Abstract
For many researchers, next generation sequencing data holds the key to answering a category of questions previously unassailable. One of the important and challenging steps in achieving these goals is accurately assembling the massive quantity of short sequencing reads into full nucleic acid sequences. For research groups working with non-model or wild systems, short read assembly can pose a significant challenge due to the lack of pre-existing EST or genome reference libraries. While many publications describe the overall process of sequencing and assembly, few address the topic of how many and what types of reads are best for assembly. The goal of this project was use real world data to explore the effects of read quantity and short read quality scores on the resulting de novo assemblies. Using several samples of short reads of various sizes and qualities we produced many assemblies in an automated manner. We observe how the properties of read length, read quality, and read quantity affect the resulting assemblies and provide some general recommendations based on our real-world data set.
Keywords: NGS, short read, quality, Phred, assembly, quantity, Velvet
1 Introduction
Current next-generation sequencing (NGS) technologies enable researchers to address myriad questions regarding biological and genetic mechanisms. NGS enables researchers to rapidly sequence genomes (Bentley et al. 2008; Li et al. 2010) or transcriptomes, to obtain snapshots of global gene expression levels in RNA-seq experiments (Wang et al. 2009; Costa et al. 2010) and to examine genome-wide protein-DNA interactions in ChIP-seq experiments (Barski et al. 2009; Park 2009) among others. Advancing technologies and economies of scale have collaborated to bring these capabilities within reach of even small research communities studying non-model or wild systems and organisms. Even so, there exist many barriers to entry into such studies that can make it difficult to initiate NGS projects or to extract meaning from the data. In this project we address some of the questions researchers may have when embarking on a new NGS project with regard to both quantity and quality of short reads needed for assembly.
There are many NGS platforms available (Metzker 2009; Voelkerding et al. 2009; Bräutigam et al. 2010; Nowrousian 2010), but the most common and prolific NGS methods available today produce extremely large quantities of very short reads (Illumina or ABI SOLiD sequencing platforms). Successfully assembling these short reads into a set of contigs representing the original sequences in the biological sample is a complex problem. This problem is easiest to solve if a reference genome or transcriptome is available to guide the assembly. However, in the absence of a reference genome, one must resort to de novo assembly of short-read data, which is more difficult. More challenging still is de novo assembly of data derived from source material that is expected to have uneven coverage, such as RNA transcripts where message abundance varies by several orders of magnitude, and there may be multiple versions of each message. For many researchers working with non-model organism transcriptomes, the last situation is the norm.
Many of these technologies are capable of producing single or paired-end reads. Paired-end read information can help to resolve repetitive regions which might otherwise be intractable to the assembly program (Narzisi et al. 2011; Wetzel et al. 2011). They also allow scaffolding of contigs which would otherwise remain fragmented and they aid in the identification of splice junctions and alternative splice forms. They can also be beneficial to further analyses downstream of assembly. Of the variety of NGS technologies available, we will focus here on RNA transcript paired-end short reads collected from the Illumina Genome Analyzer platform and assembled de novo without the aid of a reference genome or reference transcriptome. The raw sequence reads produced by this platform are commonly in the range of 60–150 bp in length. The inherent errors in this sequencing technique are generally well-understood (Dohm et al. 2008), and there exist many open-source and commercial packages designed to assemble these data into contigs (Metzker 2009; Miller et al. 2010; Nowrousian 2010). A discussion of the many available assembly algorithms is beyond the scope of this paper and has been covered before (Pop 2009; Paszkiewicz et al. 2010). Here, we have chosen to use the Velvet short-read assembler (Zerbino et al. 2008; Zerbino et al. 2009; Zerbino 2010) because it is widely used, and its abilities and limitations are relatively well understood. We will also use the Oases extension for Velvet which is designed to assemble transcript data specifically and is a much less well documented software package. While Oases is still a relatively ‘young’ program, it addresses problems inherent in packages tuned to assemble genome data. A recent publication critically assessed its performance with respect to other assembly packages and found it to be the best tool to assemble the chickpea transcriptome (Garg et al. 2011).
Over the course of one year, we produced several sets of transcript data using the Illumina Genome Analyzer platform. Over this time period we observed an increase in both the lengths of reads that could be reliably sequenced as well as the overall average quality scores reported by the sequencer software. While there were several changes to the software, firmware, and sequencing chemistry made available by Illumina, clearly factors outside the sequencing process may also have contributed to the observed differences in read quality. This real-world data set presents a variety of relative qualities and characteristics similar to data encountered by researchers both new and veteran to this field. As such it is difficult to provide strict controls for each variable within the data we have. Nevertheless we are able to make several useful observations that may serve to guide decisions regarding the amount of data needed to obtain an assembly of reasonable quality with a minimal investment.
