Abstract
Schistosoma genomes provide a comprehensive resource for identifying the molecular processes that shape parasite evolution and for discovering novel chemotherapeutic or immunoprophylactic targets. Here, we demonstrate how intra- and intergenus comparative genomics can be used to drive these investigations forward, illustrate the advantages and limitations of these approaches and review how post genomic technologies offer complementary strategies for genome characterisation. While sequencing and functional characterisation of other schistosome/platyhelminth genomes continues to expedite anthelmintic discovery, we contend that future priorities should equally focus on improving assembly quality, and chromosomal assignment, of existing schistosome/platyhelminth genomes.
Comparative genome basics
July 2009 marked a seminal date in the history of parasite genomics, helminthology and evolutionary biology. After more than 20 years of collaborative research, both Schistosoma japonicum[1] and Schistosoma mansoni[2] draft genomes were elucidated using whole-genome shotgun sequencing (Glossary). Along with Schistosoma haematobium, these parasitic trematodes are responsible for most cases of human hepatosplenic (S. japonicum and S. mansoni) and urinary (S. haematobium) schistosomiasis[3], a chronic and morbid neglected tropical disease afflicting hundreds of millions of people in sub-Saharan Africa, Asia and South America[4]. Information contained in these genomes has fuelled optimism that novel drug targets, vaccine candidates and immunomodulatory gene products will be found leading to the development of urgently needed control strategies [5, 6].
However, to comprehensively interrogate this vast amount of sequence information, comparative genome investigations are necessary. Comparative genomics can be defined on many levels, but for the purpose of this review, we will discuss how inter- and intragenus genome analyses are shaping schistosome post-genomic activities. Intergenus analyses involve cross-phylum genome comparisons in the primary search for gene loss/gene gain events. Both schistosome genome reports employed intergenus comparative genomics by contrasting the Schistosoma genomes with other genomes to discover features specific to, or shared between, parasitic and non-parasitic organisms [1, 2]. This approach is similar to that performed for other parasitic helminth genome projects (e.g. Brugia malayi [7] or Trichinella spiralis [8]) and has identified metabolic chokepoints, conserved druggable targets, expanded gene families and protein domain loss within the Schistosoma.
Intragenus comparative genome analyses involve the exploration of relatedness/differences between species (within a single genus), which in turn could provide physical markers of chromosomal evolution and highlight conserved regions (e.g. protein coding genes or gene families) of possible relevance as anti-schistosomal targets. For the apicomplexans[9] and trypanosomes[10], intragenus comparative genomics has already facilitated the discovery of novel biological observations (i.e. core genomes and lineage specific expansions) that may lead to innovative anti-protozoan treatments. To date, similar intragenus analyses have not been performed for schistosomes.
Here, for the first time, using the most recent Schistosoma genome assemblies (SJR2 for S. japonicum and SMA5.0 for S. mansoni), an intragenus comparative genomic analysis is performed. Utilising a proven strategy to compare phylogenetically related genomes[11], regions of chromosomal similarity and dissimilarity are identified between basal- (S. japonicum) and derived- (S. mansoni) schistosome species[12]. Complementing this karyotypic analysis is a review of how improvements in intergenus comparative and functional genomics can be applied to identify schistosome gene products of potential importance to parasite viability, development and host interrelationships. We suggest that the collective application of these comparative/functional genomics methodologies will lead to a better understanding of schistosome genome structure, gene function and evolution.
