Abstract
Arginase (EC 3.5.3.1) transcript level and activity were measured in soybean (Glycine max L.) embryos from the reserve deposition stage to postgermination. Using a cDNA probe for a small soybean arginase gene family, no transcript was detected in developing embryos. However, arginase transcripts increased sharply on germination, reaching a maximum at 3 to 5 d after germination. There was low but measurable in vitro arginase specific activity in developing embryos (less than 6% of seedling maximum). During germination arginase specific activity increased in parallel with the sharply increasing arginase transcript level. Seedling arginase activity was largely localized in cotyledons. Arginase activity was assayed in vivo by measuring urea accumulation in a urease-deficient mutant. No urea was detected in developing embryos, whereas accumulated urea paralleled arginase specific activity and transcript level in germinating seedlings. As in planta embryos, cultured cotyledons did not accumulate urea when arginine (Arg) was provided with other amino acids in a “mock” seed-coat exudate. Arg as the sole nitrogen source was converted to urea but did not support cotyledon growth. There appeared to be a lack of recruitment of the low-level arginase activity to hydrolyze free Arg in developing embryos, thus avoiding a futile urea cycle.
Arg, a nitrogen-rich compound, is one of the predominant amino acids in the seed and storage organs of numerous plant species (Van Etten et al., 1967; Polacco and Holland, 1993) and represents a major form of reserve nitrogen. In developing soybean (Glycine max) cotyledons, Arg is both actively synthesized (Micallef and Shelp, 1989b) and supplied from the seed-coat exudate (Rainbird et al., 1984). Arg is incorporated into protein, where it accounts for 18% of the total seed-protein nitrogen, or it remains in the free amino acid pool, constituting more than 60% of the free amino acid nitrogen in developing cotyledons (Micallef and Shelp, 1989a).
During early seedling growth, storage proteins are mobilized to provide amino acids for proteins synthesized in the expanding axes. As part of the overall reconfiguration of free and storage-protein-bound amino acid profiles to that of seedling protein (Thompson, 1980; Polacco and Holland, 1993), Arg breakdown to urea and Orn occurs by the action of arginase (l-Arg amidinohydrolase, EC 3.5.3.1). Arginase activity increases sharply during germination in several species, including pumpkin (Splittstoesser, 1969), broad bean (Kollöffel and Van Dijke, 1975), soybean (Matsubara and Suzuki, 1984; Kang and Cho, 1990), Arabidopsis (Zonia et al., 1995), and loblolly pine (King and Gifford, 1997). After arginase action, Arg nitrogen contained in urea is recycled to ammonia by the action of urease (Polacco and Holland, 1993).
There have been contradictory reports on arginase activity in developing cotyledons in contrast to those on arginase in seedlings. Micallef and Shelp (1989c) suggested that during soybean embryo development, arginase metabolizes a portion of the abundant free Arg. In developing pea cotyledons, however, no Arg-derived urea was detected, although in vitro arginase activity was present at an early stage of cotyledon development (De Ruiter and Kollöffel, 1983).
In the present study we used the cDNA clone AG1, which encodes a soybean seedling axis arginase (Goldraij et al., 1998), and the pleiotropic urease-deficient mutant eu3-e1/eu3-e1 (Meyer-Bothling et al., 1987) to investigate the expression and in vivo activity, respectively, of soybean embryo arginase from the reserve deposition stage to postgermination. In addition, functional tests of arginase were performed in in vitro-cultured cotyledons in which Arg was tested as a nitrogen source and as a precursor of urea. Our results indicate that the increase of arginase activity upon germination was due to an increase in arginase transcript, and are consistent with the absence of an arginase-catalyzed reaction in developing embryos during the reserve deposition stage.
MATERIALS AND METHODS
Plant Material and Germination/Growth Conditions
Wild-type Eu3/Eu3 soybean (Glycine max L. Merrill cv Williams 82) plants and an otherwise isogenic Eu3/eu3-e1 heterozygote were grown in a controlled-environment chamber at 27°C with a 12-h/12-h light/dark regime at 180 μmol m−2 s−1. The eu3-e1 mutation has been characterized previously (Meyer-Bothling et al., 1987). The homozygous eu3-e1/eu3-e1 mutant lacks the activities of all soybean ureases. Eu3/eu3-e1 heterozygous plants (Stebbins et al., 1991; Stebbins and Polacco, 1995) have urease levels similar to wild-type Eu3/Eu3 plants. All urease-negative eu3-e1/eu3-e1 seeds used in this study were derived from selfed Eu3/eu3-e1 plants. Urease-positive and -negative seeds were distinguished by a seed-chip urease assay (Meyer-Bothling and Polacco, 1987). After removal from pods, immature embryos were separated from the testa, frozen in liquid nitrogen, lyophilized, and stored at −70°C. The relative water content of embryos (milligrams of water per milligram of embryo fresh weight × 100) was taken as the criterion for the physiological state of the embryo.
