Abstract
We describe the gut bacterial diversity inhabiting two saprophagous syrphids and their breeding substrate (decayed tissues of the columnar cactus Isolatocereus dumortieri). We analyzed the gut microbiota of Copestylum latum (scooping larvae that feed on decayed cactus tissues) and Copestylum limbipenne (whose larvae can also feed on semiliquid tissues) using molecular techniques. DNA was extracted from larval guts and cactus tissues. The V1-V3 region of the 16S rRNA genes was amplified and sequenced. A total of 31079 sequences were obtained. The main findings are: C. limbipenne is dominated by several Enterobacteriaceae, including putative nitrogen-fixing genera and pectinolitic species and some denitrifying species, whereas in C. latum unclassified Gammaproteobacteria predominate. Decayed tissues have a dominant lactic acid bacterial community. The bacterial communities were more similar between larval species than between each larva and its breeding substrate. The results suggest that the gut bacterial community in these insects is not strongly affected by diet and must be dependent on other factors, such as vertical transmission, evolutionary history and host innate immunity.
Introduction
Copestylum is a neotropical endemic syrphid lineage that harbours one of the highest species richness, with over 400 species [1], [2], [3]. Larvae of Copestylum are saprophagous (Figure 1) and live in a large variety of microhabitats, with decaying Cactaceae and Agavaceae tissues as one of their most frequently reported breeding media [4], [5]. Saprophagous syrphids are ecologically important because of the potential role of their larvae in nutrient recycling processes [3], [6], [7]. For instance, larvae of Copestylum Macquart 1986 (Diptera: Syrphidae) are commonly bred in decayed cactus species and assist in the degradation of cactus necroses contributing to recycling processes in xeric environments [6], [7], [8].
Figure 1. Larval species (a), decayed cactus tissues (b) and columnar cactus Isolatocereus dumortieri (c).
Besides their ecological importance, this group presents interesting feeding strategies: Rotheray et al. found morphological differences among Copestylum larvae reared from Cactaceae [3]. They found two functional morphological trends: one trend is towards feeding on watery decay and the other towards feeding in firmer decay. The species that can feed on solid material have specialized grinding mills in their head skeletons to break up the tissues and scoop food, specialized armoured thoraces for gripping and protection during tunnelling, and a short posterior breathing tube. The species that can feed on watery material (straining) have reduced armature and have an elongate posterior breathing tube. The elongate breathing tube in Copestylum species enables them to obtain atmospheric oxygen from these decomposed substrates. Finally, some species are intermediate between these feeding strategies. Examples of scooping species are C. latum and C. posticum; some straining species are C. mila and C. hidalgense, and the intermediate species are C. limbipenne and C. marginatum
There are no studies about the microbial community found in the intestinal tract of Copestylum larvae. Otherwise, the roles of microorganisms are well-studied in the cactus-microorganism-Drosophila model [9], [10]. Bacteria are the first microorganisms to grow in newly injured tissue, cactophilic yeast are secondary invaders and the medium created by bacteria serve to host selection for Drosophila and stimulate oviposition. Bacteria are also important sources of nutrition for larvae.
The ecology of cactus degradation (Figure 1) is a complex process, involving many different interacting microorganisms, including both yeast and bacteria [10]. Arms and stems of columnar cacti (Cactaceae) occasionally become necrotic and serve as feeding and breeding sites for a variety of arthropods [3]. Several kinds of these rots develop when bacteria and yeast colonize tissue weakened by injury, environmental stress or senescence. The bacterial communities utilizing the necrotic tissues of columnar cacti are important components of the decayed tissues; injured cactus tissue can be infected by bacteria in the environment developing a rot pocket or necrosis [11]. In cactus necrosis, microbes lyse the plant cells, creating a wet, nutrient-rich microenvironment in the midst of the xeric environments. Necrosis provides substrates for feeding and breeding to cactophilic species such as beetles (Coleoptera) [12] and flies (Diptera) [13].
Isolatocereus dumortieri (Scheidw) Backeb (Figure 1) is a cactus species endemic of the central Mexican semiarid scrublands [14], [15]. This cactus is a common breeding medium for hoverflies [6], [7].
The characterization of the interactions in this cactus-microorganism-hoverfly system provides valuable information about host selection and feeding behaviour of the hoverflies in xeric environments, and important data about the role of each component (microorganisms and hoverflies) in decomposition processes in Mexican scrublands. Despite their central role in the cactus-microorganism-hoverfly system, the bacterial component has not been characterized. There is a complete lack of information on the microorganisms inhabiting both decaying cacti and larvae breeding on them, which is a key to understand the interactions developing between the cactus and the insect.
This study describes, for the first time, the bacterial diversity inhabiting in necrotic tissue of the columnar cacti Isolatocereus dumortieri and in the gut of two species of Copestylum by partial sequencing of 16S rRNA genes directly amplified from samples. We have chosen two species of Copestylum with two different feeding behaviours: C. latum, which can scoop decayed tissues, and C. limbipenne, which has an intermediate behaviour between scooping tissues and feeding on liquid decomposed cactus. The goals are to know whether these two different species of Copestylum larvae harbour different microbiota, what the differences in the microbial communities inhabiting cactus tissues in different degrees of decomposition are and to what extent the larval microbiota is related to that of their feeding material. The possible role of bacterial communities in the larval biology and the decomposition of the columnar cactus I. dumortieiri are discussed.