2 Methods
2.1 Transcript sequencing
Transcript RNA was prepared from 13 separate tissues/organs and life stages of the live-bearing fish, Xiphophorus maculatus Jp 163 A. The X. maculatus Jp 163 A line is maintained at the Xiphophorus Genetic Stock Center by brother-sister matings and is inbred over 100 generations (Walter et al. 2006)see http://xiphophorus.txstate.edu). RNA samples were sequenced using the Illumina Genome Analyzer platform; the 13 samples were sequenced in 3 batches, submitted at 3 separate time points over a year (Catchen, J., et al., unpublished results). Each tissue or life stage was sequenced in an individual flow cell lane as paired end reads of 36 bp or 60 bp. Overall average quality scores were calculated for each sample, ignoring any quality score values of 2 (encoded as a ‘B’ in FASTQ format), which is used as a special flag to indicate that some type of sequencing error had occurred. The percentage of reads in a sample containing the ‘B’ flag was also reported. The short reads were grouped by their time of creation into three supersets designated as L1, L2, and L3. The tissues/stages contained in each sample is as follows: L1 contains whole body RNA from 5 age and/or gender specific samples from 5 days to 15 months of age, L2 contains whole body RNA from two embryonic stages and two tissue samples, L3 contains three tissue samples.
2.2 Quality Filtration
We processed raw transcript sequence data to remove low-quality reads by developing a four-stage filtration algorithm for the FASTQ-encoded data set that is comprised of several stages of checks and modifications. Stage 1 is a check for uncalled bases; the default setting is to reject any reads with more than two uncalled bases, but this can be altered at run-time. Velvet converts uncalled bases (‘N’ characters) into ‘A’ characters (adenine bases), but in most cases, this would be no different than a random read error. So, instead of rejecting reads with 2 or fewer uncalled bases, we opted to use the default setting to err for increased read coverage. Stage 2 searches for the presence of ‘B’ characters in the quality scores; these indicate that a particular error event is likely to have occurred and therefore the remainder of the read (distal to the ‘B’ character) should not be used. Any positions after and including the first ‘B’ character are trimmed off the sequence. Stage 3 scans for low-quality regions, deleting them and leaving high-quality fragments. Stage 3 first examines each position with a quality equal to or below 20 (1 error in 100). If the mean of the quality scores of the position in question and its up and downstream neighbors is still 20 or below, that position is marked for deletion. Any position with a quality score of 10 (1 error in 10) or below is marked for deletion without the possibility of rescue by neighboring positions. All the marked positions are then deleted, possibly breaking the read into smaller fragments of high quality. In Stage 4, the largest of the fragments remaining from the original read is selected as the trimmed read and the rest is discarded.
In addition to the process of filtering reads, the tool set we developed also measures statistics on the final outcomes of each read. Reads are sorted into four main categories: 1) failure due to quantity of uncalled bases, 2) failure due to size restrictions post trimming, 3) passage with trimming, and 4) passage without trimming. The total number of reads in each of those categories is tallied post filtration. Additionally those reads that passed may have lost or retained their mate in the filtration process and this is noted. The failure rate of a set of reads indicates the fraction of reads that failed for any reason, and the average failure rates calculated for Figure 1B are the average of failure rates of each component in the L1, L2, and L3 samples.
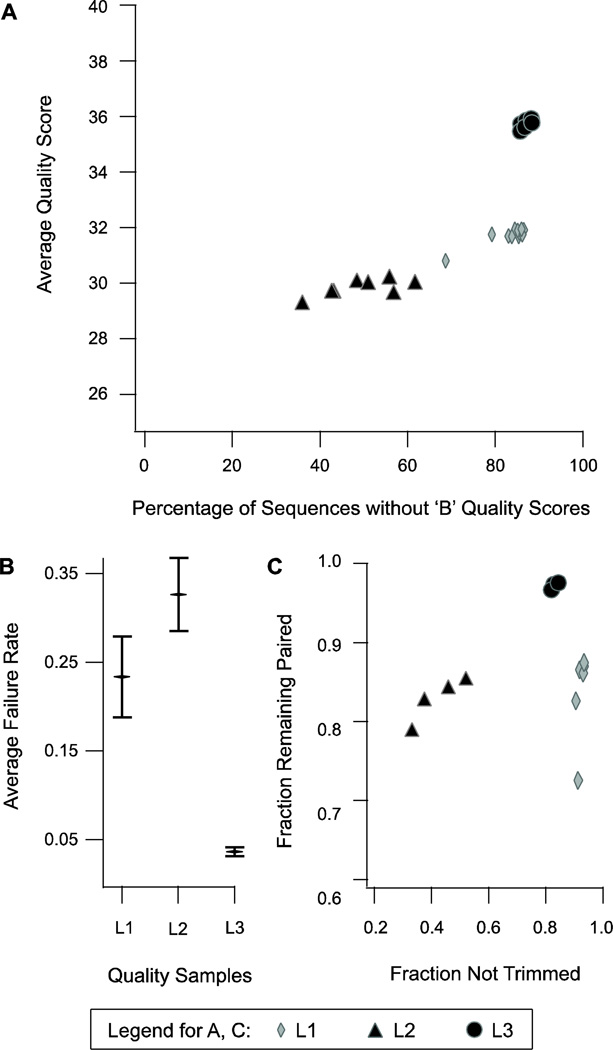
Figure 1. Raw Read Quality Statistics.
