Abstract
In humans, allelic variants in Toll-like receptors (TLRs) associate with several pathologies. However, the underlying cellular and molecular mechanisms of this association remain largely unknown. Analysis of the human TLR9 promoter revealed that the C allele of the rs5743836 polymorphism generates several regulatory sites, including an IL-6-responding element. Here, we show that, in mononuclear cells carrying the TC genotype of rs5743836, IL-6 up-regulates TLR9 expression, leading to exacerbated cellular responses to CpG, including IL-6 production and B-cell proliferation. Our study uncovers a role for the rs5743836 polymorphism in B-cell biology with implications on TLR9-mediated diseases and on the therapeutic usage of TLR9 agonists/antagonists.
Introduction
Toll-like receptors (TLRs) are pattern recognition receptors with a major role in activation and homeostasis of the immune system upon pathogen recognition [1]. However, it has become apparent that self-recognition through TLRs can also take place and that this can play a role in sterile inflammation and autoimmunity [2]–[3].
TLR9 activates the innate immune system upon recognition of unmethylated CpG DNA motifs as “danger signals”. In humans, TLR9 is mainly expressed in plasmacytoid dendritic cells and B-lymphocytes [4]–[5], playing a role on viral, fungal, mycobacterial and Helicobacter pylori infections [6]–[10] and being implicated in the pathogenesis of several autoimmune diseases [3], [11].
Despite growing evidence of a strong link between deregulated TLR9 responses and immune-mediated diseases, the underlying cellular and molecular mechanisms remain largely unknown. In psoriasis, uncontrolled expression of the antimicrobial peptide LL37 breaks innate immune tolerance by delivering self-DNA to TLR9-containing endosomes, causing TLR9-mediated type I interferon production that drives skin inflammation [3], [11]. Other mechanisms linking TLR9 deregulation and disease may involve TLR9 gain-of-function due to transcriptional deregulation. We and others have shown that the C allele of the single nucleotide polymorphism (SNP) rs5743836 (T-1237C) in TLR9, known to display minor allele frequencies ranging from 0.02 to 0.38 across distinct ethnicities, predisposes to non-Hodgkin [12] and Hodgkin lymphoma [13], as well as to several autoimmune and chronic inflammatory diseases, including asthma [14] and Crohn's disease [15].
In light of TLR9 relevance in disease, synthetic oligodeoxynucleotides that specifically inhibit or activate TLR9 have been developed [16]. These compounds can potentially be used in infectious diseases, allergy/asthma and cancer therapy to either block TLR9 self-recognition or stimulate the immune response in conditions of immune tolerance. Given the highly promising effects of CpG-based therapy, understanding the biological and functional impact of TLR9 genetic variability is pertinent as it will certainly contribute to anticipate potential side-effects associated with TLR9 agonist/antagonist therapy.
Here, we report that the C allele of rs5743836 introduced a new IL-6-dependent transcription factor binding site in the TLR9 promoter. Peripheral blood mononuclear cells (PBMCs) harboring the TC genotype of rs5743836 show higher expression of both TLR9 and IL-6 and increased B-cell proliferation in response to CpG, functionally dependent on IL-6 signaling. Our findings not only contribute to a better understanding of the functional impact of TLR9 overexpression, but also raise important questions on the therapeutic usage of CpGs in the context of the immunogenetic profile of the patient.
Results and Discussion
The C allele of rs5743836 introduces a functional IL-6-responsive element in the TLR9 promoter
Several SNPs in TLR9 have been associated with increased risk for immune-mediated diseases (International HapMap Project; http://hapmap.ncbi.nlm.nih.gov/). We hypothesized that some of these SNPs might alter the transcriptional regulation of TLR9, leading to inappropriate gain- or loss-of-function. To test this, we screened the transcription factor-binding profile of the human TLR9 promoter for alterations introduced by known SNPs. In silico analysis revealed that the C allele of rs5743836 generated several novel binding sites for different transcription factors (Fig. 1A). The potential regulatory transcription factor-binding motif with the highest score corresponded to an interleukin-6 (IL-6) response element (IL-6 RE) at position -1238 to -1234 with the consensus sequence TTCCAG (Fig. 1A). This type II IL-6 consensus motif has already been shown to play a key role on STAT3 transactivation of the human γ-fibrinogen promoter triggering up-regulation [17], but was never studied in the context of TLR9.