Materials and Methods
Sample collection
Five samples of decayed cactus tissues from different individuals of I. dumortieri (Pap of Copestylum limbipenne or PLIM in text) with larvae of C. limbipenne (CLIM) and five different samples of stems of I. dumortieri (Pap of Copestylum Latum or PLAT in text) with larvae of C. latum (CLAT) were collected in one survey in March 2009 in “Barranca de Metztitlán” Biosphere Reserve, Hidalgo, México. In these samples neither species was found together (but other research has reported that they may be found in the same stem of decayed cactus tissue) [6]. All larvae in each sample were collected and placed in 90% ethanol. Necrotic tissue in which each species grew was put in sterile containers that were frozen until further manipulation. Six larvae for each species were randomly chosen from the collected cactus samples for dissection and their complete intestinal tract was extracted using a maculating loop. All necessary permits were obtained for the described field studies. The field studies did not involve endangered or protected species.
DNA extraction
DNA was extracted from larval guts and cactus tissues as described in Latorre et al. [16]. Before DNA extraction, cactus tissues were treated as follows: they were homogeneized in PBS (containing, per litre, 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 [pH 7.2]) and centrifuged at 1,800 g for 8 min to remove plant material as far as possible; 1–4 mL of supernatants were centrifuged at 22,000 g for 5 min to pellet bacterial cells.
PCR amplification of bacterial 16S rRNA gene sequences
DNA samples from each fly species and each cactus tissue were used as templates for PCR amplification of a fragment of the 16S rRNA gene using the composite forward primer 5′-GCCTCCCTCGCGCCATCAGNNNNNNTCAGAGTTTGATCMTGGCTCAG-3′ (where the underlined sequence is that of 454 Life Sciences primer A, NNNNNN designates the unique six base barcode used to tag each PCR product, and the broad range bacterial primer B8F is in italics), and the reverse primer 5′-GCCTTGCCAGCCCGCTCAGGC TGCTGCCTCCCGTAGGAGT–3′ (where the underlined sequence is that of 454 Life Sciences primer B and the broad range bacterial primer B357R is in italics). The PCR conditions were 5 min of initial denaturation at 95°C followed by 25 cycles of denaturation (30 s at 95°C), annealing (30 s at 52°C) and elongation (60 s at 72°C), with a final extension at 72°C for 8 min.
PCR product purification and pyrosequencing
Each PCR product was purified by filtration and equal amounts of the four samples with different sample-specific barcode sequences were pooled. Then, the pooled DNA was isolated from a 0.8% agarose gel and purified. Purifications were carried out using the High Pure PCR Product Purification Kit (Roche). The pooled DNA was sent for pyrosequencing with primer A on an eight-lane picotiter plate on a Genome Sequencer FLX system (Roche).
Sequence analysis
Sequences with low average quality scores (<20) and short read lengths (<200 nt) were removed. The remaining sequences were checked for potential chimeras using the chimera.slayer and the chimera.pintail tools as implemented in the mothur software package v.1.13.0 [17].
Taxonomic affiliation
The taxonomic affiliation of partial-length sequences was determined using the Classifier tool of the Ribosomal Database Project-II (RDP) [18], [19]. This method is widely used and provides rapid taxonomic classification from domain to genus of both partial and full-length 16S rRNA gene sequences. We used a 50% bootstrap threshold, stopping the assignation at the last clear taxonomic level and leaving successive levels as unclassified (uc).
Phylotype definition
Clustering at 98% of sequence identity was carried out using cd-hit-est [20] and the resulting phylotypes were used to study sample composition at the ‘species’ level.
Adjustment of the number of reads in each sample to the smallest data set size
Re-sampling of the 4 samples to identical sequencing depth was done by randomly selecting reads in the fasta files using Daisy_chopper v0.6 (http://www.genomics.ceh.ac.uk/GeneSwytch/Tools.html).
Estimation of bacterial diversity
The Shannon diversity index (H) [21], that correlates positively with taxa richness and evenness, the Chao1 richness estimator [22], and rarefaction curves, were calculated for each sample at family, genus and phylotype levels (clusters at 98% sequence identity). Diversity and richness were estimated with both the full data sets and the data sets adjusted to equal sequence number.
Statistical comparison of sample composition
The patterns of variation in the taxonomic distributions found in our samples were explored using detrended correspondence analysis (DCA).
Diversity and richness indices and DCAs were calculated using the free-licence R package [23] and the vegan R package [24].
Nucleotide sequence accession numbers
The non-redundant sequences from this study have been deposited in the GenBank database under accession numbers JN569361 - JN570496.
