Abstract
The human 7SK small nuclear RNA (snRNA) is an abundant noncoding RNA whose function has been conserved in evolution from invertebrates to humans. It is transcribed by RNA polymerase III (RNAPIII) and is located in the nucleus. Together with associated cellular proteins, 7SK snRNA regulates the activity of the positive transcription elongation factor, P-TEFb. In humans, this regulation is accomplished by the recruitment of P-TEFb by the 7SK snRNA-binding proteins, HEXIM1 or HEXIM2, which inhibit the kinase activity of P-TEFb. P-TEFb regulates the transition of promoter proximally paused RNA polymerase II (RNAPII) into productive elongation, thereby, allowing efficient mRNA production. The protein composition of the 7SK small nuclear ribonucleoprotein (snRNP) is regulated dynamically. Whereas the La related protein LARP7 is a constitutive component, the methyl phosphate capping enzyme MePCE associates secondarily to phosphorylate the 5' end of 7SK snRNA. The release of active P-TEFb is closely followed by release of HEXIM proteins and both are replaced by heterogeneous nuclear ribonucleoproteins (hnRNPs). The released P-TEFb activates the expression of most cellular and viral genes. Regulated release of P-TEFb determines the expression pattern of many of the genes that respond to environmental stimuli and regulate growth, proliferation and differentiation of cells.
Although 7SK snRNA was first described in 1976,1 its function became appreciated only 25 years later when it was found to be the core of the 7SK snRNP, which interacts with and inhibits P-TEFb.2, 3 Since that time, the 7SK snRNP has been studied extensively and many details of its role in regulating P-TEFb have been forthcoming.4 P-TEFb, is a regulated constituent of the 7SK snRNP involved in RNA polymerase II elongation control.5, 6 7SK snRNA should be viewed as the RNA scaffold on which an elaborate P-TEFb regulatory machine is assembled and the reversible association of P-TEFb with the 7SK snRNP is an important regulatory mechanism for eukaryotic gene expression.7, 8
P-TEFb is a cyclin dependent kinase required for the transition of promoter proximally paused polymerases into productive elongation. It is inhibited by DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) and flavopiridol, which are ATP analogs that inhibit most RNAPII transcription.9 Indeed, transcription complexes are already engaged on most human and many Drosophila promoters irrespective of expression, which emphasizes the generality of RNAPII elongation control.10, 11 In humans, P-TEFb is composed of the cyclin dependent kinase Cdk9,12 Cyclin T1 (CycT1) or one of two alternatively spliced isoforms of Cyclin T2 (CycT2a or CycT2b)13 (Table 1). In Drosophila only one cyclin partner for Cdk9, Cyclin T, is found.14 Regulated release of P-TEFb from the 7SK snRNP can be considered a rheostat for cell growth and proliferation, malignant transformation as well as terminal differentiation. HIV requires P-TEFb for its replication12, 15 and this suggests that the 7SK snRNP could be targeted for silencing and/or re-activating viral gene expression in the infected host. Other diseases that are impacted by P-TEFb include cardiac hypertrophy, steroid-dependent or responsive cancers, leukemias, lymphomas, as well as inflammatory and autoimmune states.16, 17
Table 1 .
Components of 7SK snRNPs, P-TEFb and N-TEFs
| Complex | Component | Name | Size |
|---|---|---|---|
| 7SK snRNP | |||
| (P-TEFb/HEXIM) | 7SK snRNA | 7SK small nuclear RNA | 330–332 nt RNA |
| LARP7 | La-related protein 7 | 67 kDa | |
| MePCE | Methylphosphate capping enzyme | 74 kDa | |
| HEXIM1/2 | HMBA-induced mRNA 1/2 | 41 /32 kDa | |
| P-TEFb | Positive transcription elongation factor b | 150 kDa | |
| 7SK snRNP | |||
| (hnRNPs) | 7SK snRNA | 7SK small nuclear RNA | 330–332 nt RNA |
| LARP7 | La-related protein 7 | 67 kDa | |
| MePCE | Methylphosphate capping enzyme | 74 kDa | |
| hnRNPs | Heterogeneous nuclear ribonucleoproteins | ~216 kDa | |
| P-TEFb | |||
| Cdk9 | Cyclin dependent kinase 9 | 43 kDa or 55 kDa | |
| CycT1 | Cyclin T1 | 81 kDa | |
| CycT2a, 2b | Cyclin T2a, T2b | 74 kDa, 82 kDa | |
| hnRNPs | |||
| hnRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 38 kDa | |
| hnRNPA2 | Heterogeneous nuclear ribonucleoprotein A2 | 36 kDa | |
| hnRNPQ1 | Heterogeneous nuclear ribonucleoprotein Q1 | 62 kDa | |
| hnRNPR | Heterogeneous nuclear ribonucleoprotein R | 80 kDa | |