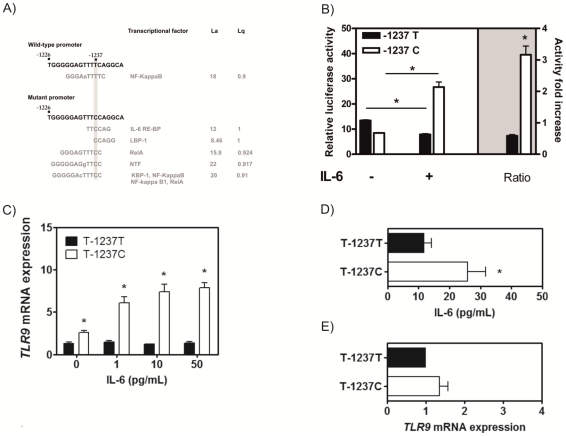
Figure 1. IL-6 increases the activity of the TLR9 promoter carrying the C allele of rs5743836.
(A) In silico analysis of the fragment of the TLR9 promoter containing the T/C substitution using the TESS interface. La, log-likelihood score, Lq, a measure of the goodness-of-fit of the DNA sequence to the consensus binding motif. (B) Luciferase reporter assay of Raji B cells transfected with plasmid vectors containing the luciferase gene under the control of a 3.2 kb fragment of the promoter sequence carrying the T or C allele. After transfection, cells were left untreated or stimulated with IL-6 (n = 3). (C) IL-6 dose-dependent stimulation of either TT (n = 27) or TC (n = 25) PBMCs. Cells were left untreated or stimulated with increasing doses of recombinant IL-6. TLR9 mRNA expression levels were determined by real-time PCR. (D) IL-6 secretion by of either TT (n = 21) or TC (n = 17) PBMCs. IL-6 in untreated cells was quantified by ELISA. (E) TLR9 expression in TT (n = 21) or TC (n = 17) PBMCs in IL-6-neutralizing conditions. Cells were cultured with an anti-IL-6 antibody and TLR9 mRNA expression was determined by real-time PCR. Results shown are the mean ± SD.
To investigate whether the TLR9 promoter carrying the C allele of rs5743836 was regulated by IL-6, we measured the ectopic expression of luciferase under control of the TLR9 promoter sequence with either the T or C allele in transiently transfected B-cells. As shown in Fig. 1B, upon stimulation with recombinant human IL-6, cells carrying the variant construct displayed a significant 3.2-fold increase in luciferase activity (p<0.001), whereas this effect was not observed for the wild-type construct. This result indicates that rs5743836 regulates the transcriptional activity of the TLR9 promoter in an IL-6-dependent manner.
We next evaluated whether the C allele of rs5743836 in its native chromosomal context also led to an IL-6-dependent increase in TLR9 mRNA expression. For this, PBMCs with the TT or TC genotypes were stimulated with increasing doses of recombinant IL-6. We observed that only TC PBMCs responded to the presence of IL-6 with increased TLR9 expression, in a dose-dependent manner (Fig. 1C).
In addition, we observed that, as compared to TT, unstimulated TC PBMCs displayed an approximately 2.4-fold increase in the basal transcription of TLR9 (p = 0.008) (Fig. 1C), due to basal production of IL-6 (Fig. 1D). Interestingly, the neutralization of endogenous IL-6 resulted in a decreased TLR9 expression to levels similar to those of TT cells (Fig. 1E). Overall, our data point towards a major impact of the rs5743836 polymorphism on the transcriptional regulation of TLR9 and support an IL-6-dependent modulation of TLR9 gene expression.