For seedling studies, seeds were germinated in the dark at 28°C in rolls of germination paper (Anchor Paper, St. Paul, MN) moistened with deionized water. Seeds routinely germinated 1 d after imbibition. Whole etiolated seedlings were frozen immediately in liquid nitrogen and stored at −70°C until use.
Arginase Activity Assay
Embryos or seedlings (1.5–2.0 g fresh weight) were ground and powdered in a mortar with liquid nitrogen. The fine powder was resuspended to a final volume of 4 mL in cold 0.1 m Tris-maleate, pH 7.0, containing 1 mm EDTA and 0.1 mm PMSF. The suspension was maintained on ice and disrupted (three 15-s cycles) in a homogenizer (Ultra Turrax, Tekmar, Cincinnati, OH) and centrifuged at 13,000g for 10 min at 4°C. The supernatant was filtered through Miracloth (Calbiochem), brought to 5 mm in MnCl2, and left for 10 min at room temperature to activate arginase.
Arginase activity was assayed by measuring the Arg-dependent production of urea. One milliliter of standard assay medium contained 160 mm l-Arg (adjusted to pH 9.7 with KOH), 33 μm phenyl phosphorodiamidate (a urease inhibitor, Liao and Raines [1985]), and about 0.1 to 0.3 mg of extract protein. The reaction mixture was incubated for 30 min at 30°C. Aliquots (400 μL) were removed and the reaction was stopped by adding 1 n H2SO4 to 600 μL. Urea released was determined colorimetrically (Schimke, 1970). Arginase activity was expressed as nanomoles of urea released per minute per milligram of protein. Protein was determined by the method of Lowry et al. (1951).
Tissue Extraction and Urea Analysis
Lyophilized embryos or whole seedlings were frozen in liquid nitrogen and ground in a mortar to a fine powder. Weighed portions (40–70 mg) were extracted three times with 250 μL of 3% (w/v) HClO4. Combined supernatants were neutralized by adding 15 μL of 2.5 m K2CO3, and 1 mm EDTA per 100 μL of extract. Samples were placed on ice for 10 min, and the potassium perchlorate pellet was removed by centrifugation. Urea was measured by adding urease to the supernatant and determining the ammonia liberated by the phenol-hypochlorite method (Weatherburn, 1967), basically as described by Zonia et al. (1995), except that control reactions were also performed without urease.
Nucleic Acid Techniques
Genomic DNA was isolated from soybean leaves according to the method of Dellaporta et al. (1983). For DNA analysis 8 μg of DNA was digested overnight with EcoRl, BamHl, XbaI, or EcoRV at 37°C, and then separated on a 1% agarose gel. DNA was blotted to a nitrocellulose membrane and hybridized at high stringency in 50% (v/v) formamide, 100 μg mL−1 sonicated salmon-sperm DNA, 100 μg mL−1 yeast RNA, 5× Denhardt's solution (1× Denhardt's is 0.02% Ficoll, 0.02% PVP, and 0.02% BSA), 50 mm NaPO4, pH 6.5, 5× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate), and 0.2% SDS at 42°C for 14 to 16 h. The filter was washed in 2× SSC, 0.2% SDS at room temperature for 15 min, followed by 0.2× SSC, 0.2% SDS at 65°C for 30 min. Low-stringency hybridization conditions were identical to high-stringency conditions except that formamide was reduced to 25%. The filter was washed in 2× SSC, 0.2% SDS at room temperature for 15 min, followed by a second wash in the same solution at 42°C for 15 min. The dried blot was exposed for 3 to 5 d to radiographic film (X-Omat, Kodak) with intensifying screens at −70°C.
Total RNA was isolated from embryos or seedlings according to the method of Murfett et al. (1994). For RNA analysis 8 μg of RNA was separated in a 16.2% formaldehyde and 1% agarose gel and then transferred to a nylon membrane. Before hybridization the membrane was stained with 0.3 m sodium acetate, pH 5.2, containing 0.03% methylene blue to reveal RNA. Hybridization conditions and washes were identical to the high-stringency conditions used for DNA analysis. The RNA blot was exposed to radiographic film as indicated above.
Cotyledon Culture
Developing urease-positive (Eu3/Eu3 and Eu3/eu3-e1) and -negative (eu3-e1/eu3-e1) cotyledons (80–100 mg fresh weight), derived from selfed Eu3/eu3-e1, were collected and their phenotype was identified by a seed-chip assay performed on excised embryonic axes (Meyer-Bothling and Polacco, 1987). The testa and axes were aseptically removed and each embryo was halved into separate cotyledons. Cotyledons derived from the same embryo were used to compare different nitrogen sources. Cotyledons were cultivated in 3 mL of culture medium as described by Thompson et al. (1977), except that vitamins were at half-strength, Gly was omitted, and 5.9 mm K2SO4 was used as the sulfur source.