| DSIF | DRB-sensitivity inducing factor | 180 kDa | |
| Spt4 | Suppressor of Ty protein 4 | 16 kDa | |
| Spt5 | Suppressor of Ty protein 5 | 160 kDa | |
| NELF | Negative elongation factor | 200 kDa | |
| NELF-A | Wolf-Hirschhorn syndrome protein WHSC2 | 66 kDa | |
| NELF-B | COBRA1, a BRCA1-associated protein | 62 kDa | |
| NELF-C/D | TH1 proteins | 60/59 kDa | |
| NELF-E | RD, multiple R, D repeats | 46 kDa | |
BIOSYSTHESIS OF 7SK snRNP containing P-TEFb
Human 7SK RNA is a highly structured, abundant snRNA that is synthesized as a 332 nt transcript (Fig. 1) containing four uridine residues at its 3’ end (…UCUUUU). Although hundreds of pseudogenes have been identified, the bona fide 7SK snRNA is transcribed by RNAPIII from a single locus on human chromosome 6. Its constitutively active promoter consists of distal and proximal promoter elements, which bind octamer-binding protein 1 (Oct1), selenocysteine tRNA gene transcription activating factor 1 (Staf1) and snRNA activating protein complex (SNAPc), respectively.18, 19 A TATA box binds TATA-binding protein (TBP) and RNAPIII. Of interest, it was this promoter architecture that helped to identify 7SK snRNA in invertebrates.20
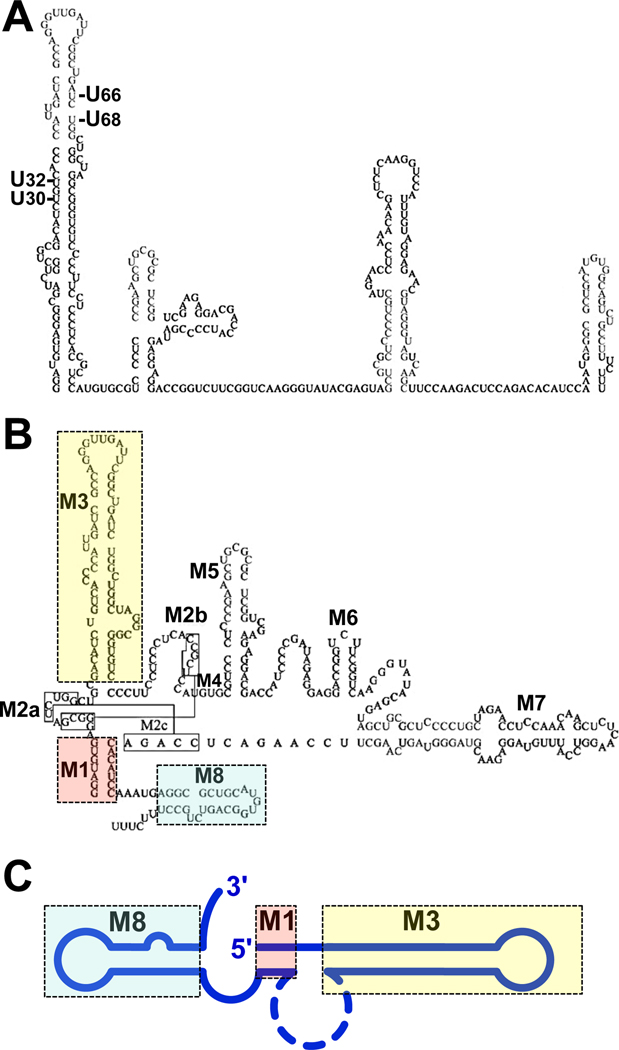
Figure 1. Conserved 7SK snRNA motifs.
A) Structure of 7SK snRNA proposed by Wassarman and Steitz.34 Residues in paired regions that were sensitive to CMCT are indicated. B) Detailed structure of the human 7SK snRNA showing possible structural pairing based on conservation of sequences across many species.38 Structural motifs (M) are numbered from M1 to M8 and correspond with the regions of highest conservation. Colored boxes indicate motifs that have been determined to be important for binding of proteins to 7SK snRNA. M1 involves base pairing between the two termini of the RNA bringing them into relatively close proximity and potentially aiding in protein interactions. M3 is a stem-loop required for the binding of HEXIM. M8 is a stem-loop near the 3' end that is necessary for P-TEFb association. C) Simplified cartoon model of 7SK RNA showing the important structural motifs corresponding to the colors of the detailed structure in B. The images in A and B are based on a figure in Marz et al.38.
Upon transcription by RNAPIII, the Lupus antigen (La) protein initially protects the 3’ end of 7SK snRNA.21 With time, nucleases remove 1, 2, or 3 U residues from the 3’ end and an adenine is added post-transcriptionally by an unidentified adenylating enzyme.22 At steady state, the sequence of the 3’ end is a mixture of …UCUA, …UCUUA, and …UCUUUA giving a total length for 7SK snRNA of 330–332 nt with 331 being slightly preferred over the other two lengths.22
Soon after synthesis of 7SK the La protein is replaced with La related protein 7 (LARP7)(Table 1).21 LARP7 is a 67 kDa protein that contains 582 residues with a Lupus Antigen Motif (LAM) at its N-terminus (amino acids 37–111) and two RNA Recognition Motifs (RRMs) in the rest of the protein (amino acids 111–189 and 461–559).23 Together, the LAM and first RRM mediate the specific binding to 3’ uridines in 7SK snRNA. In cells, 90% of 7SK snRNA is bound by LARP7. In addition, LARP7 associates primarily with 7SK snRNA.23, 24 Since the knockdown of LARP7 leads to dramatic reduction of 7SK snRNA and release of P-TEFb24, 25, it is not surprising that mutations in LARP7 have been detected in gastric, breast and cervical cancers.23, 25
At its 5’ end, 7SK snRNA is methylated by MePCE26 (Table 1). MePCE is a 74 kDa protein that contains 689 residues with overlapping RNA methylase (amino acids 448–615) and bicoid interacting motifs (amino acids 574–683) and was previously called BCDIN3.26 It is an S-adenosine methionine-dependent methyl transferase that adds a monomethyl cap onto the gamma phosphate at the 5’ end of 7SK snRNA. In addition to 7SK snRNA, it also binds U6 snRNA in cells. Nevertheless, its depletion destabilizes 7SK snRNA and leads to the release of P-TEFb.26 MePCE and LARP7 interact even in the absence of RNA.27, 28 However, this association is strengthened by 7SK snRNA. Knockdown of either MePCE26 or LARP723, 24 greatly destabilizes 7SK snRNA. Upon its degradation, active P-TEFb is released, which impacts transcription and mRNA processing in cells.23, 29 In addition, cells undergo hypertrophic and/or malignant transformation.23 Together LARP7, MePCE, and the 7SK RNA form the constitutive core of 7SK snRNP (Fig. 2).