B cells harboring the TC genotype of rs5743836 show increased proliferation upon CpG stimulation
To investigate whether the transcriptional upregulation of TLR9 caused by the TC genotype of rs5743836 led to a gain-of-function phenotype, TC PBMCs were stimulated with CpG. Although IL-6 was produced upon TLR9 activation regardless of the genotype, its production was higher in TC cells (p = 0.009) (Fig. 2A). We next questioned whether in TC cells, CpG stimulation, via IL-6 production, could lead to further up-regulation of TLR9 transcription and thus to a perpetuation loop of TLR9-mediated mechanisms. We stimulated PBMCs with CpG and measured TLR9 transcription. We observed that CpG induced a strong increase in TLR9 expression in TC (p<0.0001), but not TT cells (Fig. 2B). This up-regulation of TLR9 expression was abrogated by neutralizing the biological activity of IL-6 (Fig. 2B). In the presence of a TLR9 antagonist (TTAGGG), the up-regulation of TLR9 expression upon CpG stimulation was only partially reverted (Fig. 2B), probably due to the basal presence of IL-6 (Fig. 1C and 1D). Altogether, our data suggest that PBMCs carrying the TC genotype of rs5743836 are more responsive to CpG stimulation and that this effect is most likely perpetuated by increased TLR9 transcription and IL-6 production.
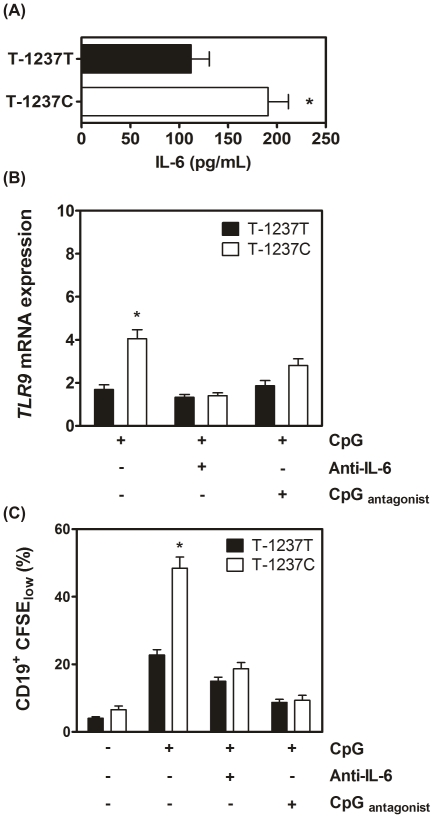
Figure 2. Human PBMCs carrying the TC genotype of rs5743836 stimulated via TLR9 show increased transcription of TLR9 and enhanced B-cell proliferation.
(A) TC PBMCs secrete higher amounts of IL-6 upon activation of TLR9 with CpG ODN 2006. IL-6 production was quantified by ELISA in the supernatants of TT (n = 22) or TC (n = 17) PBMCs following culture with CpG. (B) TLR9 activation of TC cells by CpG ODN 2006 increases TLR9 gene expression. Quantification of TLR9 mRNA expression in either TT (n = 24) or TC (n = 27) PBMCs cultured with: CpG ODN 2006 (0.05 µM); CpG ODN 2006 and anti-IL-6 antibody; CpG ODN 2006 and the TLR9 antagonist CpG ODN TTAGGG in a 1∶2 ratio. (C) Proliferation of CD19+ cells was assessed by CFSE dilution in PBMCs with different rs5743836 genotypes, TT (n = 40) and TC (n = 33), left untreated or stimulated with: CpG ODN 2006 (0.05 µM); CpG ODN 2006 and anti-IL-6 antibody; CpG ODN 2006 and CpG ODN TTAGGG in a 1∶2 ratio. Results shown are the means ± SD.
Taking into consideration that B-cells constitute the major cellular population expressing TLR9 within PBMCs (4), and given that rs5743836 affected the levels of both TLR9 and IL-6 in response to CpG, we investigated whether the presence of this variant impacted B-cell proliferation. To answer this question, PBMCs were stimulated with CpG, and CD19+ cell proliferation was assessed. CD19+ cell proliferation induced by CpG was significantly higher in TC cells as compared to TT cells (48.5% vs. 22.8%, respectively; p<0.0001) and was abrogated by co-incubation with a TLR9 antagonist (Fig. 2C). In line with the fact that TLR9 signaling is stronger in cells overexpressing TLR9 [4], [18], and that IL-6 mediates TLR9 up-regulation in TC cells, neutralization of IL-6 abrogated the increased proliferation of these cells in response to CpG to levels similar to those of wild-type cells (Fig. 2C). Again, our data support an amplification loop mediated by CpG and IL-6 that culminates with a deregulated TLR9 signaling.