A mock seed-coat exudate was made up according to the reported five main amino acids in the in vivo soybean seed coat exudate and used to provide amino acids to the cotyledon (Rainbird et al., 1984). The amino acid mixture was 12.6 mm Gln, 4.5 mm Asn, 1.1 mm Ser, 0.7 mm His, and 0.6 mm Arg (N = 40 mEq L−1). Arg was provided as the sole nitrogen source at either 10 mm (N = 40 mEq L−1) or at 0.6 mm. In both cases the results with respect to urea evolution, growth, and total protein after 6 d of culture were identical. For cotyledon protein determination, the 3% HClO4 pellet (see above) was boiled for 15 min in 1 mL of 1.5 n NaOH. The insoluble material was removed by centrifugation and the solubilized protein was determined in the supernatant by the Lowry method (Lowry et al., 1951).
In experiments with radiolabeled Arg, 1.1 × 106 dpm l-[guanido-14C]Arg (51.5 Ci/mol, DuPont NEN) were added to the culture medium. Treatment with Dowex 50 W-X8 columns separated [14C]urea in cotyledon extracts and in the culture medium from unreacted l-[guanido-14C]Arg, as indicated by Daghigh et al. (1994), and then measured by liquid scintillation counting. The eluate was confirmed to be urea by its conversion to 14CO2 with added urease.
RESULTS
Arginase Sequence and Copy Number
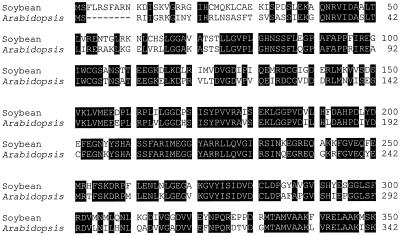
Using a full-length Arabidopsis arginase cDNA (Krumpelman et al., 1995) as a probe, we have previously reported (Goldraij et al., 1998) the cloning of a 1324-bp cDNA fragment (AG1) encoding a soybean seedling arginase. Figure 1 shows a comparison of the deduced amino acid sequences of the two enzymes. They are similar in size (36.5 and 38.6 kD for Arabidopsis and soybean, respectively) and are 78% identical. The most notable differences are in the N-terminal region. Considering that all plant arginases reported so far are mitochondrial enzymes (Polacco and Holland, 1993), this region constitutes a putative transit peptide. In addition, both sequences have characteristics common to N-terminal transit peptides, such as abundance of hydroxylated and positively charged residues (Hartl et al., 1989).
Figure 1.
Sequence alignment of plant arginases. Deduced amino acid sequence of soybean seedling arginase (Goldraij et al., 1998) was compared with that of Arabidopsis arginase (Krumpelman et al., 1995). Amino acids conserved between the two proteins are highlighted. The N-terminal region, which exhibits the weakest identity, is a putative transit peptide. Dashes are gaps for optimization of alignments.
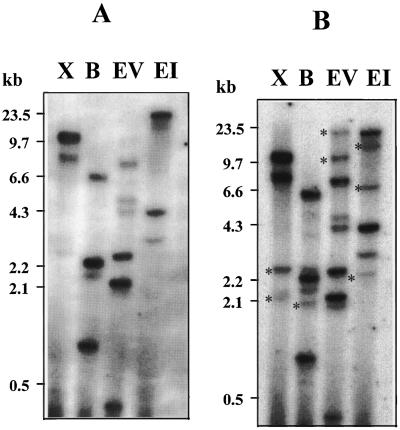
To examine the copy number of the soybean arginase gene and the presence of related genes, we used AG1 as a probe to perform DNA analysis. Under high-stringency conditions, genomic DNA digested with XbaI and EcoRI showed two and three bands, respectively, whereas BamHI and EcoRV digestions showed four or more bands (Fig. 2A). The cloned AG1 had only one restriction site for BamHI and EcoRV, indicating the presence of more than one arginase gene and/or the presence of intron restriction sites for the enzymes used in the analysis. Additional bands appeared in all DNA digestions when the hybridization was performed at low stringency (Fig. 2B), indicating the existence of more distantly related genes in the soybean genome.
Figure 2.
DNA analysis of arginase genes in the soybean genome. Autoradiogram of blot containing soybean genomic DNA (8 μg) digested with XbaI (lanes X), BamHI (lanes B), EcoRV (lanes EV), and EcoRI (lanes EI). Hybridization was carried out under high- (A) and low- (B) stringency conditions using radiolabeled AG1 cDNA as a probe. Asterisks indicate additional bands not present under high-stringency conditions. Numbers at left indicate marker sizes.