Intragenus Schistosoma comparative genomic analyses
For the 2009 S. mansoni genome assembly [2], 43% (distributed over 153 scaffolds) was unambiguously mapped onto schistosome chromosomes (7 autosomes and the Z/W sex-determination pair). Mapping was achieved by fluorescence in situ hybridisation (FISH) of bacterial artificial chromosomes (BACs) containing parasite genomic DNA fragments[13]. In 2011, with improvements in genome reassembly and additional FISH experiments, 81% of the S. mansoni genome (comprising 885 scaffolds, version 5.0) can now be explicitly mapped to chromosomes (Matt Berriman, unpublished) (Figure 1(a)). This robust physical map allowed us to align the S. japonicum whole genome assembly (Figure 1(b)) to S. mansoni chromosomes (Figure 1(c)) and subsequently generate a comparative genomic map (Figure 1d). Blocks of intragenus homologous synteny (at 100 kb and 10 kb minimum size resolutions) are illustrated with the corresponding S. japonicum protein coding genes contained within these regions described in Tables S1 and S2. At 100 kb resolution, the homologous synteny blocks (HSBs) cover approximately one quarter to one third of the Schistosoma genome (101.17 and 112.93 Mb of the S. japonicum and S. mansoni genomes, respectively) and most likely represent regions of chromosomes derived from a common ancestor[14] (Figure 2), which may be shared in all Schistosoma species. In mammalian genomes, HSBs are enriched for genes involved in developmental processes, neurogenesis, and cell-to-cell interaction[11]. Accordingly, in schistosomes, these HSBs (containing 4 443 genes at 10 kb resolution, Table S2) could encode products suitable for the identification of pan-species drug and vaccine targets (further discussed below). Our analysis shows that boundaries of adjacent HSBs found in the same S. japonicum scaffold tend to flank putative evolutionary breakpoint regions (EBRs) between S. mansoni and S. japonicum genomes. These EBRs can be derived either from large-scale interchromosomal (black arrows and numbers, Figure 2) or intrachromosomal (red arrows and numbers, Figure 2) rearrangements and are created from the non-allele joining of broken double stranded DNA ends during meiosis[15]. In mammalian genomes, EBRs contain gene dense-, transcriptionally active-, poorly methylated- and replication initiation-rich elements [16, 17]. These particular features are all associated with chromatin in a relaxed or open state and likely explain why evolutionary breakpoints occur here. As such, EBRs (and the genes contained within them) are subjected to greater genomic instabilities during normal cellular processes and are under increased evolutionary pressure to mutate[18].
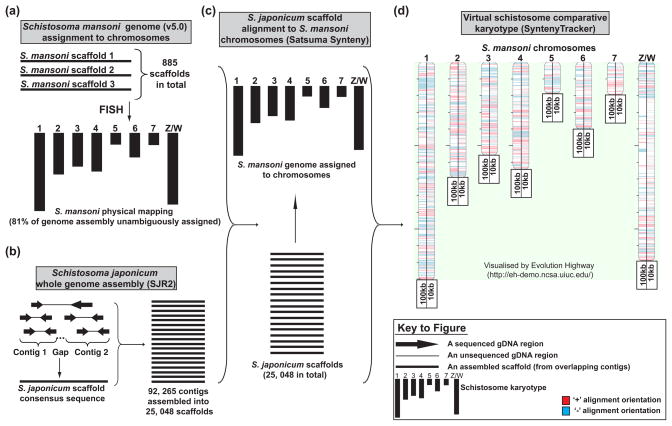
Figure 1. Intra-Schistosoma comparative genomic analysis: generation of a preliminary virtual karyotype.
An intragenus comparative genome analysis was initiated with sequence information contained in the originally published S. japonicum genome (http://www.chgc.sh.cn/japonicum/Resources.html, SJR2) as well as the recently reassembled S. mansoni genome (http://www.sanger.ac.uk/resources/downloads/helminths/schistosoma-mansoni.html, SMA5.0). (a) Version 5.0 of the S. mansoni genome was reassembled into 885 scaffolds, where 81% of them were unambiguously assigned to chromosomal positions (7 autosomes and Z/W sex-determining chimera) by FISH (Unpublished data, Matt Berriman).). (b) The original S. japonicum genome assembly was assembled into 25 048 scaffolds from 92 265 contigs[1]. (c) To facilitate intergenus genomic comparisons, S. japonicum scaffolds were aligned to S. mansoni chromosomes by the Satsuma Synteny program[91] followed by detection of >100 kb (full details found in Table S1) and >10 kb (full details found in Table S2) homologous synteny blocks (HSBs) defined by the SyntenyTracker program[92]. (d) Both 100 kb and 10 kb HSB sets are visualised on the virtual S. mansoni comparative chromosomal map (generated by Evolution Highway, http://eh-demo.ncsa.uiuc.edu/). We used the 100 kb HSB set for identifying interchromosomal evolutionary breakpoint regions (EBRs) between schistosome genomes (Figure 2) whereas the 10 kb HSB set was used to interrogate Gene Ontology (GO) enrichment in all putative EBRs found between S. mansoni and S. japonicum genomes (Figure 3).