Results
Bacterial diversity and rarefaction analysis
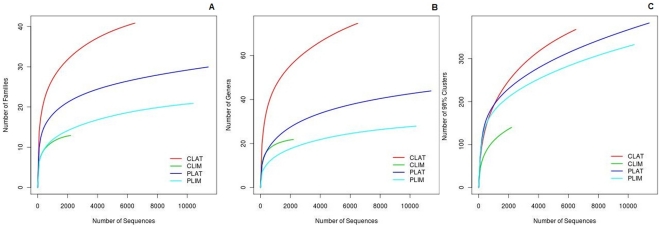
We were close to completeness of the bacterial inventory at family and genus level according to the rarefaction curves (Figure 2) and the Chao1 estimator of bacterial richness (Table 1). The curves for phylotypes (at 98% identity), which do not reach the plateau, and the comparison between observed and estimated richness, indicate some of phylotypes that have been missed.
Figure 2. Rarefaction curves calculated at family (a), genus (b) and phylotype (clustering at 98% of identity) (c) levels.
CLAT, C. latum larvae; CLIM, C. limbipenne larvae; PLAT, C. latum cactus medium; PLIM, C. limbipenne cactus medium.
Table 1. Observed richness, Chao1 richness estimator (and standard error, SE), and Shannon diversity index (H) in larval guts (CLIM: C. limbipenne, CLAT: C. latum) and in decayed cactus stems (PLAT: I. dumortieri decayed tissues with C. latum larvae, PLIM: I. dumortieri decayed tissues with C. imbipenne larvae) for the full data sets and re-sampled data sets adjusted by the smallest sample size (average value and standard deviation, SD, for three replicates).
| CLAT | CLIM | PLAT | PLIM | |||
| Full | # sequences | 6639 | 2363 | 11505 | 10572 | |
| data sets | Family | # families | 41 | 13 | 30 | 21 |
| Chao1 (SE) | 45 (4.84) | 13 (0.73) | 35 (10.17) | 23 (5.29) | ||
| Shannon H | 1.87 | 0.91 | 1.48 | 1.20 | ||
| Genus | # genera | 75 | 22 | 44 | 28 | |
| Chao1 (SE) | 86 (8.33) | 22 (1.87) | 53 (10.68) | 33 (10.17) | ||
| Shannon H | 2.16 | 1.86 | 1.50 | 1.23 | ||
| Clusters 98% | # clusters | 370 | 143 | 384 | 334 | |
| Chao1 (SE) | 464 (24.84) | 188 (19.84) | 651 (74.50) | 473 (39.93) | ||
| Shannon H | 4.84 | 3.99 | 5.09 | 4.98 | ||
| Re-sampled | # sequences | 2363 | 2363 | 2363 | 2363 | |
| data sets | Family | # families | 33 (5.51) | 13 | 22 (1.15) | 14 (1) |
| (average (SD)) | Chao1 | 65 (44.59) | 13 | 24 (2.65) | 15 (1.26) | |
| Shannon H | 1.86 (0.01) | 0.91 | 1.46 (0.02) | 1.19 (0.02) | ||
| Genus | # genera | 59 (6.66) | 22 | 28 (2.08) | 18 (2.52) | |
| (average (SD)) | Chao1 | 97 (36.84) | 22 | 39 (15.49) | 20 (3.93) | |
| Shannon H | 2.14 (0.02) | 1.86 | 1.48 (0.02) | 1.22 (0.02) | ||
| Clusters 98% | # clusters | 263 (9.45) | 143 | 236 (3.51) | 219 (4.62) | |
| (average (SD)) | Chao1 | 381 (43.01) | 188 | 324 (21.44) | 283 (33.42) | |
| Shannon H | 4.78 (0.03) | 3.99 | 5.02 (0.02) | 4.92 (0.01) |
The Shannon diversity index (Table 1), calculated at each taxonomic level (family, genus, phylotype), show the same tendency between species in larval and substrate samples: C. latum is more diverse than C. limbipenne, and PLAT is more diverse than PLIM. In a global view, C. latum is the most diverse sample, except at the phylotype level, where substrate samples are more diverse than C latum. This fact is probably due to the higher number of sequences obtained from PLAT and PLIM regarding larval samples.
As shown in Figure 2 and Table 1, the bacterial communities inhabiting both insect and cactus samples show a high level of diversity, with hundreds of different phylotypes in each sample. Among the insect samples, CLAT gut microbiota is the most complex. It displays the highest diversity indices at any of the three levels of diversity considered, whereas CLIM gut microbiota is less diverse, partly because CLIM has the lowest number of sequences. These facts could be related to the complexity of the vegetal substrates they feed on (i.e., a more diverse microbiota is expected in insects that feed on more complex substrates, composed of different polymeric substances whose degradation in anaerobic gut conditions requires a more complex microbial community).
Bacterial distribution among samples
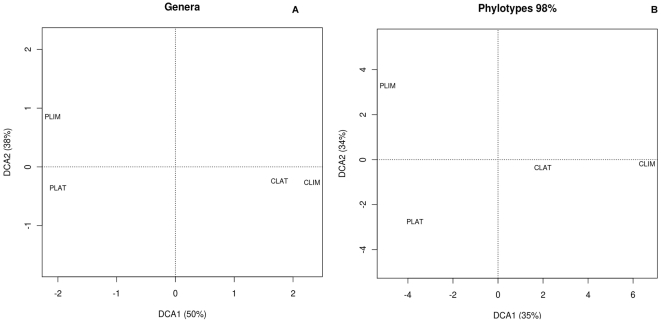
DCAs indicate that the bacterial communities present in both fly species are more related to each other than to those harboured in their plant substrates (Figure 3). The first DCA axis clearly separates insects from their substrates. It explains 50% of variance at genus level, and 35% at phylotype level. The second axis separates both types of substrates, and accounts for 38% of variance at genus level, and 34% at phylotype level. However, both insects clearly harbour different communities that include species-specific sequences, as well as bacterial groups that are widespread in other Diptera analyzed so far [25] (see below). In addition, some phylotypes are shared only among larvae and the plant they feed on, whereas every sample harbours distinctive phylotypes.