Arginase Expression in Developing Embryos and in Germinating Seeds
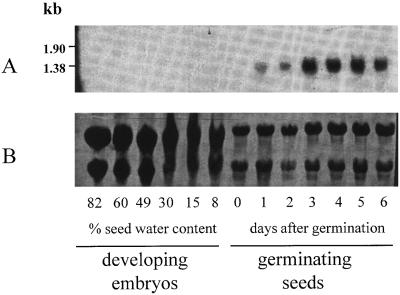
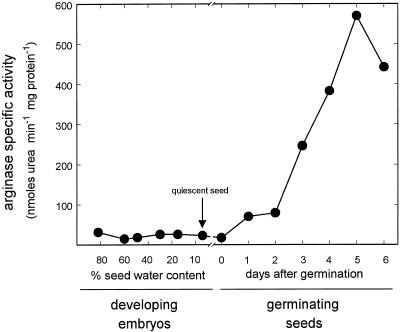
Arginase expression was studied at the transcript level in developing embryos and in germinating soybean seeds. Total RNA isolated from immature embryos, quiescent seeds, and whole 0- to 6-DAG seedlings was subjected to blot analysis and probed with AG1 (Fig. 3). Hybridization revealed a single band migrating slightly faster than the 1.38-kb standard, which is consistent with the size of the arginase cDNA. The transcript appeared at 1 DAG and stayed at the same level at 2 DAG. It accumulated to higher levels in the 3- to 5-DAG interval and then decreased slightly by 6 DAG. In contrast, no transcript was seen in developing embryos or in quiescent seeds, although the amount of RNA analyzed was nearly four times greater. A nearly identical pattern was obtained for arginase specific activity of the same samples used in the RNA blot experiment (Fig. 4). Arginase activity was barely detectable in developing embryos and in quiescent seed, but a sharp and constant increase was seen from 2 DAG until reaching a maximum at 5 DAG. Therefore, the increase of arginase activity is consistent with an increase in the transcript level, indicating that expression of the arginase gene(s) is a developmentally controlled process coincident with germination and seedling development.
Figure 3.
Soybean arginase expression in developing embryos and in germinating seeds. A, Autoradiogram of blot containing total RNA isolated from developing embryos (30 μg) and whole seedlings (8 μg). An embryo water content of 8% corresponds to quiescent seed. Hybridization was carried out using radiolabeled AG1 cDNA as a probe. B, Uniformity of RNA loading and transfer was confirmed by nylon membrane staining with methylene blue. Numbers at left indicate marker sizes.
Figure 4.
Soybean arginase specific activity in developing embryos and in germinating seeds. Arginase specific activity was measured in developing embryos at the reserve deposition stage and in germinating seeds 0 to 6 DAG. The values are averages of duplicate reactions and are representative of two separate experiments. Replicates were within 5% of the average values shown.
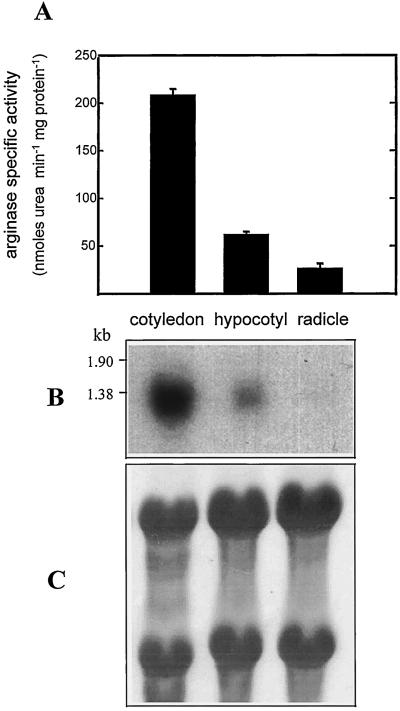
The organ distribution of arginase expression and activity was investigated in 3- to 4-DAG seedlings. The results showed that the highest arginase specific activity was in the cotyledon, followed by the hypocotyl and radicle, with only about 30% and 15% of the cotyledon specific activity, respectively (Fig. 5A). The arginase activity levels of the different seedling organs roughly corresponded to their arginase transcript levels (Fig. 5B). Because the total activity in the cotyledon was about 50 times higher than in the radicle or hypocotyl, we concluded that arginase was localized mainly in the cotyledons.
Figure 5.
Organ distribution of arginase expression and specific activity in etiolated 3- to 4-DAG soybean seedlings. A, Arginase specific activity. Results are means ± sd of two separate experiments. B, Autoradiogram of blot containing total RNA (8 μg) isolated from the three organs. Hybridization was similar to that in Figure 3. C, Uniformity of RNA loading and transfer was confirmed by nylon membrane staining with methylene blue. Numbers at left indicate marker sizes.