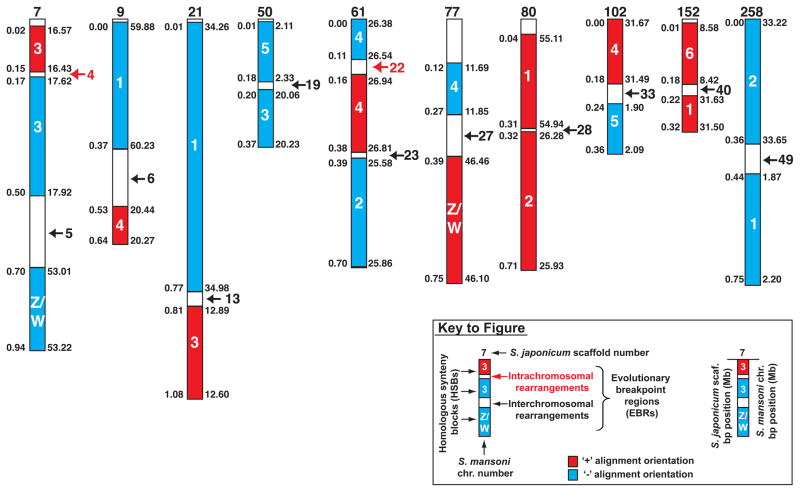
Figure 2. Schistosoma comparative genome analysis reveals interchromosomal evolutionary breakpoint regions (EBRs).
The whole genome alignment output from Satsuma Synteny[91] was translated into homologous synteny blocks (HSBs, >100 kb) using the SyntenyTracker program[92]. Combined, these HSBs (red blocks, ‘+’ alignment orientation; blue blocks, ‘−‘ alignment orientation) cover 112.93 Mb (31%) of the S. mansoni- and 101.17 Mb (25%) of the S. japonicum- genomes, respectively. Numbers inside the HSBs indicate S. mansoni chromosome designations (version 5.0) and numbers above HSBs correspond to S. japonicum scaffold IDs (SJR2). Numbers (indicated as Mb ranges) to the right side of the HSBs are S. mansoni chromosomal positions and the numbers to the left side of the HSBs are coordinates derived from S. japonicum scaffolds. Black arrows represent positions of all putative interchromosomal rearrangements between the S. mansoni and S. japonicum genomes with the EBR identifiers (numbered 5, 6, 13, 19, 23, 27, 28, 33, 40 and 49) included in Table S1. Red arrows indicate positions of two putative intrachromosomal rearrangements (out of 42 identified in this analysis) between the S. mansoni and S. japonicum genomes. Details of all intrachromosomal rearrangements can also be found in Table S1.
Whereas all schistosome chromosomes, except chromosome 7, contain HSBs interrupted by interchromosomal EBRs at 100 kb resolution, closer interrogation of these EBRs (Figure 2, Table S1 and Table S2) revealed that Schistosoma genomes evolve preferentially by intra- rather than interchromosomal rearrangements (42 intrachromosomal EBRs versus 10 interchromosomal EBRs). This is consistent with nematode genomes [7, 19] and illustrates a mechanism of chromosome evolution conserved between worm phyla. Furthermore, a ninefold increase in the total number of all genome rearrangements was found when the 10 kb set (477 EBRs comprising 410 intrachromosomal rearrangements and 67 interchromosomal rearrangements) was compared to the 100 kb set (52 EBRs). Numerical differences may indicate that: (i) a significant number of rearrangements missing in the 100 kb set are due to the scattered nature of the S. japonicum genome, (ii) errors in scaffold-to-chromosome assignments exist in one or both genomes and (iii) true small-scale synteny differences in S. mansoni and S. japonicum genomes are prevalent. We anticipate that resolution of these possibilities will be facilitated in the near future by advanced, intra-Schistosoma comparative genomics.