A comparison of quality metrics regarding quality filtration of raw reads in the samples L1, L2, and L3. A) The samples L1, L2, and L3 have 6, 4 and 3 components respectively, each of which was sequenced in a separate lane in a flow cell and each of which has a set of forward reads and paired reverse reads in separate files. Each forward and reverse read file is represented by a marker indicating the sample set to which it belongs, and is at a position indicating the average quality score and percentage of sequences without a ‘B’ quality score of the reads in that file. B) The failure rates in the components of each sample set are shown by a marker at the mean with error bars indicating one standard deviation above and below. C) The fraction of reads that remain paired and those that did not need to be trimmed are indicated for each component in a sample set. The legend at the bottom indicates the meanings of marker types in A and C.
2.3 Assembly and Analysis
We chose to use the Velvet short-read assembler because it is widely used, well-documented, and expertise in its use exists in many institutes. Many parameters can be fine-tuned to improve aspects of a de novo assembly, but it is difficult to generalize their use into an algorithm that gives the best assembly for all data sets. In the manual refinement of an assembly, intuition and trial and error can be important components of the process. However, in order to avoid introducing user-bias into the assembly process, we used a script included with the Velvet package called VelvetOptimiser that optimizes two of the most important settings based on predefined rules.
Version 1.0.14 of Velvet (Zerbino et al. 2008) was installed on a Dell R910 rack-mount server with 1TB of physical memory and 8 quad-core Xeon processors to carry out assemblies. All samples were assembled independently using VelvetOptimiser version 2.1.7 set to select the k-mer size in the range of 21 to 55 bp (a range of 21 to 35 bp was used for the sample with 36 bp reads) that produced the best N50 (contig length-weighted median) value; and then to select a coverage-cutoff that maximized the number of base pairs in large contigs. A further assembly step was carried out by submitting the same read set and kmer size determined by VelvetOptimiser to the Oases (version 0.1.18) extension to Velvet (REF) with an insert length parameter of 200bp given.
Determining the quality of an assembly can become very involved after several rounds of refinement, but initial efforts can be guided by some basic metrics describing the general size characteristics of the assembled sequences. A target number of assembled transcripts is very difficult to provide a priori. While there is no published Xiphophorus maculatus genome, Oryzias latipes is a closely related species with a well annotated genome that contains approximately 20,000 genes. However, even with this number as an estimate, alternative splice forms and tissue-specific gene inactivity make it impossible to determine from this number of transcripts we can expect to see. We will instead use generic metrics based on the number and sizes of transcripts to assess the quality of a transcript assembly and observe how these are changed over an experimental range. The N50 value is a measure of contiguity in the assembly and low values generally indicate a more fragmented assembly. The N50 value can be inflated if lower coverage transcripts are selected against due to assembly parameters such as very high k-mer values. The size of the largest contig can also be an indicator of contiguity, when comparing versions of an assembly a spike in the size of the largest contig can indicate a chimeric transcript or other mis-assembled contig.
Basic metrics of assembly quality were calculated for each assembly, including overall N50, size of the longest contig, and number of contigs 500 bp or longer. Assembled sequences 500 bp or longer were queried against the NCBI non-redundant (nr) database through BLASTX. The BLAST searches were run on a cluster using MPI-BLAST (http://www.mpiblast.org) keeping 10 hits per query with an expect value threshold of e-10. To automate the process of filtering BLAST hits for tens of thousands of sequences, we employed the Blast2GO program (Conesa et al. 2005; Conesa et al. 2008; Götz et al. 2008) http://blast2go.org) to select likely matches and generate useful descriptions for each sequence. We did not proceed to any further steps of the Blast2Go program before exporting the matches it made and analyzing those results with a custom-written Perl script.
2.4 Other Bioinformatic Analysis
Besides using the Velvet, MPI-BLAST, and Blast2GO software packages, we developed a suite of bioinformatic tools as Perl, Bash shell, and Make scripts. These scripts serve many functions, including filtering short reads, calculating and reporting their statistics prior to and subsequent to filtration, preparing reads in an appropriate format for Velvet and other computational tools, and analyzing the results downstream of assembly. Our software tools are available upon request.
3 Results and Discussion
3.1 Ab initio Short Read Quality
Our raw data included FASTQ-encoded short reads produced by Illumina sequencing of 13 separate tissues/organs and life stages of the live-bearing fish, Xiphophorus maculatus Jp 163 A. These 13 component samples were grouped by time of creation into three supersets termed L1, L2, and L3. Each superset was sequenced at a different time period in 2009–2010 and each component occupied an individual flow cell lane. Before the short reads were filtered, some basic statistics were obtained regarding quality scores normally reported by the Illumina pipeline software. First overall average quality scores were calculated for each sample ignoring any quality score values of 2 (encoded as a ‘B’ in FASTQ format). Also, the percentage of sequences containing the ‘B’ flag was recorded. A plot of the quality scores and % containing B flags shows that the components of the three supersets L1, L2, and L3 cluster neatly together (Figure 1A). This indicates some categorically-unique factors are acting in each of the three supersets; however, whether these are due to variations in the sample preparation, sequencing instruments, or downstream processing is unknown. We do know that over the year long time period, Illumina updated firmware, processing software, and reagent mixes on several occasions. Any of these modifications may have contributed to the overall increase over time in average quality observed among these three sample sets.