Figure 3. Detrended correspondence analysis based on a) genus and b) phylotype distributions.
CLAT, C. latum larvae; CLIM, C. limbipenne larvae; PLAT, C. latum cactus medium; PLIM, C. limbipenne cactus medium.
An overall description of the sequences found in the analyzed samples is shown in Table 2, where a list of the taxonomic affiliation of the sequences down to genus level, together with their relative frequency in each of the 4 analyzed samples, is provided. Larva and cactus microbiota differ not only in genus composition but also in the relative frequency of shared genera. In good agreement with the diversity data discussed above, there are many genera that are only present in C. latum, such as the putative tethatrionate oxidyzing Tetrathiobacter, which accounts for almost 5% of CLAT sequences. Most of those genera belonged to the classes Actinobacteria and Alphaproteobacteria. Another distinctive feature of C. latum is the high prevalence of Gammaproteobacteria, most of them non-characterized below the class taxonomic level (52% of all sequences). In contrast, C. limbipenne is characterized by a high prevalence of Enterobacteriaceae (76% of all sequences), most of them within the Enterobacter, Citrobacter and Pectobacterium genera. The lactic acid bacteria Lactobacillus and Leuconostoc are the most frequently retrieved genera in the cactus tissues, although they are also found in the larval samples.
Table 2. Taxonomic composition of the samples (percentage of sequences belonging to each bacterial genus).
| Phylum | Class | Order | Family | Genus | CLAT | CLIM | PLAT | PLIM |
| Acidobacteria | Acidobacteria Gp1 | Acidobacteria Gp1 | Acidobacteria Gp1 | Gp1 | 0.0001 | |||
| Actinobacteria | Actinobacteria | Actinomycetales | Actinomycetaceae | uc Actinomycetaceae | 0.0003 | |||
| Cellulomonadaceae | Cellulomonas | 0.0002 | ||||||
| Corynebacteriaceae | Corynebacterium | 0.0003 | ||||||
| Dietziaceae | Dietzia | 0.0020 | ||||||
| Microbacteriaceae | Agreia | 0.0002 | ||||||
| Microbacterium | 0.0002 | |||||||
| uc Microbacteriaceae | 0.0002 | |||||||
| uc Actinomycetales | uc Actinomycetales | 0.0051 | 0.0001 | 0.0001 | ||||
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 0.0012 | 0.0008 | ||
| Porphyromonadaceae | Butyricimonas | 0.0003 | ||||||
| Dysgonomonas | 0.0434 | 0.0719 | 0.0002 | 0.0003 | ||||
| Parabacteroides | 0.0051 | 0.0067 | 0.0012 | |||||
| Proteiniphilum | 0.0015 | |||||||
| uc Porphyromonadaceae | 0.0048 | 0.0106 | 0.0001 | |||||
| Prevotellaceae | Prevotella | 0.0003 | ||||||
| uc Prevotellaceae | 0.0017 | |||||||
| Rikenellaceae | Alistipes | 0.0005 | 0.0013 | 0.0002 | ||||
| uc Bacteroidales | uc Bacteroidales | 0.0023 | ||||||
| Flavobacteria | Flavobacteriales | Flavobacteriaceae | Wautersiella | 0.0038 | 0.0002 | |||
| uc Flavobacteriaceae | 0.0002 | 0.0017 | 0.0003 | |||||
| uc Flavobacteriales | uc Flavobacteriales | 0.0006 | ||||||
| Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | Parapedobacter | 0.0005 | 0.0003 | |||
| uc Bacteroidetes | uc Bacteroidetes | uc Bacteroidetes | uc Bacteroidetes | 0.0380 | 0.0013 | 0.0003 | ||
| Cyanobacteria | Cyanobacteria | Cyanobacteria | Chloroplast | Streptophyta | 0.0022 | |||
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | Facklamia | 0.0015 | 0.0059 | ||
| Enterococcaceae | Enterococcus | 0.0008 | 0.0030 | 0.0009 | ||||
| Vagococcus | 0.0012 | 0.0068 | 0.0002 | |||||
| uc Enterococcaceae | 0.0003 | 0.0245 | 0.0003 | |||||
| Lactobacillaceae | Lactobacillus | 0.0524 | 0.0161 | 0.3955 | 0.6222 | |||
| uc Lactobacillaceae | 0.0002 | 0.0009 | 0.0031 | |||||
| Leuconostocaceae | Leuconostoc | 0.0033 | 0.0008 | 0.4075 | 0.0760 | |||
| Streptococcaceae | Lactococcus | 0.0068 | 0.0221 | |||||