Though we have yet to fully detail the molecular features (e.g. DNA methylation status[20]) found within these Schistosoma EBRs, long terminal repeat (LTR) and non-LTR retrotransposable elements are found in a significantly greater proportion here (Table S3). As these elements drive evolutionary processes[21] and have been associated with EBRs of mammalian genomes[22], their enrichment in schistosome EBRs is not surprising. The extent and role of these (functional or recently active) LTR and non-LTR retrotransposons in shaping schistosome evolution has yet to be determined, but is an area of obvious interest. Further Gene Ontology (GO) analysis (Table S2) suggests that, like mammalian EBRs, these parasite genomic features are gene-rich and, therefore, likely composed of open chromatin. This conclusion is supported by the finding that ‘cell part’ and ‘catalytic activity’ GO categories, belonging to the ‘cellular component’ and ‘molecular function’ high level GO terms (populated by gene products involved in transcription and DNA replication/repair), are significantly enriched within the Schistosoma EBRs (Figure 3 and Table S2). It is tempting to speculate that lineage-specific S. mansoni or S. japonicum markers are also found within these EBRs and may be mined from within the 140 and 854 gene products identified at 100 kb (Table S1) and 10 kb resolution (Table S2), respectively.
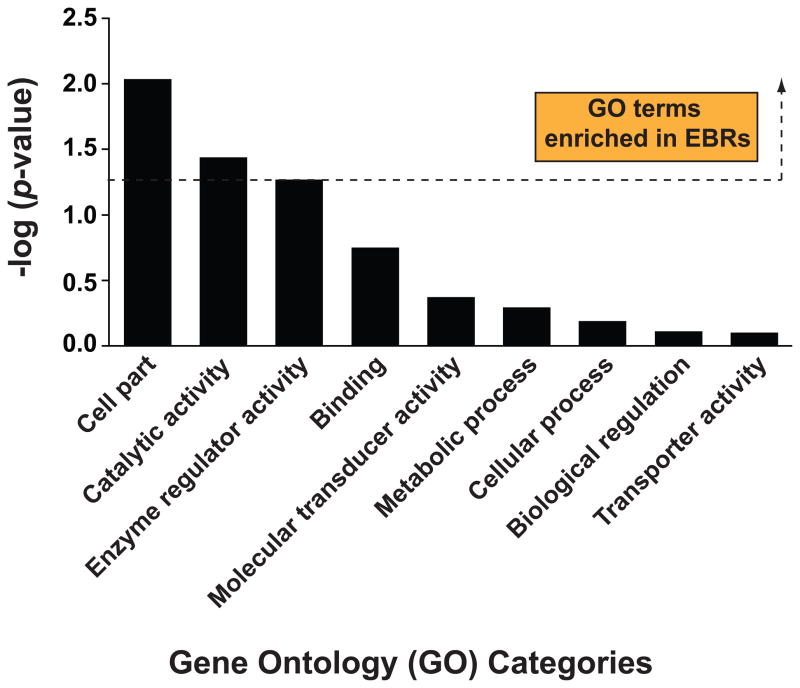
Figure 3. Schistosome EBRs and surrounding intervals (<10 kb) are enriched for specific gene ontology (GO) categories.
GO categories (across molecular function, biological process and cellular component terms) were assigned to S. japonicum genes using an in-house script that attempted to assign gene products to GO names and synonyms from version 1.2 (release date 15/03/2011 CVS revision number 1.1836) of the ontology (http://www.geneontology.org). The script was able to calculate a score for each GO ID by matching words in the gene product to words in the GO name or synonym. Gene products that scored above a minimum score (or confidence value) were allocated to their highest scoring GO ID. The script then followed the links through the GO hierarchy to identify the corresponding top-level GO categories for each allocated GO ID. These top-level GO categories were subsequently used in the analysis. In the case of a gene product for which no GO name or synonym scored above the confidence value, no GO category was assigned. S. japonicum scaffolds were divided into 10 kb windows and the number of genes from each GO category that had >100 assigned genes was counted in each 10 kb window. Next, the average number of genes for each GO category was calculated separately for the windows located within EBRs and the remainder of S. japonicum scaffolds. The average number of genes was calculated for 10 kb windows located within the EBRs and compared with the average number of genes in 10 kb windows found outside the EBRs using a t-test with unequal variances as described previously [93]. The significance threshold (p-value = 0.05) is indicated by a horizontal dashed line. All GO categories above this threshold are more likely to occur in EBRs when compared to the rest of the genome whereas GO categories below this threshold do not demonstrate any preferential localization in EBRs compared to other regions of the genome.
