3 locus. Immunogenetics. 2006;58:614–627. doi: 10.1007/s00251-006-0130-5. [DOI] [PubMed] [Google Scholar]
- 46.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, Trowsdale J, Moffett A. Molecular characterization of KIR3DL3. Immunogenetics. 2006;57:904–916. doi: 10.1007/s00251-005-0060-7. [DOI] [PubMed] [Google Scholar]
- 47.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 48.Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, Moretta A. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–228. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- 49.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 50.Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127:26–39. doi: 10.1111/j.1365-2567.2008.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, van der Merwe PA, Kumagai I, Maenaka K. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci U S A. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharkey AM, Gardner L, Hiby S, Farrell L, Apps R, Masters L, Goodridge J, Lathbury L, Stewart CA, Verma S, Moffett A. Killer Ig-like receptor expression in uterine NK cells is biased toward recognition of HLA-C and alters with gestational age. J Immunol. 2008;181:39–46. doi: 10.4049/jimmunol.181.1.39. [DOI] [PubMed] [Google Scholar]
- 53.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2011;39:D1171–1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Rood JJ, van Leeuwen A. Leukocyte grouping. A method and its application. J Clin Invest. 1963;42:1382–1390. doi: 10.1172/JCI104822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert E, Amos DB, Bodmer WF, Ceppellini R, Dausset j, Kissmeyer-Nielsen F, Mayr W, Payne R, Van Rood JJ, Terasaki PI, Trnka Z, Walford RL. Nomenclature for factors of the HLA system--1977. Tissue Antigens. 1978;11:81–86. doi: 10.1097/00007890-197805000-00011. [DOI] [PubMed] [Google Scholar]
- 56.Wan AM, Ennis P, Parham P, Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986;137:3671–3674. [PubMed] [Google Scholar]
- 57.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158:4026–4028. [PubMed] [Google Scholar]
- 62.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 63.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 Domain of KIR3D Acts as a Major Histocompatibility Complex Class I Binding Enhancer. J Exp Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, Pando M, Thananchai H, Dong T, Rowland-Jones S, Brodsky FM, Parham P. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;51:639–646. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- 66.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 67.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 68.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 69.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens. 2003;62:79–86. doi: 10.1034/j.1399-0039.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 70.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, Middleton D, Carrington M, Trowsdale J. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2009;19:737–751. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 72.Williams F, Maxwell LD, Halfpenny IA, Meenagh A, Sleator C, Curran MD, Middleton D. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum Immunol. 2003;64:729–732. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- 73.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, Parham P. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 76.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 77.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 78.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 81.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, Marsh SG, Miller JS, Parham P, Geraghty DE. Different patterns of evolution in the centromeric and telomeric regions of group a and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stewart CA, Laugier-Anfossi F, Vâely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagnâe F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185:4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 86.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 87.Peruzzi M, Parker KC, Long EO, Malnati MS. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol. 1996;157:3350–3356. [PubMed] [Google Scholar]
- 88.Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585–1590. doi: 10.1084/jem.184.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 91.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med. 1996;183:1817–1827. doi: 10.1084/jem.183.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 94.Taner SB, Pando MJ, Roberts A, Schellekens J, Marsh SG, Malmberg KJ, Parham P, Brodsky FM. Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J Immunol. 2011;186:62–72. doi: 10.4049/jimmunol.0903657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parsons MS, Boulet S, Song R, Bruneau J, Shoukry NH, Routy JP, Tsoukas CM, Bernard NF. Mind the gap: lack of association between KIR3DL1*004/HLA-Bw4-induced natural killer cell function and protection from HIV infection. J Infect Dis. 2010;202(Suppl 3):S356–360. doi: 10.1086/655966. [DOI] [PubMed] [Google Scholar]
- 96.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 97.Santourlidis S, Graffmann N, Christ J, Uhrberg M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180:418–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 98.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 99.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trompeter HI, Gomez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, Uhrberg M. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. 2005;174:4135–4143. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]
- 101.Presnell SR, Zhang L, Ramilo CA, Chan HW, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int Immunol. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 102.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 103.van Bergen J, Stewart CA, van den Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. 2005;35:2191–2199. doi: 10.1002/eji.200526201. [DOI] [PubMed] [Google Scholar]
- 104.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 105.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 107.Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185:2009–2012. doi: 10.4049/jimmunol.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, Anderson SK, Miller JS. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mrozek E, Anderson P, Caligiuri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 110.Pascal V, Yamada E, Martin MP, Alter G, Altfeld M, Metcalf JA, Baseler MW, Adelsberger JW, Carrington M, Anderson SK, McVicar DW. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 111.Murphy WJ, Pevzner PA, O’Brien SJ. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 112.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Yang H. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, Kim RW, Huntley JJ, Luo S, Schroth GP, Wu TD, Kim H, Yang KS, Park WY, Church GM, Lee C, Kingsmore SF, Seo JS. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. 2010;185:4238–4251. doi: 10.4049/jimmunol.1001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]