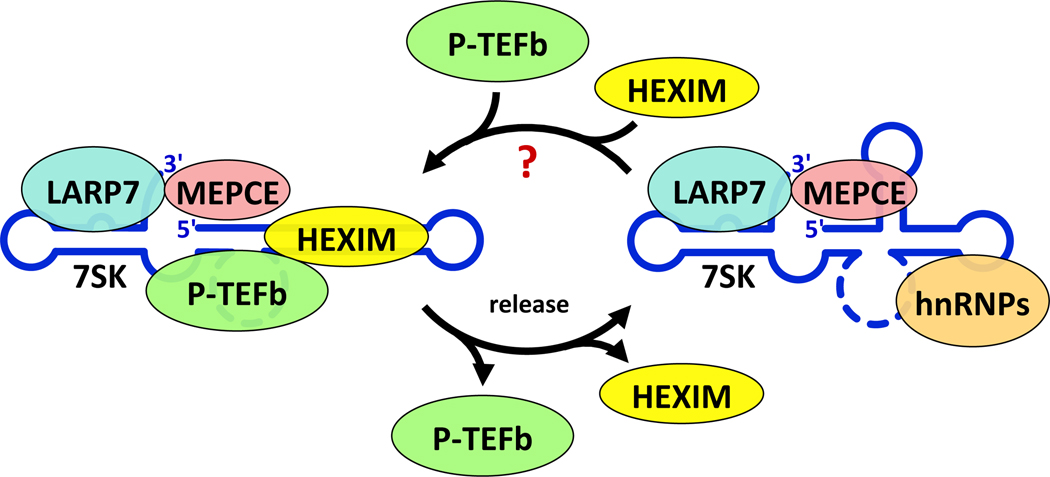
Figure 2. Reversible association of P-TEFb and HEXIM with the 7SK snRNP.
Cyclic model for interactions between P-TEFb and the 7SK snRNP. Left: The 7SK snRNP containing the indicated proteins is acted upon directly by Tat and Brd4,36 or by UV light, actinomycin D, any P-TEFb inhibitor, HMBA, or SAHA (release). P-TEFb is extracted from the snRNP, which causes the subsequent release of HEXIM. Right: After release of P-TEFb and HEXIM, there is a conformational change in the RNA that is stabilized by hnRNPs. LARP7 and MePCE remain associated with 7SK snRNA. The release of P-TEFb and HEXIM is not directly reversible and the mechanism involved in re-sequestration of HEXIM and then P-TEFb remains unknown.
The incorporation of P-TEFb into the 7SK snRNP starts with the binding of a HEXIM dimer to a region near the 5’ end of 7SK snRNA. This RNA•protein interaction causes a conformational change in HEXIM, which unmasks its P-TEFb-binding surface.27, 30, 31 F208 and Y271 of HEXIM1 are thought to occlude the ATP pocket in Cdk9 and thus inactivate P-TEFb.30 This inhibition would parallel that described for the inhibition of CycA•Cdk2 by p27.32 Moreover, the complete assembly of the inactivated P-TEFb bound 7SK snRNP in cells also requires the presence of a stem loop motif near the 3’ end of 7SK snRNA.33 Thus, the complete 7SK snRNP contains MePCE, LARP7 as well as two HEXIM and P-TEFb subunits (Fig. 2).
STRUCTURE OF 7SK snRNA
Complex protein interactions that lead to the sequestration of P-TEFb are facilitated by the structural nature of the RNA. To date, modest progress has been made on the structure of 7SK snRNA and how this influences protein interactions. Initially, nuclease sensitivity and chemical footprinting were used to probe the secondary structure of 7SK snRNA from 7SK snRNP, which was isolated from HeLa cells.34 The ensuing Wassarman and Steitz model contained four RNA hairpins or stem loops (Fig. 1A). There were some discrepancies between the actual data and the proposed structure. For example, residues U28, U30, U66, and U68 were depicted as being in paired regions, but the experimental results indicated that they were sensitive to chemical modification (Fig. 1A). Moreover, the structure differed from that obtained by predictive RNA-folding programs.35 Some of these differences can be reconciled by the conformational change of 7SK snRNA upon the release of HEXIM and P-TEFb.36 Indeed, the presence of two 7SK snRNA conformers in the population of 7SK snRNPs could explain the original mapping data.36 In support of this conformational change, the RNA secondary structure prediction program mFold 37 yields two families of highly related human 7SK snRNA structures with similar stabilities.36 An extensive examination of 7SK snRNA sequences from other organisms supports this and other modifications to the original Wassarman and Steitz structure (Fig. 1B).38
The known binding sites of the proteins in the 7SK snRNP give insights into how the structure of the RNA itself may lead to regulated release of P-TEFb. Though many details are unclear, several regions of the RNA have importance in protein binding. The sites of association of HEXIM dimers have now been mapped with reasonable accuracy to the main 5’ motif (M3) (Fig. 1B). This determination was difficult because HEXIM1 also binds dsRNA in a sequence-independent manner.39 Nevertheless, HEXIM1 can be cross-linked to U at position 30 in the lower half of M3 in 7SK snRNA 40. Further detailed analyses concluded that binding of HEXIM1 was directed by the upper half and strengthened by the lower half of M3.41 Since HEXIM1 exists as an obligate dimer and binds 7SK snRNA as such, 30, 42, 43 it is likely that both sites are occupied simultaneously. NMR techniques demonstrated that two GAUC motifs (42–45 paired to 64–67) are the critical recognition sequence, which is opened and stabilized by short peptides derived from the arginine rich motif (ARM) in HEXIM.44 Proline and serine residues in the middle of basic residues are also essential for this recognition.