Though this analysis has detected a core set of HSBs containing ~25% of the ancestral Schistosoma genome, we expect that the number of EBRs in the two genomes is underestimated because a comparable physical S. japonicum genome map is not yet available. Progress in intra-Schistosoma comparative genomics will depend on improving genome assemblies mapped to chromosomes (e.g. S. japonicum) and de novo sequencing and draft assemblies of other schistosome genomes (e.g. S. haematobium). Indeed, recent intragenus comparative genome analysis of Caenorhabditis elegans and Caenorhabditis briggsae have improved defective gene models, detected potential new genes and identified missing orthologous relationships within this free-living nematode genus[23]. Further iterations of the present analysis will help identify molecular drivers of schistosome evolution (e.g. functional retrotransposable elements within EBRs) and highlight conserved regions (e.g. protein encoding genes or gene families within HSBs) of interest as pan-specific, anti-schistosomal targets. Comparative genomics between the Platyhelminthes will also be possible in the near future due to the completion or ongoing sequencing of Schmidtea mediterranea[24], Taenia solium[25], Echinococcus sp. and Hymenolepsis microstoma genomes (sequencing/draft assembly in progress for the last two helminths at the Wellcome Trust Sanger Institute (WTSI)). These efforts will help us to understand the origin and nature of parasitism within the phylum as well as highlight the chromosomal events associated with the evolution and divergence of parasitic trematodes and cestodes from free-living turbellarians.
Intergenus comparative genomics to predict Schistosoma drug targets
A major goal consequent on the arrival[1, 2] and continuing refinement[26] of annotated schistosome genomes is the identification of new drug targets. In this regard, intergenus comparative genomics implements a series of user-defined criteria to derive short lists of prioritized and actionable gene targets [27–30]. Short-listed, high-quality targets are derived from sequence comparisons between schistosome(s) and comparator(s) genomes in order to identify genes for which experimental data indicate that their products are essential to parasite survival or viability. Actionable entails having the experimental tools at hand to interrogate short-listed targets. So far, target validation in schistosomes has predominantly used transient RNA interference (detailed below) or more recently, employing vector-based small hairpin RNAs (shRNA) [31, 32]. The alternative or complementary approach of chemical validation relies on small molecules most often developed against a distinct (usually human) target with the implication, and sometimes formal demonstration [33, 34], that the anti-schistosomal effect is associated with binding to the intended schistosome ortholog(s). Based on the presented intra-Schistosoma comparative genomic analysis, an additional criterion for short-list inclusion and target validation would be chromosomal position. For example, 252 eukaryotic protein kinases (ePKs) have recently been identified in S. mansoni and many of these may be potential drug targets[35]. Our analysis (Figures 1 and 2) indicates that some ePKs (e.g. Sjp_0015710.1, Sjp_0019720.1, Sjp_0027360.1, Sjp_0056860.1) are physically positioned within intra-schistosomal EBRs (Table S2). As EBRs are actively evolving, the selection of pan-Schistosoma chemotherapeutic targets physically positioned within these regions should proceed cautiously.
Online tools are under development to utilise comparative genomics for short-listing possible drug targets. For example, the TDR Targets Database (http://tdrtargets.org/) provides a variety of ‘tunable’ and Boolean-capable filters to generate user-defined lists of potential targets for 11 different pathogens, including S. mansoni and several other helminths[29, 30]. In addition, SchistoDB (http://schistodb.net/)[36] will expand considerably in the next year with improved query functionality, graphical user interfaces and other features from the EuPathDB family of genome databases[37] (personal communications; Oliveira, Wei, Kissinger and Roos). Utilising these resources, intergenus comparative genomics can be used to identify pathogen-specific proteins or those that are sufficiently different so as to be functionally absent from the host[38]. This is in an attempt to limit potential drug toxicity arising from chemical cross-reactivity between the orthologous parasite and host targets. Yet, the alternative (i.e. to explore orthologous genes as a source of potential drug targets) is also useful. Here, genes that are shared among species are likely to be essential and thus more attractive as targets due to the severity of phenotypes produced upon perturbation [38, 39]. Sufficient physiological, parasitological and/or protein structural circumstances often exist that can offset possible host toxicity[28] and, therefore, this concept has been actively exploited in recent comparative genomic studies involving S. mansoni[28, 29]. This approach might also focus on those genes that are expressed in both juvenile and adult worms[40–42] considering that the current schistosomicide, praziquantel, is less effective against immature parasites[43].