Altogether, our data indicate that TLR9 signaling is amplified by a positive feedback loop through IL-6 in TC carriers of rs5743836 (Fig. 3), an example on how functional genomic studies may give insights on the regulation of gene activity under different situations, and can contribute to our knowledge on molecular and genetic mechanisms underlying disease susceptibility. In summary, we present evidences that shed light on how a certain genetic profile associated with excessive immune/inflammatory activation may contribute to human disease. Our findings not only identify a new regulatory element in the promoter region of TLR9, leading to its transcriptional activation through the IL-6 signaling pathway, with implications on cellular physiology, but also demonstrate the possibility to abolish the cellular effects of this SNP thus strongly supporting new potential individually-targeted treatments. In addition to the broad implications of our study in immune-mediated diseases, another important issue raised by our data is related to the therapeutic application of CpG-based compounds. As we now show, individual genetic variations can affect the outcome of the TLR9 signalling pathway. Re-interpreting CpG therapy trial results, taking into consideration the genotype of the participants, may therefore be of interest, since the global results may have been masked by the presence of the relatively common rs5743836 SNP. Ultimately, the therapeutic applications of CpG should be individually tailored as they may prove to be either detrimental or more effective for individuals harbouring the TC genotype of rs5743836.
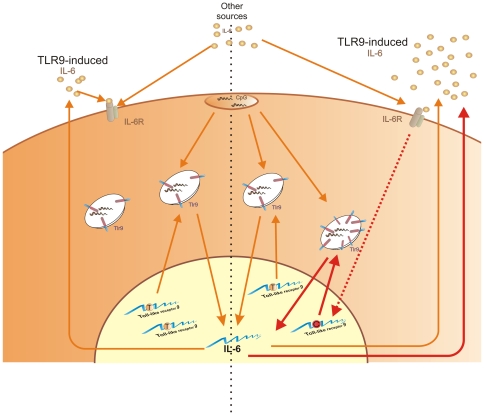
Figure 3. Proposed model of the effect of rs5743836 in B-lymphocyte activation and proliferation.
TLR9 activation by CpG DNA induces the production of IL-6, which upon binding to its receptor, promotes translocation of STAT3 to the nucleus. This transcription factor can bind to and trans-activate promoters containing STAT3 binding elements, thus initiating a specific transcriptional program. In cells harboring the TC genotype of rs5743836, STAT3 will bind to a new IL-6 RE site, created by the T/C substitution within the TLR9 promoter. As a result, TLR9 expression increases in response to IL-6. Therefore, in these cells, a loop of TLR9/IL-6 signaling amplification is created, leading to a deregulation in B-cell activation and proliferation upon CpG stimuli. Of note, the IL-6 needed to generate this loop may be TLR9-independent.
Materials and Methods
Ethics Statement
Study protocols were approved by Instituto Português de Oncologia de Lisboa, Francisco Gentil, EPE and Instituto Português de Oncologia de Coimbra, Francisco Gentil, EPE, Portugal. All study participants provided written informed consent prior to biospecimen collection.
In silico analysis of TLR9 promoter allelic variants
Allelic variants within TLR9 promoter were analyzed to predict alterations in transcription factor-binding sites, using the TESS interface (http://www.cbil.upenn.edu/cgi-bin/tess/). Parameters used in motif prediction included La, log-likelihood score, and Lq, measure of the goodness-of-fit of the DNA sequence to the consensus binding motif; the best possible Lq value was 1.000.
Plasmid constructs and mutagenesis
A plasmid vector containing the luciferase gene under control of the wild-type TLR9 promoter [19] was used for site-directed PCR mutagenesis. Briefly, the substitution at position -1237 was introduced by all-round PCR amplification using the primers 5′-TATGAGACTTGGGGGAGTTTC CAGGCAGAGGGAACAGCACA-3′ and 5′-TGTGCTGTTCCCTCTGCCTGGAAACTCCCCCAAGTCTCATA-3′. The correct construct was confirmed by direct sequencing.