Humans express two HEXIM paralogs, HEXIM1 and HEXIM2. Expressed ubiquitously from adjacent genes on the human chromosome 17, these 41 and 32 kDa proteins contain 359 and 286 residues, respectively (Table 1). In the central region of both HEXIM proteins are found conserved basic and acidic residues that are autoinhibitory in the absence of 7SK snRNA. 43, 45 Only upon the binding of their ARMs to 7SK snRNA is their conformation changed so that they can interact with and inhibit P-TEFb.39 At their C-termini are two coiled coil regions that are used for dimerization and association with the cyclin boxes in CycT1 and CycT2.31 The formation of HEXIM1 oligomers is a prerequisite for RNA-binding.30 Fully assembled 7SK snRNP measures 550–600 kDa (Table 1). HEXIM1 is more abundant than HEXIM2 in HeLa cells, but mixed oligomers are thought to exist.30 In addition, when HEXIM1 is knocked down, HEXIM2 replaces it on 7SK snRNA with no loss of function.43, 45
The interaction of P-TEFb, and therefore its regulation, is the fundamental reason for the existence of the 7SK snRNP. Though details of the interaction with the RNA are unresolved, it has been shown that CycT1 from P-TEFb may bind to M8 possibly through its Tat•TAR Recognition Motif (TRM) (Fig. 1B). Whether P-TEFb directly binds to this region of 7SK snRNA or through another protein is unclear.
When P-TEFb and HEXIM dissociate from the 7SK snRNP, another set of factors enter the complex. They are the heterogeneous ribonucleoprotein particle (hnRNP) proteins, hnRNPA1, hnRNAPA2, hnRNPQ1 and hnRNPR (Table 1). They bind M1 and M7 (hnRNPQ1, hnRNPR), or M7 only (hnRNPA1 and hnRNPA2)(Fig. 1 and 2). They were identified in proteomic screens by 7SK snRNA affinity chromatography.46–48
The 7SK RNA undergoes a conformational change when P-TEFb is released from the 7SK snRNP. Chemical (CMCT: N-Cyclohexyl-N-(β-[N-methylmorpholino]ethyl)carbodiimide p-toluenesulfonate) footprinting, which modifies unpaired uridines, revealed that the release of P-TEFb and HEXIM1 is accompanied by a change in the conformation in 7SK snRNA.36 (Fig. 2) Since 7SK snRNA binds HEXIM tightly in vitro, this conformational change could contribute to the loss of affinity between 7SK snRNA and HEXIM1 in cells, where about half of the 7SK snRNP lacks HEXIM1 even though HEXIM1 is present in about a 5-fold molar excess in HeLa cells.24, 45 Of note, it is also difficult to equilibrate exogenously introduced HEXIM1 with endogenous HEXIM1 even under conditions that disrupt and reform the 7SK snRNP in transient transfection assays. In this scenario, hnRNPs could stabilize a HEXIM-excluded conformation of 7SK snRNA. (Fig. 2)
As described above, LARP7 interacts with the 3’ end and MePCE methylates and, therefore, likely interacts with the 5’ end of 7SK. It is interesting that all metazoan 7SK snRNAs contain a highly conserved motif (M1) that pairs the exact 5’ end with a region just upstream of the 3’ stem and loop (M8).38 This pairing brings both termini of the RNA into close proximity (Fig. 1B and C). This arrangement may help position MePCE for a functional interaction with the 5’ end of 7SK snRNA, because LARP7 is associated with the 3’ end and has a direct interaction with MePCE.28
Another interesting comparison that has been made is between HIV transactivation response (TAR) RNA hairpin and 7SK snRNA. HIV transcriptional transactivator Tat binds the 5’ U bulge in TAR, which is similar to two U bulges in M3 of 7SK snRNA.44, 49 Moreover, the ARMs that bind RNA in Tat and HEXIM1 are almost identical.50 Tat and HEXIM1 also bind cyclin boxes in CycT1. Thus, it is not surprising that in vitro Tat competes with HEXIM1 for P-TEFb in the 7SK snRNP.51, 52 Tat also binds the top of M3 in 7SK snRNA,41, 52 however, the impact of this interaction on HIV transcription and replication is not clear.