3.2 Quality Filtration
The incidence of errors in this type of data increases with each additional nucleotide base call such that the distal ends are much more likely to have erroneous calls than the initial positions (Dohm et al. 2008). With the improvements made by Illumina, however, lengths of reads reliably produced by NGS have increased from an average of 36 bp up to 150 bp over just the past two years. Even so, a filtration step can be beneficial to downstream analysis(Zhao et al. 2010). Each erroneous base call can produce up to k erroneous k-mers which at best increases the memory requirements of assembly programs toward untenable sizes, and at worst confounds assembly programs into producing misassemblies. The details of our filtration algorithm are given in the methods section, and statistics describing various parameters measured by the filter are presented in Figure 1B and 1C. These results underscore variations among the three supersets.
Sample L1 consisted of 6 sequencing runs from separate life stages as 36 bp, paired-end reads. It had an average Phred quality score of just under 32 and, out of an original 65,733,076 read pairs, retained 42,170,233 (86.2%) paired and 16,244,743 (33.2%) singleton reads post-filtration. Sample L2 was composed of 4 sequencing runs from separate tissues/life stages as 60 bp, paired-end reads. It had an average Phred quality score of about 30, but contained many sequences with B-flags. Of an initial 79,876,151 read pairs, 44,586,554 (55.8%) pairs and 18,388,380 (23.0%) singleton reads remained post-filtration. Sample L3 was composed of only 3 sequencing runs from separate tissues as 60 bp, paired-end reads. The average Phred quality score for this sample was just over 35. Very few of these sequences were trimmed, and most remained paired post-filtration. This sample began with 107,086,149 read pairs and post-filtration, included 100,276,422 (93.6%) pairs and only 5,809,259 (5.4%) singleton reads.
The most recent sample, L3, was composed of three tissues and performed exceptionally well, with an average 96% overall read pass rate (Fig 1B). Of those reads that passed filtration, over 95% remained paired (Fig 1C, circle). A high percentage of retained read pairs is very important for a longer and more accurate sequence assembly (Zerbino et al. 2009). The earlier two supersets, L1 and L2, had average failure rates of 23% and 33%, respectively (Fig 1B). Though these failure rates are high, it is important that over 80% of the reads that passed filtration remained paired for most components of L1 and L2 (Fig 1C, diamond and triangle). The relatively low percentage of trimmed reads in L1 may be, in part, a result of the relatively short original read size (36 bp). The quality filter would only pass a trimmed read if it was 31 bp or longer; thus, if a 36-bp read needed to be trimmed at all, it likely became too short to keep. The components of superset L2, by contrast, had the highest incidences of read trimming (Fig 1C, triangle); this was likely due to the increased number of sequences that contained one or more terminal bases with a ‘B’ flag (Fig 1A), which the filter automatically trims.
3.3 Assembly of Quality Set
In order to compare the performance of each of these samples in an assembly, we reduced the effects of differences in quantity by randomly sampling each post-filtered set to give each set an equal number of paired and single-end reads. Each sub-sampled set contained 42,170,233 pairs and 5,809,259 singleton reads; to distinguish these from the original supersets, we define these as: L_s1, L_s2, and L_s3. The de novo assembly results of each of these sub-samples are summarized in Figure 2.
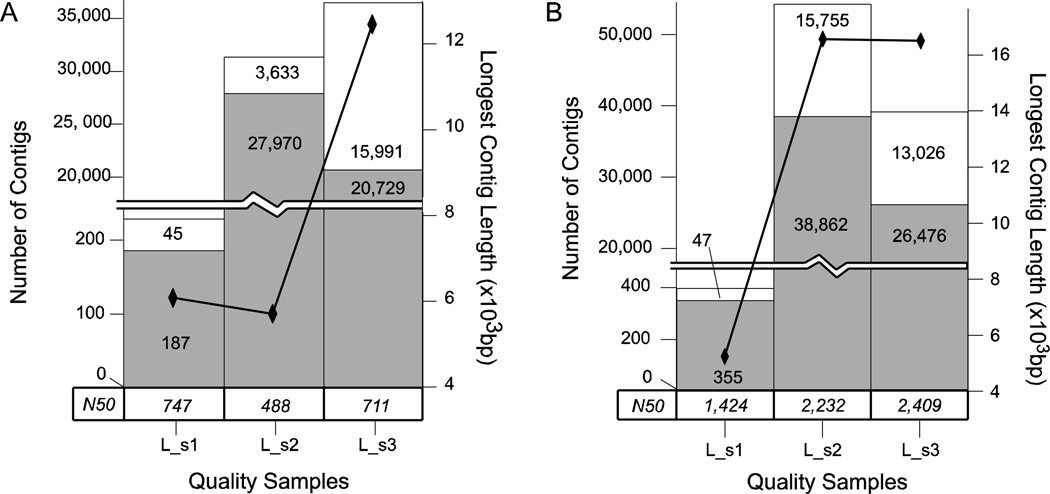
Figure 2. Quality Set Assembly.