The Arginase Reaction Is Inoperative in Developing Embryos
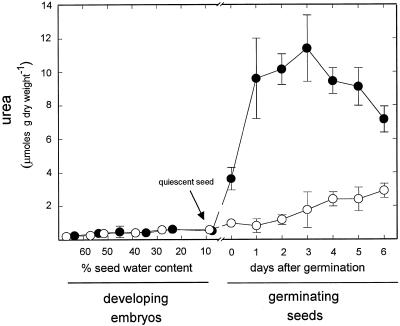
The arginase expression patterns (Figs. 3 and 4) suggest that the role of arginase is largely confined to germination and seedling development. We tested this suggestion by an in-vivo approach exploiting mutant eu3-e1/eu3-e1, which lacks all urease activity (Meyer-Bothling et al., 1987). The quantity of urea accumulated by the mutant is a useful indicator of in vivo arginase activity (Stebbins and Polacco, 1995). Virtually identical, low levels of urea were found in developing embryos of the mutant and its urease-positive siblings (all progeny of selfed Eu3/eu3-e1 plants) (Fig. 6). The lack of urea accumulation in the mutant suggests that the arginase-catalyzed reaction is inoperative in developing embryos.
Figure 6.
Urea accumulation in developing embryos and in germinating seeds. Urea was measured in urease-positive (Eu3/Eu3 or Eu3/eu3-e1) (○) and urease-negative (eu3-e1/eu3-e1) (•) developing embryos at the reserve deposition stage and in germinating seeds 0 to 6 DAG. Each value represents the mean ± sd of two separate experiments. Bars are shown only when they are bigger than the symbol.
In contrast, there was a sharp increase in urea upon germination of urease-negative seeds, reaching a peak at 3 DAG. This pattern is roughly in agreement with the pattern for arginase expression and arginase specific activity, albeit with an earlier peak. Based on studies using labeled precursors and an inhibitor (allopurinol) of purine conversion to ureides (Stebbins and Polacco, 1995), we demonstrated previously that the vast majority of urea generated in soybean seedlings comes from Arg rather than from ureides. As expected, much less urea accumulated in urease-positive seedlings, although the levels were somewhat higher than previously reported.
Arginase Is Conditionally Operative in in Vitro-Cultured Cotyledons
Developing soybean cotyledons were cultured in vitro in defined media to evaluate Arg as a nitrogen source and to test for a functional arginase using the criterion of urea accumulation in the urease-deficient mutant eu3-e1/eu3-e1. We previously determined that Arg uptake was not altered in the urease-deficient mutant compared with the wild type (results not shown). Arg was supplied to immature cotyledons as the sole nitrogen source or included in an amino acid mixture approximating the seed-coat exudate (Rainbird et al., 1984) that nourishes the developing embryo (see Methods).
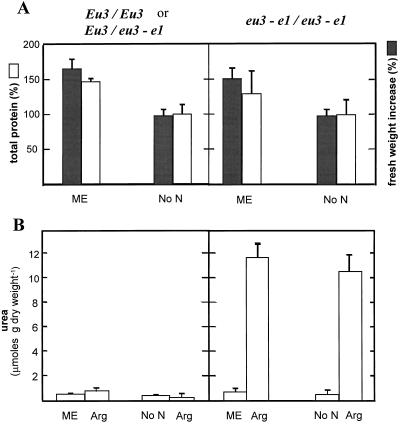
In both the urease-positive seedlings and in the isogenic urease-deficient sibling, the growth and total protein in cotyledons cultured with Arg as the sole nitrogen source were almost identical to values obtained in cotyledons cultured without nitrogen (Fig. 7A), even though urea derived from Arg accumulated in the urease-deficient mutant (Fig. 7B). Arg is the precursor of urea when it is the sole nitrogen source, as confirmed by the conversion of l-[guanido-14C]Arg to [14C]urea (Table I). Therefore, the products derived from Arg breakdown (urea and Orn) did not support growth and protein deposition beyond developing cotyledons cultured without nitrogen.
Figure 7.
In vitro culture of urease-positive (Eu3/Eu3 or Eu3/eu3-e1) and urease-negative (eu3-e1/eu3-e1) immature cotyledons. After detaching the two cotyledons from a given embryo, one was cultivated on mock exudate (ME) or nitrogen-free medium (No N), whereas the other cotyledon was cultivated on Arg. Fresh weight, total protein, and total urea were determined in each cotyledon after 6 d of culture. A, Fresh weight increase (gray bars) and total protein (white bars) relative to values of paired cotyledon cultured on Arg, established as 100%. B, Total urea accumulation in cotyledon and culture medium. Results are means ± sd of three separate experiments.
Table I.
Total urea accumulation in cotyledons cultivated with l-[guanido-14C]Arg
| Soybean Genotype | Urea
|
|
|---|---|---|
| Arg | MEa | |
| nmol cotyledon−1 | ||
| Eu3/Eu3 or Eu3/eu3-e1 | 29 ± 20 | 6 ± 0.9 |
| eu3-el/eu3-e1 | 1220 ± 150 | 8 ± 3 |
Results are averages ± sd of two different experiments.