RELEASE OF P-TEFb FROM 7SK snRNP
How is P-TEFb liberated from 7SK snRNP? One important mechanism has been mentioned, in which Tat competes with HEXIM1 for P-TEFb binding (Fig. 3). The same situation pertains to bromo domain-containing protein 4 (Brd4), which binds CycT1 via its extreme C-terminus.36, 53 Indeed, much of the active P-TEFb exists in complex with Brd4, which also brings P-TEFb to acetylated histones and helps to store P-TEFb on chromatin during mitosis.54–56 Signaling pathways, stress, UV light and other external cues also lead to the release of P-TEFb. Compounds that inhibit transcription directly, such as actinomycin D, or indirectly such as the P-TEFb inhibitors DRB, flavopiridol and roscovitine all lead to the release of P-TEFb from the 7SK snRNP.3, 57, 58 Differentiation agents, such as HMBA and SAHA (suberoylanilide hydroxamic acid, Vorinostat or Zolinza), which is a pan-histone deacetylase inhibitor (HDACi), also lead to the release of P-TEFb before increasing the synthesis of HEXIM1.59–61. Most likely, signaling via growth factor, T and B cell antigen receptors, also affects levels and the equilibrium of active and inactive P-TEFb complexes in cells.
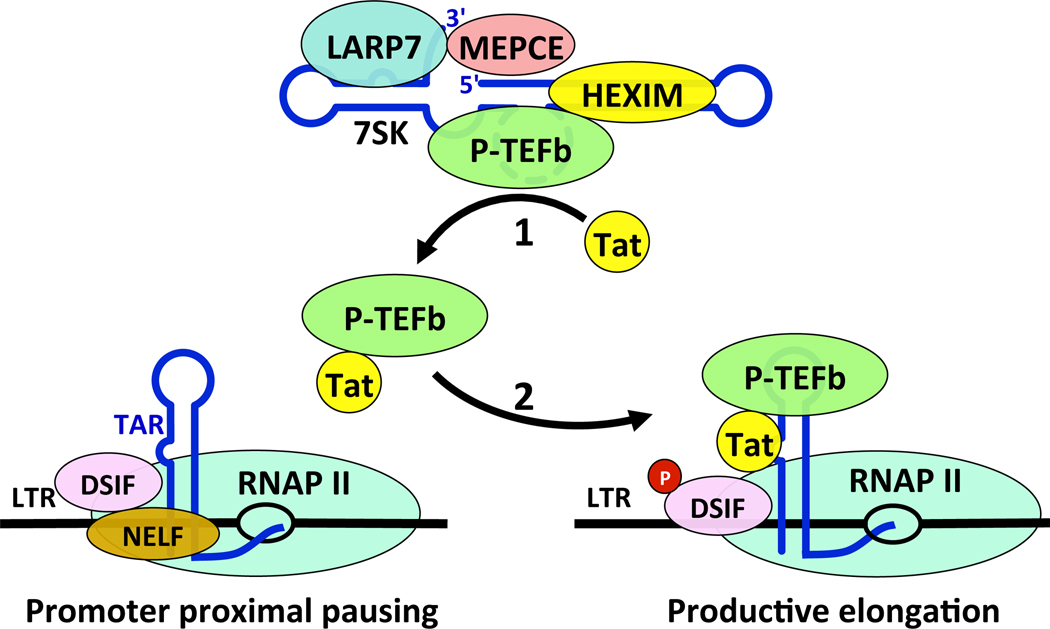
Figure 3. Control of HIV transcription by Tat.
1: The binding between Tat and P-TEFb releases P-TEFb from the 7SK snRNP and 2: a Tat•P-TEFb complex is delivered to the promoter proximal paused RNAPII on the HIV LTR through an interaction with TAR, which is present in all nascent viral transcripts. Upon delivery of P-TEFb, the negative factor NELF is phosphorylated and removed. DSIF is also phosphorylated (P) and facilitates the transition of RNAPII into productive elongation.
Indeed, the absence of transcription by RNAPII triggers signaling cascades that release P-TEFb. However, they have not been studied in great detail, but T186 in the T loop of Cdk9 remains phosphorylated and no major changes in HEXIM proteins have been detected. In contrast, HMBA engages a series of kinases and phosphatases to modify Cdk9 and HEXIM proteins. In one study, PP1a and PP2a were activated by HMBA, which resulted in the dephosphorylation of T186 and the release of an inactive P-TEFb from 7SK snRNP.59 In the other, HMBA activated the PI3K pathway so that Akt phosphorylated HEXIM1 on S270 and T278, which released P-TEFb from 7SK snRNA.60 Since SAHA, which is a potent pan-HDACi, replicated effects of HMBA, the most likely scenario for both effects is that cytoplasmic HDAC6 is involved.61 By inhibiting the deacetylation of tubulin and HSP90.62, these structures aggregate with their associated client proteins, which include S/T phosphatases, PI3K and Akt. This aggregation leads to their activation, which results in these cumulative effects that dissociate P-TEFb from the 7SK snRNP. Importantly, the most likely mechanism of action of SAHA as a therapy for cutaneous T cell lymphoma involves this transient release of P-TEFb, subsequent synthesis of HEXIM1 and increased assembly of 7SK snRNP.
A class III HDACi has also been implicated in the release of P-TEFb. SirT1 deacetylates CycT1 on K380, K386 and K390.63 Thus, combined actions of nicotinamide adenine dinucleotide (NAD) and Trichostatin A (TSA), which block SirT1, lead to the release of P-TEFb from 7SK snRNP. These residues are acetylated first by p300/CBP, which most likely represents another signaling intermediate in the regulation of P-TEFb. Importantly, as effects of HDACis have been thought to be directly on chromatin, these studies suggest that some of their anti-proliferative effects could well be due to the increased synthesis of HEXIM1 that follow transient releases of P-TEFb.