Cell lines and cell culture
The Raji B-cell line (ATCC; wild-type for rs5743836) were cultivated in RPMI culture medium with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mM HEPES, 1.0 mM sodium pyruvate and 10% fetal bovine serum - FBS] without antibiotics.
Transient transfection and luciferase reporter gene assay
Raji cells (5×106 cells/ml) were transfected using the Microporator system (NanoEnTek Inc, Korea) following manufacturer instructions. Transfection efficiency was normalized by co-transfection of 0.5 µg of the control β-galactosidase vector (pCMV-βgal). After recovery, human recombinant IL-6 (100 ng/ µl) was added to 1×106 cells for 12 hours. Cells were lysed with 1× Passive Lysis Buffer (Promega, USA) and luciferase activity was measured using the Luciferase Assay System (Promega). Firefly, luciferase activity was normalized against β-galactosidase activity, determined with the β-galactosidase Enzyme Assay System (Promega). Data are expressed as relative luciferase units.
PBMC culture
PBMCs of healthy blood donors were isolated using Histopaque-1077 (Sigma, USA). Given the very low frequency of individuals with the CC genotype, PBMCs with TT and TC genotypes were used throughout. Cell viability was determined using trypan blue exclusion. 1×106 PBMC/ml were seeded in 24-well flat bottom plates and cultivated for 5 days with 0.05 µM CpG ODN 2006 (5′-TCGTCGTTTTGTCGTTTTGTCGTT-3′) (InvivoGen, France). Cells either left unstimulated or stimulated with control CpG ODN 2006 (5′-TGCTGCTTTTGTGCTTTTGTGCTT-3′) (InvivoGen) were included as controls throughout the study.
CFSE proliferation assay
PBMCs (2.5×106 cells/ml) were resuspended in serum-free Hank's Balanced Salt Solution (HBSS) and labeled with CFSE (Molecular Probes, USA) at a concentration of 5 µM. Labeling was quenched by the addition of 1/5 of the total volume of FBS. After washing with complete RPMI medium, cells (2×106 cells/ml) were cultured in 24-well plates for 5 days in medium alone or with CpG ODN plus or minus rabbit anti-IL-6 antibody (Sigma-.Aldrich) or CpG antagonist. Cells were washed, resuspended in staining buffer (0.5% BSA, 0.04% EDTA, 0.05% sodium azide in PBS) and stained with APC-conjugated anti-human CD19 (Becton Dickinson, San Jose, CA, USA) and analyzed on a BD FACSCalibur flow cytometer (Becton Dickinson).
Real-time RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Germany) and reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, France). For RT-PCR, 1 µl of cDNA was used as template using the QuantiTect Sybr Green PCR Kit (Qiagen). RT-PCR primers were as follows: β-actin, sense: 5′-GCCGTCTTCCCCTCCATCGTG-3′, antisense: 5′-GGAGCCACACGCAGCTCATTGTAGA-3′; TLR9, sense: 5′-CAGCAGCTCTGCAGTACGTC-3′, antisense: 5′-CTCAGGCCTTGGAAGA AGTG-3′. RT-PCR was conducted on a LightCycler System (Roche Applied Science, Switzerland) and the thermal cycling conditions included 50 cycles of 10 s at 95 °C, 20 s at 55 °C, and 20 s at 72 °C, after a 15-min initial step of enzyme activation at 95 °C, a melting step of 55–95 °C (0.5 °C/s) and a final cooling step at 40 °C. Expression of β-actin was used as internal control. Expression of TLR9 is indicated as n-fold increase relative to the level of TLR9 expression in untreated wild-type cells, using the 2-ΔΔCt method.
IL-6 quantification
IL-6 was quantified in supernatants from PBMC cultures either in untreated conditions or stimulated for 5 days with CpG ODN 2006 using human IL-6 Quantikine ELISA kit (R&D Systems, Minneapolis, MN, USA).