ME, Mock exudate.
The cotyledons used in the experiment shown in Figure 7 received 10 mm Arg. Identical results, including urea accumulation (data not shown), were obtained when Arg was reduced to 0.6 mm. With a mock seed-coat exudate (containing 0.6 mm Arg, see Methods) as the nitrogen source, growth and protein yield were increased about 40% to 50% in both genotypes with respect to cotyledons cultured with Arg (Fig. 7A), whereas no evolution of urea was detected in the urease-deficient mutant (Fig. 7B). As expected, no urea accumulation was seen in urease-positive cotyledons in any of the three treatments assayed. In preliminary experiments, when l-[guanido-14C]Arg was provided in a mock exudate, the label was rapidly incorporated into protein, as previously reported (Micallef and Shelp, 1989c). Thus, in agreement with the lack of Arg degradation by developing embryos in vivo (Fig. 6), there was no arginase-catalyzed degradation of Arg by cultured cotyledons provided with a mock seed-coat exudate (Fig. 7).
DISCUSSION
We investigated the expression and functioning of soybean arginase in developing embryos and in germinating seeds. We used the cDNA clone AG1, which encodes a soybean seedling arginase (Goldraij et al., 1998) that is 78% identical to that of Arabidopsis (Fig. 1), the only other plant arginase whose sequence is currently available. High-stringency genomic hybridization indicated that arginase may be a member of a small gene family, whereas low-stringency conditions indicated the presence of more distantly related homologs in soybean (Fig. 2).
Arginase activity increased 20-fold upon germination to a maximum at 5 DAG, whereas RNA blots probed with AG1 revealed a transcript peak at 3 to 5 DAG. In contrast, arginase activity in developing embryos was basal and no transcript signal was detected at any time during the reserve deposition stage. Therefore, the increased arginase activity during germination and seedling development was most likely to be due to de novo enzyme biosynthesis and not to activation of pre-existing enzyme.
Germination as the signal for the synthesis of arginase is in agreement with a catabolic role. Its substrate, Arg, is a major nitrogenous storage compound in plants. For 379 species analyzed, Arg averaged 7.7 mol % of seed amino acids and 21.1% of total amino acid nitrogen, the highest contribution of any amino acid (Van Etten et al., 1967; Polacco and Holland, 1993). In soybean, Arg contains 18% of seed protein-bound nitrogen (Micallef and Shelp, 1989a). Arg degradation by arginase during germination is consistent with the sharp increase in urea in urease-deficient mutant seedlings (Fig. 6; Stebbins and Polacco, 1995). Urease has been proposed to function coordinately with arginase in the utilization of seed protein reserves during germination (Thompson, 1980). In Arabidopsis, urease functions to recycle urea nitrogen derived from Arg breakdown during seedling development (Zonia et al., 1995).
The preponderance of arginase activity in seedling cotyledons (Fig. 5) indicates that Arg degradation might occur mainly in the cotyledon, prior to the delivery of its nitrogen to the growing points. This is in agreement with observations in pumpkin cotyledons (Chou and Splittstoesser, 1972), in which Arg constitutes 30% of amino acid nitrogen but is only a minor component of the amino acid nitrogen (1.7%) transported out of the cotyledon.
In contrast to the abundant arginase activity and urea reaction product in developing seedlings, the enzyme does not appear to be active in developing embryos. This conclusion is based mainly on the absence of urea accumulation in urease-deficient developing embryos (Fig. 6). At this stage arginase activity was extremely low but detectable. The absence of any transcript detectable by the seedling axis clone AG1 is consistent with either a lack of arginase transcript in the developing embryo or with a poorly cross-hybridizing minor arginase mRNA.
In vitro cotyledon culture was used to evaluate the capacity of Arg to support growth and protein synthesis in immature soybean cotyledons. A mixture of amino acids containing Arg and resembling the seed-coat exudate (mock exudate) stimulated an increase in fresh weight and protein deposition similar to that obtained with Gln alone (2.5 times the initial fresh weight and 95 mg of protein per cotyledon after 6 d culture), which was previously shown to be the best nitrogen source (Thompson et al., 1977). Increases in fresh weight and protein supported by the mock exudate were not, however, accompanied by Arg breakdown caused by arginase, because no urea accumulated in urease-deficient cotyledons (Fig. 7).
The situation with Arg as the sole nitrogen source (at 0.6 or 10 mm) was exactly reversed. Arg did not support growth or protein deposition in urease-positive or -negative cotyledons, but was actively broken down by arginase, as evidenced by urea accumulation in the mutant. We observed no induction of arginase by Arg. The in vitro arginase specific activity of cultured cotyledons (<11 nmol urea min−1 mg−1 protein) was lower than the specific activity observed in embryos in planta. Because the other amino acids in the mock exudate appear to prevent arginase action, they could prevent uptake of Arg into the cotyledon or into the mitochondrion, the site of all or most plant arginases (Polacco and Holland, 1993). Inhibition of uptake into the cotyledon appears unlikely, because we observed no differences in Arg uptake whether it was provided alone or included in the mock exudate. In the latter case, Arg was abundantly incorporated (35% of total Arg uptake) into perchloric-acid-precipitable material (A. Goldraij and J. Polacco, unpublished results).