P-TEFb AND THE CONTROL OF TRANSCRIPTIONAL ELONGATION
The process impacted by 7SK snRNA regulates transcriptional elongation and co-transcriptional processing of nascent transcripts by RNAPII. P-TEFb is the kinase that phosphorylates subunits of N-TEF and serine 2 in the YSPTSPS repeats in the CTD of the large subunit of RNAPII to transition RNAPII from initiation to elongation of transcription.7 Thus, P-TEFb acts after the preinitiation complex (PIC) has been assembled on promoters, RNAPII has cleared the promoter, Cdk7 in TFIIH has phosphorylated S5 and S7 in the CTD and nascent mRNAs had been capped. At this point, N-TEF, which consists of the negative elongation factor (NELF) and DRB-sensitivity inducing factor (DSIF) is recruited to RNAPII, which stalls the transcription complex a short distance 3’ of transcription start sites (Table 1). Only short, capped but not polyadenylated transcripts are synthesized. At this point, P-TEFb must be recruited to the stalled RNAPII, which can occur via DNA-bound activators such as c-Myc, CIITA, MyoD, NFκB, steroid hormone receptors, the chromatin-interacting protein Brd4, or the RNA-bound activator Tat.7 In the case of HIV, although the 7SK snRNP can be recruited to the LTR64, it is the free, active Tat•P-TEFb complex that carries out these phosphorylation reactions.
There are several targets of P-TEFb phosphorylation. Most important for the transition into productive elongation is the phosphorylation of the Spt5 subunit of DSIF, which converts DSIF into a positive elongation factor.65 It also phosphorylates the RD, the NELF-E subunit of NELF, which dissociates NELF from double-stranded RNA.66 After P-TEFb function DSIF travels with the elongating RNAPII.10 P-TEFb also phosphorylates Ser2 in the CTD of RNAPII which facilitates the association of proteins that are involved in co-transcriptional splicing and polyadenylation of nascent transcripts.7 Recent evidence suggests that additional CTD-kinases participate in this process during transcriptional elongation. They include CycL•Cdk11, CycK•Cdk12 and CycK•Cdk13, which might be recruited to the elongating RNAPII via nascent RNA.67, 68 After polyadenylation, CTD phosphatases, which include Fcp1, dephosphorylate the CTD which allows formation of preinitiation complexes.69
Additional proteins that bind P-TEFb to affect elongation by RNAPII include AFF4, AF9, ENL and ELL2 which form what is referred to as the super elongation complex (SEC).70 These proteins had been demonstrated to affect transcriptional elongation in in vitro reconstituted systems.71, 72 Of interest, they are also fusion partners of the mixed lineage leukemia (MLL) protein from transposed chromosomes (histone H3 lysine 4–specific methyltransferase from Ch11 linked to various effector proteins), which bring P-TEFb to Hox and possibly other genes to cause leukemic transformation of white blood cells.73, 74 Thus, it is not surprising that HMBA and SAHA, which elevate levels of HEXIM, would counteract effects of these fusion proteins.75 Although in one study, Tat was also found in a different 7SK snRNP, which contained P-TEFb, but lacked HEXIM1,41, 76 its levels must be low compared to the dominant complex between Tat and P-TEFb.52
Besides the equilibrium between 7SK snRNP and active P-TEFb, levels of CycT1 are also vanishingly low in resting cells.77 Thus, circulating mononuclear cells contain abundant Cdk9 but little CycT1, despite abundant transcripts for both proteins. In part, this situation is due to miR27b and miR198 that inhibit the translation of CycT1 mRNAs in monocytes and T cells, respectively.78
Early on, P-TEFb was identified as the co-activator of Tat.12, 15, 79 Tat is a unique activator in that it binds the transactivation response (TAR) RNA rather than DNA. TAR measures 59 nt and is found at the very 5’ end of all viral transcripts. Following proviral integration, RNAPII is recruited to the HIV long terminal repeat (LTR), copies TAR and stalls.80, 81 As shown in Figure 3 cooperative interactions between Tat with the 5’ bulge and CycT1 with the central loop in TAR then bring P-TEFb to RNAPII.82, 83 Thus recruited, P-TEFb modifies transcription complexes for efficient elongation. P-TEFb can also be brought to the HIV LTR via NF-kB and Brd4 55, 84. Since this early example, the recruitment of P-TEFb by additional DNA-bound activators was found on many cellular genes.7, 8
P-TEFb, which is needed to activate genes, most certainly comes from the 7SK snRNP and when P-TEFb’s job is done it must be safely re-sequestered in the 7SK snRNP. Tat and Brd4 can extract P-TEFb from this inactive complex directly (Fig. 2 and 3).51, 52 Where does this release happen for HIV LTR and other cellular genes? Although the 7SK snRNP can associate with the PIC on the HIV LTR.64, its other biochemical properties indicate that it is not tightly associated with chromatin.58 Indeed, 7SK snRNP is the only snRNP that is extracted readily from detergent treated nuclei at a very low salt concentration. In another study using a large promoter array, P-TEFb, but not 7SK snRNA moved to activated promoters.85 The 7SK snRNP can be considered a storage form of P-TEFb that is maintained in relatively high concentration throughout the nucleoplasm. Gene specific extraction of P-TEFb near genes where it is needed must be tightly regulated. Importantly, release of P-TEFb by the transcription apparatus also must be accompanied by immediate re-sequestration by the 7SK snRNP. In support of this idea, 7SK snRNA has been demonstrated to localize to a promoter array only when genes were being shut down.85 Because most human genes have promoter proximally paused polymerases that are waiting for P-TEFb, the re-uptake of P-TEFb by the 7SK snRNP would help avoid accidental activation of many genes. Because release of P-TEFb is accompanied by loss of HEXIM proteins, the rebinding of P-TEFb must be preceded by the incorporation of HEXIM proteins (Fig. 2). Release of P-TEFb (and HEXIM) occurs spontaneously, but the rebinding of HEXIM is blocked by the change in the structure of 7SK snRNA.36 Therefore, it is likely that incorporation of HEXIM into the 7SK snRNP is an important regulatory step in the cycle of release and rebinding of P-TEFb (Fig. 2).