Analysis of the assemblies of the three quality samples L_s1, L_s2, and L_s3 by A) Velvet and B) Oases. The total number of contigs of length equal to or greater than 500 bp for each quality sample is represented as a stacked bar graph. The white bar indicates non-annotated contigs and is stacked atop a light gray bar which indicates annotated contigs – each portion of the bar graph is labeled with the numeric quantity represented. The longest contig in an assembly is indicated by black diamonds connected by a black line measured against the right axis. Finally, the N50 for each sample is given in a table along the bottom of the chart just above the sample labels.
The Velvet assembly of the L_s1 sample was surprisingly spare, producing only 232 sequences over 500 bp; 187 of these were annotated (gray bar Figure 2A). Two main differences distinguish this sample from the two other subsets. First, the read size in L_s1 is 36 bp (compared to 60 bp for L_s2 and L_s3), so even though the number of fragments is equivalent in these three sets, L_s1 provided only about half the total cumulative length of short reads. Second, L_s1 is composed of 6 tissues or life stages (compared to 4 in L_s2 and 3 in L_s3), so there may be some dilution of transcripts that are weakly expressed or alternately-spliced in one or more of those component stages.
The L_s2 sample assembly displayed much better performance than expected, comprising just over 31,000 contigs larger than 500 bp (stacked bar graph Figure 2A). Of these, 27,970 contigs were annotated (gray bar Figure 2A) in the BLAST search. This level of apparent assembly success was unexpected, considering the high failure and trimming rates of the raw reads passing through the quality filter. However, a comparison of the longest contigs (black diamonds Figure 2A) and overall N50 statistics (both parameters indicating a reduced overall contiguity in that sample) revealed that the L_s2 sample had much lower values than the assemblies of L_s1 and L_s3. The assembly of sample L_s3 contained the most assembled contigs at 36,710 with a size of 500 bp or greater (stacked bar graph Figure 2A), and the largest assembled contig at just over 12 kb (black diamonds Figure 2A). BLAST searches allowed annotation of a more modest set of 20,729 sequences from this assembly.
In general the contig assemblies resulting from Oases are much more contiguous and there are more contigs in each assembly (Figure 2B). The N50 values are 2–5 times higher indicating that more of the total sequence length is found in longer contigs than in the Velvet assembly. These numbers must be tempered, however, by the fact that Oases also performs some scaffolding and may include regions of uncalled bases encoded as ‘N’ in the fasta sequences. The L_s2 assembly seems to have benefited greatly from the use of Oases, having gained 23,014 contigs. Where the L_s2 Velvet assembly had a relatively weak N50 value compared to the other assemblies, the Oases assembly brings its N50 value up to a range comparable to other Oases assemblies. It is interesting to note that the largest contigs in the Oases assemblies of L_s2 and L_s3 are approximately the same size. The number of annotated sequences remains higher in L_s2 than in L_s3. We speculate that this may be the result of many more species of RNA messages in the L_s2 sample than in the L_s3. In summary, the short read length, medium quality L_s1 sample gave the poorest assembly, while the longer read length yet poorer quality L_s2 sample produced a relatively decent assembly with high annotation rate but poor N50 under Velvet but was improved significantly by Oases. The long read length and high quality L_s3 sample gave the Velvet assembly with the most contigs, a strong N50, the longest contig, and a moderate annotation rate, and an Oases assembly with nearly 40,000 contigs and the highest N50 of this set. While Phred quality scores did seem to have some influence on the assembly quality metrics used here under Velvet, the read length appears to have a much greater effect. The alternate contig assembly scheme of Oases significantly increased the contiguity and number of contigs resulting from L_s2. Since L_s2 contains whole-body RNA extracts compared to the three tissues in L_s3, one likely explanation is that there are many more alternate splice forms and transcript species in L_s2.
3.4 Assembly of Quantity Set 1
We next set out to examine the effects of read quantity on an assembly. The full complement of reads in the three tissue samples comprising set L3 is well over 100 million read pairs and 5 million singleton reads. That set also had the highest, and rather uniform, group of quality scores, and so we randomly sampled and assembled five read sets designated quantity set 1 as a whole and consisting of the subsets N1_s1, N1_s2, N1_s3, N1_s4 and N1_s5 which contained 20%, 40%, 60%, 80%, and 100% of the reads in L3 respectively (Table 1). For each subset we included an equal fraction of both paired and single short reads. Figure 3 summarizes the assemblies of quantity set 1 samples.
Table 1. Quantity of reads in quantity sets 1 and 2.