| uc Lactobacillales | uc Lactobacillales | 0.0008 | 0.0080 | 0.0173 | 0.0501 | |||
| uc Bacilli | uc Bacilli | uc Bacilli | 0.0047 | 0.0004 | 0.0694 | 0.1860 | ||
| Clostridia | Clostridiales | Lachnospiraceae | uc Lachnospiraceae | 0.0001 | ||||
| Ruminococcaceae | uc Ruminococcaceae | 0.0003 | 0.0001 | 0.0003 | ||||
| Veillonellaceae | Allisonella | 0.0001 | ||||||
| uc Clostridiales | uc Clostridiales | 0.0003 | ||||||
| Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Erysipelothrix | 0.0003 | 0.0001 | |||
| uc Erysipelotrichaceae | 0.0003 | 0.0010 | 0.0002 | |||||
| uc Firmicutes | uc Firmicutes | uc Firmicutes | uc Firmicutes | 0.0002 | 0.0142 | 0.0436 | ||
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Aurantimonadaceae | uc Aurantimonadaceae | 0.0002 | |||
| Brucellaceae | Ochrobactrum | 0.0203 | ||||||
| uc Brucellaceae | 0.0003 | |||||||
| Hyphomicrobiaceae | Devosia | 0.0104 | ||||||
| Xanthobacteraceae | Xanthobacter | 0.0003 | ||||||
| uc Xanthobacteraceae | 0.0023 | |||||||
| uc Rhizobiales | uc Rhizobiales | 0.0002 | ||||||
| Rhodobacterales | Rhodobacteraceae | Haematobacter | 0.0009 | |||||
| Ketogulonicigenium | 0.0002 | |||||||
| Paracoccus | 0.0491 | |||||||
| Rhodobacter | 0.0002 | |||||||
| uc Rhodobacteraceae | 0.0020 | |||||||
| Rhodospirillales | Acetobacteraceae | Acetobacter | 0.0003 | 0.0120 | 0.0076 | |||
| Betaproteobacteria | Burkholderiales | Alcaligenaceae | Achromobacter | 0.0005 | ||||
| Alcaligenes | 0.0054 | 0.0013 | ||||||
| Bordetella | 0.0008 | |||||||
| Castellaniella | 0.0015 | 0.0001 | ||||||
| Kerstersia | 0.0054 | 0.0001 | ||||||
| Pigmentiphaga | 0.0011 | |||||||
| Pusillimonas | 0.0015 | 0.0001 | ||||||
| Tetrathiobacter | 0.0499 | |||||||
| uc Alcaligenaceae | 0.0640 | 0.0025 | 0.0001 | |||||
| Comamonadaceae | Comamonas | 0.0193 | 0.0005 | |||||
| uc Comamonadaceae | 0.0117 | 0.0003 | ||||||
| Oxalobacteraceae | Oxalicibacterium | 0.0039 | ||||||
| uc Oxalobacteraceae | 0.0002 | |||||||
| uc Burkholderiales | uc Burkholderiales | 0.0041 | 0.0006 | |||||
| uc Betaproteobacteria | uc Betaproteobacteria | uc Betaproteobacteria | 0.0003 | |||||
| Deltaproteobacteria | Bdellovibrionales | Bdellovibrionaceae | Bdellovibrio | 0.0003 | ||||
| Epsilonproteobacteria | Campylobacterales | Campylobacteraceae | Campylobacter | 0.0002 | ||||
| Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Citrobacter | 0.0050 | 0.0677 | 0.0007 | ||
| Enterobacter | 0.0027 | 0.2603 | ||||||
| Erwinia | 0.0057 | 0.0025 | 0.0003 | |||||
| Klebsiella | 0.0002 | 0.0004 | ||||||
| Morganella | 0.0002 | |||||||
| Pectobacterium | 0.0036 | 0.0478 | 0.0255 | 0.0019 | ||||
| Providencia | 0.0030 | 0.0102 | 0.0003 | |||||
| Salmonella | 0.0002 | |||||||
| Serratia | 0.0001 | |||||||
| uc Enterobacteriaceae | 0.0090 | 0.3737 | 0.0007 | 0.0003 | ||||
| Oceanospirillales | uc Oceanospirillales | uc Oceanospirillales | 0.0030 | |||||
| Pasteurellales | Pasteurellaceae | uc Pasteurellaceae | 0.0170 | |||||
| Xanthomonadales | Xanthomonadaceae | Luteimonas | 0.0009 | |||||
| Stenotrophomonas | 0.0001 | |||||||
| uc Gammaproteobacteria | uc Gammaproteobacteria | uc Gammaproteobacteria | 0.5192 | 0.0901 | 0.0017 | 0.0007 | ||
| uc Proteobacteria | uc Proteobacteria | uc Proteobacteria | uc Proteobacteria | 0.0002 | ||||
| Synergistetes | Synergistia | Synergistales | Synergistaceae | Aminiphilus | 0.0001 | |||
| Tenericutes | Mollicutes | Acholeplasmatales | Acholeplasmataceae | Acholeplasma | 0.0010 | |||
| uc Mollicutes | uc Mollicutes | uc Mollicutes | 0.0001 | |||||
| uc Bacteria | uc Bacteria | uc Bacteria | uc Bacteria | uc Bacteria | 0.0002 | 0.0003 | 0.0004 |
CLAT: Copestylum latum, CLIM: Copestylum limbipenne, PLAT: C. latum cactus breeding medium, PLIM: C. limbipenne cactus breeding medium.