Other studies have examined the role of arginase in developing cotyledons of pea (De Ruiter and Kollöfel, 1983) and soybean (Micallef and Shelp, 1989b, 1989c). In both species Arg was reported to be actively incorporated into protein and was also the major constituent of the free amino acid pool. After injection of l-[guanido-14C]Arg into excised cotyledons, little urea accumulation or 14CO2 release was detected in pea despite the in vitro arginase activity detected in developing seeds. Contrary to our results, Micallef and Shelp (1989c) found that approximately 20% of injected Arg was routed to urea and Orn in soybean. Perhaps this activity was the result of arginase being released from mitochondria by mechanical disruption caused by Arg injection.
We propose that arginase is not operative in vivo in developing soybean embryos. Although very low, in vitro arginase activity could be detected and was sufficient to generate large amounts of urea when Arg was the sole nitrogen source in cultured cotyledons. The presence of arginase and a large free Arg pool without a reaction taking place indicates that a regulatory mechanism impedes the reaction. Considering that all plant arginases reported so far are mitochondrial or particulate enzymes (for review, see Polacco and Holland, 1993), this regulation may be achieved by different intracellular locations of substrate and enzyme. A similar mechanism has been suggested for pea (De Ruiter and Kollöfel, 1983).
The inoperative arginase in developing soybean embryos containing abundant free Arg avoids a futile, energy-wasting urea cycle. Unimpeded action of arginase in the developing embryo would result in a functional urea cycle with a wasteful conversion of ammonia and α-amino acid nitrogen to urea in the overall reaction of the Arg/urea cycle:
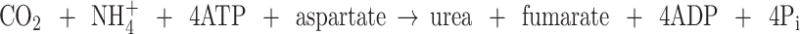 |
Urea is then reassimilated by urease, which is active in developing cotyledons:
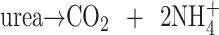 |
The combined action of urease and an Arg/urea cycle would lead to a very expensive deamination of α-amino acids:
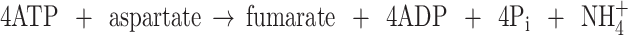 |
Nature uses a variety of strategies to avoid arginase engaging in a futile cycle with Arg biosynthesis in tissues/organisms lacking a functional urea cycle (Vissers et al., 1986; Jenkinson et al., 1996). In developing embryos of soybean, we propose that the low activity of arginase, its mitochondrial location, or both, keeps biosynthetic and catabolic phases of the urea cycle spatially and/or temporally separated. The identification of the putative Arg transporter in mitochondrial membrane and the time in which it becomes active may be the key toward a more thorough understanding of Arg metabolism in soybean seeds.
ACKNOWLEDGMENTS
We thank Dale Blevins and Krystyna Lukaszewska for critical reading of the manuscript.
Abbreviation:
- DAG
days after germination
Footnotes
This work was supported by the Missouri Agricultural Experimental Station and by the U.S. Department of Agriculture (grant no. 97-35 305-4629 to J.C.P.). A.G. was supported by a postdoctoral fellowship from the Interdisciplinary Plant Group. This is journal contribution no. 12,804 from the Missouri Agricultural Experimental Station.