The number of paired and single reads are held in the same proportion as in the source from which each subset is sampled. Quantity set 1 is sub-sampled from the full read set of 3 tissues comprising L3 while quantity set 2 is sub-sampled from one of the tissues therein. The sub-sampling is done in increments of 20% of the total: 20%, 40%, 60%, 80%, 100%.
| Quantity Set 1 | Quantity Set 2 | ||||
|---|---|---|---|---|---|
| sample | pairs | singles | sample | pairs | singles |
| N1_s1 | 21,417,230 | 1,161,852 | N2_s1 | 6,780,130 | 467,751 |
| N1_s2 | 42,834,460 | 2,323,704 | N2_s2 | 13,560,260 | 935,502 |
| N1_s3 | 64,251,689 | 3,485,555 | N2_s3 | 20,340,389 | 1,403,253 |
| N1_s4 | 85,668,919 | 4,647,407 | N2_s4 | 27,120,519 | 1,871,004 |
| N1_s5 | 107,086,149 | 5,809,259 | N2_s5 | 33,900,649 | 2,338,755 |
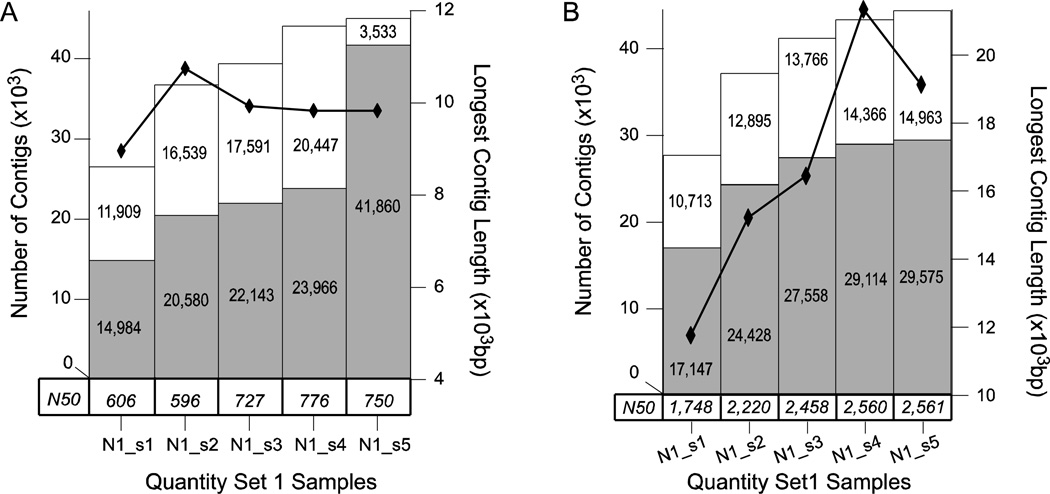
Figure 3. Quantity Set 1 Assembly.
Analysis of the assemblies of the five quantity set 1 samples N1_s1, N1_s2, N1_s3, N1_s4 and N1_s5 by A) Velvet and B) Oases. The total number of contigs of length equal to or greater than 500 bp for each quality sample is represented as a stacked bar graph. The white bar indicates non-annotated contigs and is stacked atop a light gray bar which indicates annotated contigs – each portion of the bar graph is labeled with the numeric quantity represented. The longest contig in an assembly is indicated by black diamonds connected by a black line measured against the right axis. Finally, the N50 for each sample is given in a table along the bottom of the chart just above the sample labels.
When looking at the overall N50 and largest contig size in the Velvet assembly (Figure 3A), the assemblies seemed to plateau beginning with N1_s3 (60% of L3 sequences or 64,251,689 pairs and 3,485,555 single reads). The total number of assembled sequences made sizeable increases at each step from N1_s1 to N1_s4, but the number of assembled sequences did not significantly increase from N1_s4 to N1_s5 (stacked bar graph Figure 3A). Another notable increase occurred in the number of annotated sequences from N1_s4 to N1_s5 (gray bars Figure 3A). This sudden increase in annotation frequency, without a concurrent increase in the number of contigs, was an interesting result. However, in the absence of a well-annotated genome to allow further investigation of annotations, the significance is hard to determine.
The results of the Oases assemblies of these subsets share some of the same trends as the Velvet assembly only with much longer sequences and more of them. The total number of contigs, number of annotated contigs, and N50 each increase (Figure 3B) almost in concert with each other as read quantity increases and begin to plateau also around 60% – 80% of total reads. Clearly, Oases has significantly improved the contiguity of the assemblies compared to Velvet. In any case, it is clear that there are benefits to having more reads, but the benefits gained from adding more data begin to wane at approximately 60 million read pairs in this data set.