Discussion
This is the first attempt to describe the gut bacterial communities in Copestylum larvae that breed in decomposed cacti. It is also the first report about bacterial species in Isolatocereus dumortieri (columnar cactus). Foster and Fogleman [26] reported bacteria in columnar cactus rotten tissues from Stenocereus thurberi (pipe cactus), Carnegieae gigantean (saguaro) and Lophocereus schotii (senita cactus). In contrast to our study, they found Pseudomonas, Staphylococcus, Enterococcus and Xantomonas, and similar to us, Erwinia.
C. latum and C. limbipenne samples display a high relative abundance of Gammaproteobacteria, mostly Enterobacteriaceae in C. limbipenne. Two enterobacterial genera (Enterobacter and Klebsiella) are found only in larval samples. Enterobacteria are heterotrophic facultative anaerobes and have frequently been found in insect microbiota using both culture and molecular techniques [27, 28 29; 30 31, 32; 33]. Some of these Enterobacteria are diazotrophs (i.e. nitrogen fixing), which would provide the insect with an obvious advantage in an environment depleted in fixed oxygen. In our case, some of the enterobacterial genera detected (Citrobacter, Enterobacter, Erwinia, Klebsiella) include diazotrophic species not present in the vegetal substrate microbiota. Given the selective advantage that the availability of a fixed nitrogen source would provide for the insect host, one could speculate that these bacteria are harboured in the larvae due to a vertical transmission, as postulated for the fruit fly [33]. In addition to these putatively nitrogen-fixing bacteria, both cacti and larvae include the pectinolytic and phytopathogenic genus Pectobacterium, also found in the fruit-fly.
Interestingly, other genera also involved in the nitrogen cycling have been found in association with C. latum larvae in a relatively high abundance. Such is the case of Paracoccus and Comamonas, which include some denitrifying species [25].
Compared to previously published studies on insect microbial diversity, there are some remarkable differences. An example is the absence of bacteria from the genera Spiroplasma, Wolbachia and Bacillus, frequently found in association with other insects [27]; [34]; [35]; [36]; [30]; [29], but absent from both C. latum and C. limbipenne larva. The acetic acid bacteria Acetobacter, which has recently been described as a newly emerging symbiont of insects, is mostly absent from the CLAT and CLIM larvae, although it is relatively abundant in the cactus tissue colonized by CLAT. Conversely, these species harbour bacteria that have not previously been found associated with insects, such as Dysgonomonas, Ochrobactrum and Devosia, for example [25]. Lactic acid bacteria that ferment sugars are found frequently as plant-commensal microbiota and also as part of insect-associated bacteria, where it has been speculated that they play a role in the larval digestive tract [27].
The results obtained here indicate that the insect microbiota is not the same as that found in its corresponding vegetable substrate, since there are many bacterial groups in the insects that have not been found in their substrates. However, substrates could act as a reservoir for newly acquired species, which can eventually become part of the commensal community. Furthermore, the gut bacterial community in these insects could be partially inherited by vertical transmission from mother to offspring. Thus, in the fruit fly-bacteria association, Ben-Yosef et al. [33] found that the microbiota is vertically transmitted and colonizes the plant surface after hatching. According to these authors, the larva would carry a “survival pack” of bacteria, including nitrogen fixing and pectinolytic genera, which would help in the first stages of plant colonization. In fact, as discussed above, CLAT and CLIM also harbour putatively pectinolytic and diazotrophic Enterobacteriaceae. Other factors shaping the specific commensal/mutualistic bacteria, such as the host innate immunity and evolutionary history-events (constrains, isolation, horizontal transmission, etc.), cannot be ruled out. The gut microbiota of the two Copestylum species is relatively similar, as one could expect in two phylogenetically related insect species living in similar ecological niches. On the other hand, the differences in the microbiota between the two substrates should correspond to the bacterial succession that is taking place during the decomposition process of the cactus.
Another factor that could affect the studied communities is the presence of plant allelochemicals that could restrict the growth of some cactophilic yeast and bacterial species [37]. For instance, Starmer et al. [38] recognized that some species of yeast were inhibited by some triterpene glycosides found in some columnar cacti. One possibility is that some bacteria are better adapted to the cactus necrotic niche and are more tolerant to potential toxic secondary plant compounds, because many columnar cacti have triterpene glycosides and isoquinone alkaloids [39]. Kinoshita [40] found one triterpenoid saponin called dumortierninoside A, but the role of this triterpene in dipteran-cactus relationships is unknown.