LITERATURE CITED
- Chou KH, Splittstoesser WE. Changes in the amino acid content and the metabolism of γ-aminobutyrate in Cucurbita moschataseedlings. Physiol Plant. 1972;26:110–114. [Google Scholar]
- Daghigh F, Fukuto JM, Ash DE. Inhibition of a rat liver arginase by an intermediate in NO biosynthesis, NG-hydroxy-l-Arg: implications for the regulation of nitric oxide biosynthesis by arginase. Biochem Biophys Res Commun. 1994;202:174–180. doi: 10.1006/bbrc.1994.1909. [DOI] [PubMed] [Google Scholar]
- Dellaporta S, Woods H, Hicks J. A plant DNA mini-preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- De Ruiter H, Kollöffel C. Arg catabolism in the cotyledons of developing and germinating pea seeds. Plant Physiol. 1983;73:525–528. doi: 10.1104/pp.73.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij A, Coello P, Polacco JC. Nucleotide sequence of a cDNA encoding a soybean seedling axes arginase (accession no. AF035671) (PGR 98-016) Plant Physiol. 1998;116:867. [Google Scholar]
- Hartl FU, Pfanner N, Nicholson DW, Neupert W. Mitochondrial protein import. Bochim Biophys Acta. 1989;988:1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Jenkinson CP, Grody WW, Cederbaum SD. Comparative properties of arginases. Comp Biochem Physiol. 1996;114B:107–132. doi: 10.1016/0305-0491(95)02138-8. [DOI] [PubMed] [Google Scholar]
- Kang JH, Cho YD. Purification and properties of arginase from soybean (Glycine max) axes. Plant Physiol. 1990;93:1230–1234. doi: 10.1104/pp.93.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JE, Gifford DJ. Amino acid utilization in seeds of loblolly pine during germination and early seedling growth I. Arg and arginase activity. Plant Physiol. 1997;113:1125–1135. doi: 10.1104/pp.113.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollöffel C, Van Dijke HD. Mitochondrial arginase activity from cotyledons of developing and germinating seeds of Vicia fabaL. Plant Physiol. 1975;55:507–510. doi: 10.1104/pp.55.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumpelman PM, Freyermuth SK, Cannon JF, Fink GR, Polacco JC. Nucleotide sequence of Arabidopsis thaliana arginase expressed in yeast (accession no. U15019) Plant Physiol. 1995;107:1479–1480. doi: 10.1104/pp.107.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CFH, Raines SG. Inhibition of soil urease activity by amido derivatives of phosphoric and thiphosphoric acids. Plant Soil. 1985;85:149–152. [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Matsubara S, Suzuki Y. Arginase activity in the cotyledons of soybean seedlings. Physiol Plant. 1984;62:309–314. [Google Scholar]
- Meyer-Bothling LE, Polacco JC. Mutational analysis of the embryo-specific urease locus of soybean. Mol Gen Genet. 1987;209:439–444. doi: 10.1007/BF00331147. [DOI] [PubMed] [Google Scholar]
- Meyer-Bothling LE, Polacco JC, Cianzio SR. Pleiotropic soybean mutants defective in both urease isozymes. Mol Gen Genet. 1987;209:432–438. doi: 10.1007/BF00331146. [DOI] [PubMed] [Google Scholar]
- Micallef BJ, Shelp BJ. Arginine metabolism in developing soybean cotyledons I. Relationship to nitrogen nutrition. Plant Physiol. 1989a;90:624–630. doi: 10.1104/pp.90.2.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef BJ, Shelp BJ. Arginine metabolism in developing soybean cotyledons II. Biosynthesis. Plant Physiol. 1989b;90:631–634. doi: 10.1104/pp.90.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef BJ, Shelp BJ. Arginine metabolism in developing soybean cotyledons III. Utilization. Plant Physiol. 1989c;91:170–174. doi: 10.1104/pp.91.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- Polacco JC, Holland MA. Roles of urease in plant cells. Int Rev Cytol. 1993;145:65–103. [Google Scholar]
- Polacco JC, Sparks RB., Jr Patterns of urease synthesis in developing soybeans. Plant Physiol. 1982;70:189–194. doi: 10.1104/pp.70.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbird RM, Thorne JH, Hardy RW. Role of amides, amino acids and ureides in the nutrition of developing soybean seeds. Plant Physiol. 1984;74:329–334. doi: 10.1104/pp.74.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke RT. Arginase (rat liver) Methods Enzymol. 1970;17:313–317. [Google Scholar]
- Splittstoesser WE. Metabolism of Arg by aging and 7 day old pumpkin seedling. Plant Physiol. 1969;44:361–366. doi: 10.1104/pp.44.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins NE, Holland MA, Cianzio SR, Polacco JC. Genetic tests of the roles of the embryonic ureases of soybean. Plant Physiol. 1991;97:1004–1010. doi: 10.1104/pp.97.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins NE, Polacco JC. Urease is not essential for ureide degradation in soybean. Plant Physiol. 1995;109:169–175. doi: 10.1104/pp.109.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JF (1980) Arg synthesis, proline synthesis and related processes. In BJ Miflin, ed, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 375–402
- Thompson JF, Madison JT, Muenster AE. ) In vitro culture of immature cotyledons of soya bean (Glycine maxL. Merr.) Ann Bot. 1977;41:29–39. [Google Scholar]
- Van Etten CH, Kwolek WF, Peters JE, Barclay AS. Plant seeds as protein sources for food or feed. J Agric Food Chem. 1967;15:1077–1085. [Google Scholar]
- Vissers S, Legraine C, Wiame JM. Control of a futile urea cycle by Arg feed back inhibition of ornithine carbamoyltransferase in Agrobacterium tumefaciens and Rhizobia. Eur J Biochem. 1986;159:507–511. doi: 10.1111/j.1432-1033.1986.tb09915.x. [DOI] [PubMed] [Google Scholar]
- Weatherburn MB. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- Zonia LE, Stebbins NE, Polacco JC. Essential role of urease in germination of nitrogen-limited Arabidopsis thalianaseeds. Plant Physiol. 1995;107:1097–1103. doi: 10.1104/pp.107.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]