Conclusion
The regulation of P-TEFb by 7SK snRNA represents one of the most tractable systems for the study of eukaryotic transcription. Most components are known as is the impact of active P-TEFb on the transition from initiation to elongation of RNAPII on protein coding genes. Concurrent structural studies are also revealing how activators interact with P-TEFb and how HEXIM stabilizes the 7SK snRNP. However, much remains to be discovered. For example, what is/are the role/s of free HEXIM proteins and 7SK snRNP that is bound by hnRNPs in the cell? Is the interaction of HEXIM1 with TAR RNA52 physiologically relevant? How is P-TEFb inhibited in the 7SK snRNP, i.e. does a tyrosine in HEXIM occlude the ATP-binding pocket in Cdk9? How is the dissociation and reassembly of the 7SK snRNP regulated, especially the reassociation of HEXIM proteins, and what post-translational modifications in its subunits transmit the plethora of external and internal cues that regulate P-TEFb?
With the growing list of CTD kinases, which include Cdk7, Cdk8, Cdk9, Cdk11, Cdk12 and Cdk13, how is their division of labor orchestrated? Do they phosphorylate distinct residues or combinations of serines, threonines and tyrosines in the 52 heptapeptide repeats in the CTD so as to transmit precise instructions to RNAPII for specific co-transcriptional processes, as a kind of central processing unit of transcription? For that matter, how are they and their cyclin partners regulated in cells? For CycT1 and Cdk9, we know that miRNAs and HSP70/90 complexes play major roles. How is 7SK snRNA regulated, do its levels vary between resting and activated cells? How does its conformation affect its binding partners and is it regulated by different hnRNPs in cells?
Finally, we would like to know the role of the 7SK snRNP in health and disease. Intriguing clues come from worms and flies, where embryonic pluripotency is maintained by inhibiting P-TEFb globally.86, 87 What is the situation in higher eukaryotic systems? Next, how does 7SK snRNA help to regulate growth and differentiation from embryogenesis to the adult organism? How much does P-TEFb contribute to inflammation and malignant transformation? Given examples from leukemias and solid tumors, manipulations of P-TEFb are expected to slow down the progression of tumors and/or induce apoptosis of malignant cells. To this end, structure-based studies and high throughput screening for compounds that might block Cdk9 or induce the synthesis of HEXIM seem warranted. Importantly, interfering with the activity of P-TEFb directly or via 7SK snRNA promises to become an important pharmacological tool in the treatment of diseases that range from autoimmunity to cancer and AIDS. Although a structure of EIAV-Tat fused to equine cyclin T1 complexed with EIAV TAR had been determined,88 the structure of HIV Tat bound to P-TEFb89 is the only HIV/host cell protein complex solved to date. Structural studies may prove useful in resolving the details of LARP7 and MePCE interactions with 7SK and HEXIM1 interactions with 7SK and TAR.
ACKNOWLEDGEMENTS
We apologize to those whose work we were unable to cite due to lack of space. Research in Peterlin (GMO82250, AI049104) and Price (GM35500, AI074392) laboratories is funded by the NIH.
Contributor Information
B. Matija Peterlin, Email: matija.peterlin@ucsf.edu, Departments of Medicine, Microbiology and Immunology, Rosalind Russel Medical Research Center, University of California, San Francisco, CA 94143-0703, USA. Department of Virology, Haartman Institute, University of Helsinki, Haartmaninkatu 3, 00290 Helsinki, Finland.
John E. Brogie, Biochemistry Department, University of Iowa, Iowa City, IO 52242, USA
David H. Price, Email: david-price@uiowa.edu, Biochemistry Department, University of Iowa, Iowa City, IO 52242, USA.