Finally, the availability of specific nutrients and dipteran adaptations for cactus species can be related to the specificity in the microbiota. It has been shown that some Drosophilids have the ability to metabolize volatiles, such as ethanol vapor, as an adaptation for survival in volatile-rich columnar cactus rots [41]. We can speculate that this ability is provided by the microbiota. These evolutionary trends have not yet been proved in Copestylum species. Moreover, this information is necessary to understand the evolution of Dipteran species in cactus necrosis. In this research, we only analyze decomposed stems of I. dumortieri cactus, but central Mexican scrublands have other cactus species used as breeding places for Copestylum larvae [4], [5], [8], [3], [6], [7]. Therefore, the complexity of this system needs to be investigated as in the case of the cactus-Drosophila-microorganism system. More details about the differences in the bacterial communities from the first decayed cactus stems to rotten tissues and the differences with other feeding strategies (e.g. straining larvae) will be of interest to understand the role of bacteria in the decomposition process and in the colonization of syrphid species.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was funded by AECID (project A/020305/08), FOMIX CONACYT-Hidalgo (project 95828) and SEP-CONACYT (project 84127) to M. A. M.-G., and BFU2009-12895-C02-01 from the Ministerio de Ciencia e Inovación to A.L. We extend our gratitude to the management of the Barranca de Metztitlán Biosphere Reserve for assisting us in this research. We appreciate discussions with Dr. Graham Rotheray and P.E. Cruz-Dominguez. C Aguilar-Miguel and M.C. García-Chávez assisted with fieldwork. A.P.M. acknowledge the scholarship provided by The Alβan programme, the European Union Programme of High Level Scholarships for Latin America, No. E07D401138MX and CONACYT program (207522) for doctoral fellowship. A.D. is recipient of a fellowship from the Instituto de Salud Carlos III, Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Thompson FC. A contribution to a generic revision of the Neotropical Milesiinae (Diptera:Syrphidae). Arquivos de Zoologia. 1972;23:73–215. [Google Scholar]
- 2.Rotheray GE, Hancock EG, Marcos-García MA. Neotropical Copestylum (Diptera:Syrphidae) breeding in bromelids (Bromeliaceae) including 22 new species. Zoological Journal of Linnean Society. 2007;150:267–317. [Google Scholar]
- 3.Rotheray GE, Marcos-García MA, Hancock G, Pérez-Bañón C, Maier CT. Neotropical Copestylum (Diptera, Syrphidae) breeding in Agavaceae and Cactaceae including seven new species. Zoological Journal of Linnean Society. 2009;156:697–749. [Google Scholar]
- 4.Marcos-García MA, Pérez-Bañón C. Immature stages of Copestylum tamaulipanum and Copestylum lentum (Diptera: Syrphidae). Eur J Entomol. 2001;98:375–385. [Google Scholar]
- 5.Marcos-García MA, Pérez-Bañón C. Life cycle, adult and immature stages of a new species of Copestylum (Diptera: Syrphidae) from Mexico reared from Cactaceae. Annals of the Entomological Society of America. 2002;95:432–440. [Google Scholar]
- 6.Martínez-Falcón, AP, Marcos-García MA, Díaz-Castelazo, Rico Gray V C. Seasonal changes in a cactus-hoverfly (Diptera: Syrphidae) network. Ecological Entomology. 2010;35:754–759. [Google Scholar]
- 7.Martínez-Falcón, AP, Marcos-García, MA, Moreno C. Temporal shifts and niche overlapping shifts and niche overlapping in Copestylum (Diptera, Syrphidae) communities reared in cactus species in a central Mexican scrubland. 2011. Ecological Research. 2011;26:341–350. [Google Scholar]
- 8.Martínez-Falcón AP, Marcos-García MA, Moreno CE. Entomologia Mexicana vol. 7 (ed. by EG. Estrada, A. Equihua, JR. Padilla and A. Mendoza), pp. 176-181. Colegio de la Frontera Sur, México; 2008. Diversity of Copestylum (Diptera: Syrphidae) associated to decaying cactus in a shrub forest with cattle grazing management. [Google Scholar]
- 9.Fogleman JC, Starmer W, Heed W. Larval selectivity for yeast species by Drosophila mojavensis in natural substrates. Proceedings of National Academy of Science. 1981;78:4435–4439. doi: 10.1073/pnas.78.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogleman JC, Danielson, PB Chemical interactions in the cactus-microorganism-Drosophila model system of the Sonoran desert. American Zoologist, 41, 2001;877-889 [Google Scholar]
- 11.Fogleman JC, Foster LM. Microbial colonization of injured cactus (Stenocereus gummosus) and its relationships to the ecology of cactophilic Drosophila mojavensis. Applied Environmental Microbiology. 1989;55:100–105. doi: 10.1128/aem.55.1.100-105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castrezana S, Markow TA. Arthropod diversity in necrotic tissue of three species of columnar cacti (Cactaceae). Canandian Entomologist. 2001;133:301–309. [Google Scholar]
- 13.Bravo-Hollis H, Sánchez-Mejorada H. I. Universidad Nacional Autónoma de México, México, D. F; 1978. Las cactáceas de México, vol.743 [Google Scholar]
- 14.Heed WB, Mangan RL. Community ecology of the sonorant desert Drosophila311-345. In: Ashburner M, Carson HL, Thompson JN, editors. The Genetics and Biology of Drosophila, Vol. 3e . Academic Press, N.Y; 1986. [Google Scholar]
- 15.Jiménez-Sierra C, Figueroa-Jiménez L. Cactáceas y Suculentas Mexicanas 49, 96; 2004. Isolatocereus dumortieri (Scheidw.) Backeb. (Stenocereus dumortieri). [Google Scholar]
- 16.Latorre A, Moya A, Ayala FJ. Evolution of mitochondrial DNA in Drosophila subobscura. Proceedings of National Academy of Science. 1986;83:8649–53. doi: 10.1073/pnas.83.22.8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied Environmental Microbiology. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Research. 2007;35:D169–172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Research. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 21.Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- 22.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–91. [PubMed] [Google Scholar]
- 23.R Development Core Team. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL; 2009. R: A language and environment for statistical computing. [Google Scholar]
- 24.Oksanen J, Kindt R, Legendre P, O'Hara RB, Simpson GL, et al. Vegan: community ecology package. R package version 1.15-1. 2008;25 http://cran.r-project.org/. Accessed 2011 May. [Google Scholar]
- 25.Euzéby JP. 2011. List of Prokaryotic names with Standing in Nomenclature ( http://www.bacterio.cict.fr). Accessed 2011 April 3 2011.