Statistical analysis
Data were analyzed by GraphPad Prism 4.03 program (USA). Unpaired Student's T-test with Bonferroni's adjustment was used to determine statistical significance (p<0.05). All experiments were performed at least in triplicate and repeated at least twice.
Acknowledgments
We are grateful to all persons that in any way contributed to the development of this work.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: A. Carvalho, N.S. Osório, M. Teixeira-Coelho, A.J. Almeida, M. Saraiva and C. Cunha were financially supported by Fundação para a Ciência e Tecnologia (FCT), Portugal. This study was supported by FCT - Grant Number PIC/IC/83313/2007. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Deane JA, Bolland S. Nucleic acid-sensing TLRs as modifiers of autoimmunity. J Immunol. 2006;177:6573–6578. doi: 10.4049/jimmunol.177.10.6573. [DOI] [PubMed] [Google Scholar]
- 3.Gilliet M, Lande R. Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr Opin Immunol. 2008;20:401–407. doi: 10.1016/j.coi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani, A The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 5.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 6.Berrington WR, Hawn TR. Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter? Immunol Rev. 2007;219:167–186. doi: 10.1111/j.1600-065X.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho A, Cunha C, Carotti A, Aloisi T, Guarrera O, et al. Polymorphisms in Toll-like receptor genes and susceptibility to infections in allogeneic stem cell transplantation. Exp Hematol. 2009;37:1022–1029. doi: 10.1016/j.exphem.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho A, Pasqualotto AC, Pitzurra L, Romani L, Denning DW, et al. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis. 2008;197:618–621. doi: 10.1086/526500. [DOI] [PubMed] [Google Scholar]
- 9.Ng MT, Van't Hof R, Crockett JC, Hope ME, Berry S, et al. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9 -1237T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect Immun. 2010;78:1345–1352. doi: 10.1128/IAI.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pine SO, McElrath MJ, Bochud PY. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–2395. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho A, Cunha C, Almeida AJ, Osorio NS, Saraiva M, et al. Genes Immun; 2011. The rs5743836 polymorphism in TLR9 confers a population-based increased risk of non-Hodgkin lymphoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mollaki V, Georgiadis T, Tassidou A, Ioannou M, Daniil Z, et al. Polymorphisms and haplotypes in TLR9 and MYD88 are associated with the development of Hodgkin's lymphoma: a candidate-gene association study. J Hum Genet. 2009;54:655–659. doi: 10.1038/jhg.2009.90. [DOI] [PubMed] [Google Scholar]
- 14.Lazarus R, Klimecki WT, Raby BA, Vercelli D, Palmer LJ, et al. Single-nucleotide polymorphisms in the Toll-like receptor 9 gene (TLR9): frequencies, pairwise linkage disequilibrium, and haplotypes in three U.S. ethnic groups and exploratory case-control disease association studies. Genomics. 2003;81:85–91. doi: 10.1016/s0888-7543(02)00022-8. [DOI] [PubMed] [Google Scholar]
- 15.Torok HP, Glas J, Tonenchi L, Bruennler G, Folwaczny M, et al. Crohn's disease is associated with a toll-like receptor-9 polymorphism. Gastroenterology. 2004;127:365–366. doi: 10.1053/j.gastro.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 16.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 17.Duan HO, Simpson-Haidaris PJ. Functional analysis of interleukin 6 response elements (IL-6REs) on the human gamma-fibrinogen promoter: binding of hepatic Stat3 correlates negatively with transactivation potential of type II IL-6REs. J Biol Chem. 2003;278:41270–41281. doi: 10.1074/jbc.M304210200. [DOI] [PubMed] [Google Scholar]
- 18.Jahrsdorfer B, Muhlenhoff L, Blackwell SE, Wagner M, Poeck H, et al. B-cell lymphomas differ in their responsiveness to CpG oligodeoxynucleotides. Clin Cancer Res. 2005;11:1490–1499. doi: 10.1158/1078-0432.CCR-04-1890. [DOI] [PubMed] [Google Scholar]
- 19.Takeshita F, Suzuki K, Sasaki S, Ishii N, Klinman DM, et al. Transcriptional regulation of the human TLR9 gene. J Immunol. 2004;173:2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]