References
- 1.Zieve G, Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976;8:19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 4.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 5.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 6.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 7.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 10.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. The Journal of biological chemistry. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 15.Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krystof V, Chamrad I, Jorda R, Kohoutek J. Pharmacological targeting of CDK9 in cardiac hypertrophy. Med Res Rev. 2010;30:646–666. doi: 10.1002/med.20172. [DOI] [PubMed] [Google Scholar]
- 17.Peterlin BM. Transcription elongation takes central stage: the P-TEFb connection. Cell Cycle. 2010;9:2933–2934. doi: 10.4161/cc.9.15.12698. [DOI] [PubMed] [Google Scholar]
- 18.Kleinert H, Bredow S, Benecke BJ. Expression of a human 7S K RNA gene in vivo requires a novel pol III upstream element. EMBO J. 1990;9:711–718. doi: 10.1002/j.1460-2075.1990.tb08164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd DC, Turner PC, Watkins NJ, Gerster T, Murphy S. Functional redundancy of promoter elements ensures efficient transcription of the human 7SK gene in vivo. J Mol Biol. 1995;253:677–690. doi: 10.1006/jmbi.1995.0582. [DOI] [PubMed] [Google Scholar]
- 20.Gruber AR, Koper-Emde D, Marz M, Tafer H, Bernhart S, Obernosterer G, Mosig A, Hofacker IL, Stadler PF, Benecke BJ. Invertebrate 7SK snRNAs. J Mol Evol. 2008;66:107–115. doi: 10.1007/s00239-007-9052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs) Biochim Biophys Acta. 2010;1799:365–378. doi: 10.1016/j.bbagrm.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha KM, Gu J, Chen Y, Reddy R. Adenylation of small RNAs in human cells. Development of a cell-free system for accurate adenylation on the 3'-end of human signal recognition particle RNA. J Biol Chem. 1998;273:6853–6859. doi: 10.1074/jbc.273.12.6853. [DOI] [PubMed] [Google Scholar]
- 23.Biewenga P, Buist MR, Moerland PD, Ver Loren van Themaat E, van Kampen AH, ten Kate FJ, Baas F. Gene expression in early stage cervical cancer. Gynecol Oncol. 2008;108:520–526. doi: 10.1016/j.ygyno.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 24.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 31.Dames SA, Schonichen A, Schulte A, Barboric M, Peterlin BM, Grzesiek S, Geyer M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci U S A. 2007;104:14312–14317. doi: 10.1073/pnas.0701848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo AA, Tong L, Lee JO, Jeffrey PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 33.Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol. 2006;26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol. 1991;11:3432–3445. doi: 10.1128/mcb.11.7.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5' end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol Biol Evol. 2009;26:2821–2830. doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belanger F, Baigude H, Rana TM. U30 of 7SK RNA forms a specific photo-cross-link with Hexim1 in the context of both a minimal RNA-binding site and a fully reconstituted 7SK/Hexim1/P-TEFb ribonucleoprotein complex. J Mol Biol. 2009;386:1094–1107. doi: 10.1016/j.jmb.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010;6:e1001152. doi: 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dulac C, Michels AA, Fraldi A, Bonnet F, Nguyen VT, Napolitano G, Lania L, Bensaude O. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J Biol Chem. 2005;280:30619–30629. doi: 10.1074/jbc.M502471200. [DOI] [PubMed] [Google Scholar]
- 43.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 44.Lebars I, Martinez-Zapien D, Durand A, Coutant J, Kieffer B, Dock-Bregeon AC. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 46.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 50.Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol Cell Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Matija Peterlin B, Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho WK, Zhou M, Jang MK, Huang K, Jeong SJ, Ozato K, Brady JN. Modulation of the Brd4/P-TEFb interaction by the human T-lymphotropic virus type 1 tax protein. J Virol. 2007;81:11179–11186. doi: 10.1128/JVI.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amente S, Gargano B, Napolitano G, Lania L, Majello B. Camptothecin releases P-TEFb from the inactive 7SK snRNP complex. Cell Cycle. 2009;8:1249–1255. doi: 10.4161/cc.8.8.8286. [DOI] [PubMed] [Google Scholar]
- 58.Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–1469. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, Peterlin BM. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284:6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18:291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 63.Cho S, Schroeder S, Kaehlcke K, Kwon HS, Pedal A, Herker E, Schnoelzer M, Ott M. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009;28:1407–1417. doi: 10.1038/emboj.2009.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17:815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sachs NA, Vaillancourt RR. Cyclin-dependent kinase 11(p110) activity in the absence of CK2. Biochim Biophys Acta. 2003;1624:98–108. doi: 10.1016/j.bbagen.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Majello B, Napolitano G. Control of RNA polymerase II activity by dedicated CTD kinases and phosphatases. Front Biosci. 2001;6:D1358–D1368. doi: 10.2741/majello. [DOI] [PubMed] [Google Scholar]
- 70.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes & development. 2011;25:661–672. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 72.Johnstone RW, Gerber M, Landewe T, Tollefson A, Wold WS, Shilatifard A. Functional analysis of the leukemia protein ELL: evidence for a role in the regulation of cell growth and survival. Mol Cell Biol. 2001;21:1672–1681. doi: 10.1128/MCB.21.5.1672-1681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000249. e1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santini V, Gozzini A, Ferrari G. Histone deacetylase inhibitors: molecular and biological activity as a premise to clinical application. Curr Drug Metab. 2007;8:383–393. doi: 10.2174/138920007780655397. [DOI] [PubMed] [Google Scholar]
- 76.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5:e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 80.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 81.Selby MJ, Bain ES, Luciw PA, Peterlin BM. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- 82.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujinaga K, Taube R, Wimmer J, Cujec TP, Peterlin BM. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci U S A. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 85.Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hubner MR, Spector DL. Nuclear Organization and Dynamics of 7SK RNA in Regulating Gene Expression. Mol Biol Cell. 2010;21:4184–4196. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F, Barboric M, Blackwell TK, Peterlin BM. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. 2003;17:748–758. doi: 10.1101/gad.1068203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ghosh D, Seydoux G. Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics. 2008;178:235–243. doi: 10.1534/genetics.107.083212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anand K, Schulte A, Vogel-Bachmayr K, Scheffzek K, Geyer M. Structural insights into the cyclin T1-Tat-TAR RNA transcription activation complex from EIAV. Nature structural & molecular biology. 2008;15:1287–1292. doi: 10.1038/nsmb.1513. [DOI] [PubMed] [Google Scholar]
- 89.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]