- 26.Foster JL, Fogleman JC. Identification and ecology of the bacterial communities associated with the necroses of three species of cacti. Applied and Environmental Microbiology. 1993;59:1–6. doi: 10.1128/aem.59.1.1-6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, et al. Microbial Ecology publication online; 2011. Bacterial diversity in Solenopsis invicta and Solenopsis geminate ant colonies characterized by 16S amplicon 454 Pyrosequencyng. [DOI] [PubMed] [Google Scholar]
- 28.Lauzon CR, Sjogren RE, Wright SE, Prokopy RJ. Attraction of Rhagoletis pomonella (Diptera: Tephritidae) to odors of bacteria: apparent confinement to specialized members of Enterobacteriaceae. Environmental Entomology. 1998;27:853–857. [Google Scholar]
- 29.Tóth EM, Hell E, Kovács G, Borsodi AK, Marialigeti K. Bacteria Isolated from the different Developmental stages and larval organs of the obligate parasitic fly, Wohlfahrtia magnifica (Diptera: Sarcophagidae) Microbial Ecology. 2006;51:13–21. doi: 10.1007/s00248-005-0090-6. [DOI] [PubMed] [Google Scholar]
- 30.Lindh JM, Terenius O, Faye I. 16S rRNA Gene-Based identification of midgut bacteria from field-caught Anopheles gambiae Sensu Lato and A. Funestus mosquitoes reveals new species related to known insect symbionts. Applied and Enviromental Microbiology. 2005;71:72–73. doi: 10.1128/AEM.71.11.7217-7223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacchetti P, Granchietti A, Landini S, Viti C, Giovannetti L, et al. Relationships between the olive fly and bacteria. Journal of Applied Entomology. 2008;132:682–689. [Google Scholar]
- 32.Behar A, Jurkevith E, Yuval B. Bringing back the fruit into fruit fly-bacteria interactions. Molecular Ecology. 2008;17:1375–1386. doi: 10.1111/j.1365-294X.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Yosef M, Jurkevith E, Yuval B. Effect of bacteria on nutritional status and reproductive success of the Mediterranean fruit fly Ceratitis capitata. Physiological Entomology. 2008;33:145–154. [Google Scholar]
- 34.Jaenike J, Unckless R, Cockburn SN, Boelio AM, Perlman SJ. Adaptation via symbiosis: Recent spread of Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 35.Lindh J, Lehane MJ. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek. 2011;99:711–720. doi: 10.1007/s10482-010-9546-x. [DOI] [PubMed] [Google Scholar]
- 36.Zahner V, Lucarotti CJ. Application of 16S rDNA-DGGE and plate cultura to characterization of bacterial communities associated with the sawfly, Acantholyda erythrocephala (Hymenoptera, Pamphilidae) Current Microbiology. 2008;57:564–569. doi: 10.1007/s00284-008-9243-4. [DOI] [PubMed] [Google Scholar]
- 37.Fogleman JC, Heed WB. Columnar cacti and desert Drosophila: the chemistry of host plant specificity, p 1-24. In: Schmidt J, editor. Species biotic relationships in the arid southwest. University of New Mexico Press, Albuquerque; 1989. [Google Scholar]
- 38.Starmer WT, Phaff HJ, Miranda M, Miller MW, Heed WB. The yeast flora associated with decaying stems of columnar cacti and Drosophila in North America. Evolutionary Biology. 1982;14:269–294. [Google Scholar]
- 39.Fogleman JC, Kircher HW. Differential effects of fatty acids chain length on the viability of two species of cactophilic Drosophila. Lipids. 1989;21:92–99. [Google Scholar]
- 40.Kinoshita K, Koyama K, Takahashi K, Kondo N, Yuasa H. A New Triterpenoid Saponin from Isolatocereus dumortieri. Journal of Natural Products. 2000;63:701–703. doi: 10.1021/np9903907. [DOI] [PubMed] [Google Scholar]
- 41.Etges W. Divergence in cactophilic Drosophila: The evolutionary significance of adult ethanol metabolism. Evolution. 1989;43:1316–1319. doi: 10.1111/j.1558-5646.1989.tb02579.x. [DOI] [PubMed] [Google Scholar]