3.5 Assembly of Quantity Set 2
A question that arose in the first quantity set assembly was whether or not the previously observed sudden increase in the fraction of sequences that were annotated would be present in the full set of reads from one of the component tissue samples in L3. We postulated that, since each of the N1 samples was a randomly selected set derived from three tissues, there could be some dilution effects on uniquely expressed transcripts or splice forms that are only alleviated when a large enough number of reads are present. To investigate this possibility, we chose one tissue from L3 (containing 33,900,649 pairs and 2,338,755 singleton reads) and randomly sampled it in five samples of 20%, 40%, 60%, 80%, and 100% (holding proportion of paired to unpaired reads equal) of the total reads as in section 3.4, with the set designated quantity set 2 and consisting of subsamples N2_s1, N2_ s2, N2_s3, N2_s4, and N2_s5 (Table 2).
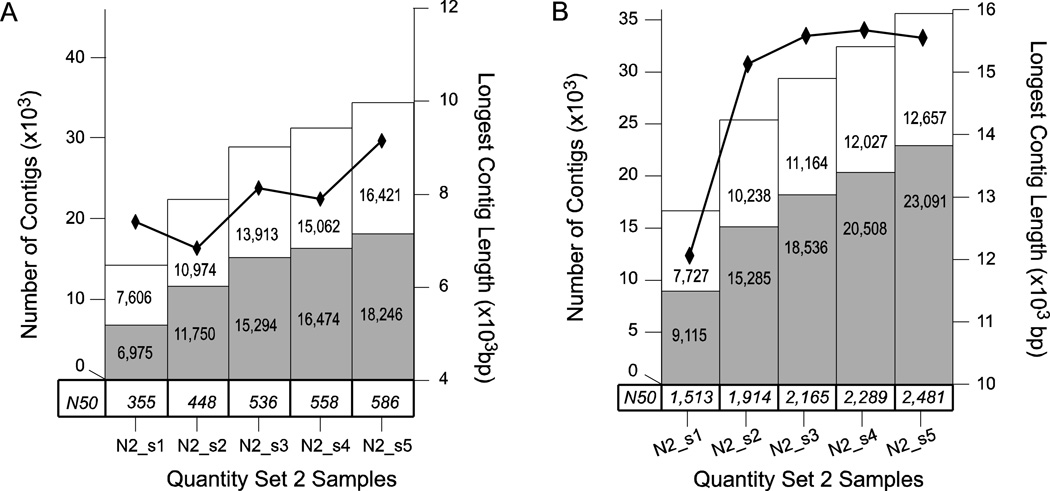
These results are summarized in Figure 4 and clearly do not indicate a spike in BLAST annotation frequency. It is possible that the annotation spike is due to some property of one of the other tissue samples in L3, but it is more likely that it is an anomaly resulting from the tendency of Velvet to produce fragmented transcript assemblies (Gibbons et al. 2009; Surget-Groba et al. 2010). Most of the assembly metrics have a general upward trend over this set in the Velvet assembly, and this trend is even more visible in the results of the Oases assembly. This behavior is clearly different from that of the larger N1 set of assemblies, in which the metrics tended to plateau or stabilize as more reads were added (Fig 3). This suggests that it may be possible to estimate the degree to which adding more short-read data could improve an assembly by simply sub-sampling a full set of data and examining the trends in the resulting trial assemblies.
Figure 4. Quantity Set 2 Assembly.
Analysis of the assemblies of the five quantity set 2 samples N2_s1, N2_s2, N2_s3, N2_s4 and N2_s5 by A) Velvet and B) Oases. The total number of contigs of length equal to or greater than 500 bp for each quality sample is represented as a stacked bar graph. The white bar indicates non-annotated contigs and is stacked atop a light gray bar which indicates annotated contigs – each portion of the bar graph is labeled with the numeric quantity represented. The longest contig in an assembly is indicated by black diamonds connected by a black line measured against the right axis. Finally, the N50 for each sample is given in a table along the bottom of the chart just above the sample labels.
4 Conclusions
In our results, the assembly of L_s1 as compared to L_s2 or L_s3 suggests that the length of reads is a significant factor in successful de novo NGS assemblies and this has been shown previously in synthetic data sets. It is also clear that the quantity of reads plays a key role. Higher numbers of reads increase depth of coverage, which makes it easier for the assembly program to assemble low-abundance transcripts and also serve to increase the signal-to-noise ratio of correctly assembled transcripts to incorrect ones. In the ranges we examined, the quality score of the reads had the least impact on the downstream de novo assembly. However, it should be kept in mind the Phred score scale is logarithmic, so if average quality scores drop, an expected increase in base calling errors could play a major factor in assembly success. We also noted in the experiments addressing read quantity that many of the metrics began to plateau as the number of reads passed sixty-million paired-end reads. Although further study is warranted, it may be a useful practice to subsample and assemble read sets in this way to estimate the likely return in assembly quality when considering the addition of further sequencing runs to supplement existing data for an assembly. It is also strongly recommended that a program designed to assemble transcript data like Oases be employed when assembling RNASeq data. We expect these findings are applicable across short-read assembly programs, though each sequencing technology, data set, and assembly algorithm will likely introduce unique elements to be considered.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Sara Volk for her critical reading of this work. This work was supported by Texas State University and the National Institutes of Health, National Center for Research Resources grant #R24-RR024790 including an American Recovery and Reinvestment act supplement to this award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This paper is based on a presentation given at the 5th Aquatic Annual Models of Human Disease conference: hosted by Oregon State University and Texas State University-San Marcos, and convened at Corvallis, OR, USA September 20–22, 2010.
References
- Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, {Keira Cheetham} R, Cox AJ, Ellis DJ, Flatbush MR, Gormley Na, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DMD, Bancarz IR, Banerjee S, Barbour SG, Baybayan Pa, Benoit Va, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham Ja, Brown RC, Brown Aa, Buermann DH, Bundu Aa, Burrows JC, Carter NP, Castillo N, {Chiara E Catenazzi} M, Chang S, {Neil Cooley} R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, {Fuentes Fajardo} KV, {Scott Furey} W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri Pa, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, {Huw Jones} Ta, Kang G-D, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent Ma, Lawley CT, Lee SE, Lee X, Liao AK, Loch Ja, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, {Ling Ng} B, Novo SM, O'Neill MJ, Osborne Ma, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, {Chris Pinkard} D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, {Chiva Rodriguez} A, Roe PM, Rogers J, {Rogert Bacigalupo} MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith Ma, {Ernest Sohna Sohna} J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Gowik U. What can next generation sequencing do for you? Next generation sequencing as a valuable tool in plant research. Plant Biol (Stuttgart, Germany) 2010;12:831–841. doi: 10.1111/j.1438-8677.2010.00373.x. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S. Blast2GO: A Comprehensive Suite for Functional Analysis in Plant Genomics. Int J Plant Genomics. 2008;2008 doi: 10.1155/2008/619832. 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (Oxford, England) 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Costa V, Angelini C, {De Feis} I, Ciccodicola A. Uncovering the Complexity of Transcriptomes with RNA-Seq. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/853916. 853916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucl Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Tyagi AK, Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol. 2009;26:2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucl Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, Zhang Z, Zhang Y, Wang W, Li J, Wei F, Li H, Jian M, Li J, Zhang Z, Nielsen R, Li D, Gu W, Yang Z, Xuan Z, Ryder OA, Leung FC-C, Zhou Y, Cao J, Sun X, Fu Y, Fang X, Guo X, Wang B, Hou R, Shen F, Mu B, Ni P, Lin R, Qian W, Wang G, Yu C, Nie W, Wang J, Wu Z, Liang H, Min J, Wu Q, Cheng S, Ruan J, Wang M, Shi Z, Wen M, Liu B, Ren X, Zheng H, Dong D, Cook K, Shan G, Zhang H, Kosiol C, Xie X, Lu Z, Zheng H, Li Y, Steiner CC, Lam TT-Y, Lin S, Zhang Q, Li G, Tian J, Gong T, Liu H, Zhang D, Fang L, Ye C, Zhang J, Hu W, Xu A, Ren Y, Zhang G, Bruford MW, Li Q, Ma L, Guo Y, An N, Hu Y, Zheng Y, Shi Y, Li Z, Liu Q, Chen Y, Zhao J, Qu N, Zhao S, Tian F, Wang X, Wang H, Xu L, Liu X, Vinar T, Wang Y, Lam T-W, Yiu S-M, Liu S, Zhang H, Li D, Huang Y, Wang X, Yang G, Jiang Z, Wang J, Qin N, Li L, Li J, Bolund L, Kristiansen K, Wong GK-S, Olson M, Zhang X, Li S, Yang H, Wang J, Wang J. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies — the next generation. Nat Revs Genet. 2009;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95 doi: 10.1016/j.ygeno.2010.03.001. 327–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narzisi G, Mishra B. Comparing de novo genome assembly: the long and short of it. PLoS ONE. 2011;6:e19175. doi: 10.1371/journal.pone.0019175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M. Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryotic Cell. 2010;9:1300–1310. doi: 10.1128/EC.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkiewicz K, Studholme DJ. De novo assembly of short sequence reads. Brief Bioinformat. 2010;11:457–472. doi: 10.1093/bib/bbq020. [DOI] [PubMed] [Google Scholar]
- Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinformat. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos J. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20:1432–1440. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Walter RB, Ju Z, Martinez A, Amemiya C, Samollow PB. Genomic resources for Xiphophorus research. Zebrafish. 2006;3:11–22. doi: 10.1089/zeb.2006.3.11. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews. Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J, Kingsford C, Pop M. Assessing the benefits of using mate-pairs to resolve repeats in de novo short-read prokaryotic assemblies. BMC Bioinformat. 2011;12:95. doi: 10.1186/1471-2105-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] Chapter 11, Unit 11.15. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, McEwen GK, Margulies EH, Birney E. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PloS one. 2009;4:e8407. doi: 10.1371/journal.pone.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Palmer LE, Bolanos R, Mircean C, Fasulo D, Wittenberg GM. EDAR: An efficient error detection and removal algorithm for next generation sequencing data. J Comput Biol. 2010;17:1549–1560. doi: 10.1089/cmb.2010.0127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.