Though comparative genomics offers a straightforward strategy to pre-validate potential drug targets, there are limitations, both inherent and contextual. For the latter, perhaps most obvious is the dearth of comprehensive genome, transcriptome and proteome information for S. haematobium which is more prevalent than S. mansoni in Africa and is often co-endemic[44]. With the current operational requirement in sub-Saharan Africa for a single anti-schistosomal drug, this lack of annotated sequence information represents a critical knowledge gap only partially filled by the 0.5 to 1.0× coverage genome sequence that is currently available (http://www.cebio.org/projetos/schistosoma-haematobium-genome). Even if highly processed/assembled S. haematobium genomic data were at hand, it might be difficult to predict how even subtle sequence differences between S. mansoni and S. haematobium targets would impact the successful development of a single drug. Also, as comparative genomics incorporates experimental loss-of-function data from model organisms, the approach may miss potential targets modulated through gain-of-function, as occurs with many of today’s anthelmintics[28]. Looking much further forward, comparative genomics and the underlying (but increasingly challenged) drug discovery philosophy of ‘one gene, one target, one disease’[45] may over-simplify the process of identifying new drug targets. Rather, a better understanding of the dynamics of how biological pathways and networks are perturbed by small molecules (polypharmacology) and/or gene disruption would provide the sophistication necessary to predict new therapeutic targets [45–47], especially with a view to limiting the establishment of drug resistance. Finally, based on the findings presented here, selecting compounds that target loci contained within HSBs (Figure 2, Table S1 and Table S2) presents new opportunities to identify therapeutic small molecules.
Comparative schistosome vaccinomics/immunomics: identifying immunoprophylactic targets within the tegument
Apart from potential drug targets, the schistosome genome[1, 2], various transcriptomes[48, 49] and proteomes of the tegument and excretory/secretory (ES) products (reviewed in [50]) have also provided researchers with a ‘molecular haystack’ in which to find a handful of ‘immunoprophylactic needles’. A major challenge is to efficiently and comparatively mine this information. DNA microarrays have partially addressed this issue with several recent reports[40, 41] identifying gene products differentially expressed throughout the parasite’s lifecycle, including the tissue migrating schistosomulum - a major target of protective immune responses[51]. As many of these transcripts (e.g. tetraspanins, cathepsins, serpins, and tegumental-associated antigens) are encoded in both S. mansoni and S. japonicum genomes (i.e. found within HSBs, Table S2) and are also expressed in all definitive host lifecycle stages, they represent key vaccine candidates.
Given the acoelomate body plan of the parasite, schistosome tissues and organs cannot easily be manually dissected in order to probe for orthologous gene products (identified by comparative genomics) of possible relevance as vaccine antigens. To overcome this challenge, laser microdissection microscopy (LMM) has helped to elucidate gene expression in specific tissues of both S. mansoni and S. japonicum. Thus far, the gastrodermis and reproductive tissues have been scrutinized using a combination of LMM, RNA extraction and DNA microarray analysis [52, 53]. This novel comparative transcriptomics approach aided the assembly of a schistosome gene atlas for the organs under study[53]. Though this methodology offers an unprecedented opportunity to identify tissue-enriched transcriptional profiles (between species), it has its limitations. For example, the schistosome tegument, a tissue often targeted by vaccinologists[54] (being situated at the host-parasite interface[55]) does not readily lend itself to LMM. This is because the tegumental cell bodies (and their nuclei) are buried deeply within the musculature.
Alternative approaches to ‘dissect’ the tegument have utilised comparative proteomics to discover and characterise proteins of interest at this host-parasite interface [50, 56–60]. This approach has revealed that several tegumental proteins are homologous between schistosome species [56, 59, 60] with some being localised to the outer, host-interacting, surface membrane [60, 61]. It is these surface-exposed proteins that are attractive vaccine targets (Figure 4). For example, many tetraspanin genes (contained within the S. japonicum and S. mansoni HSBs, Figure 1 and Table S2) are highly expressed in the intra-mammalian schistosome stages[40, 41], and some of these are located in tegumental membranes[60, 61]. The tetraspanins have been experimentally proven to be protective in small and large animal models of schistosomiasis [62, 63] and Sm-TSP-2 is also selectively recognized by naturally resistant humans [54, 62]. Thus, (comparative) proteomics has been a successful platform in the identification and short-listing of candidate protein vaccines.
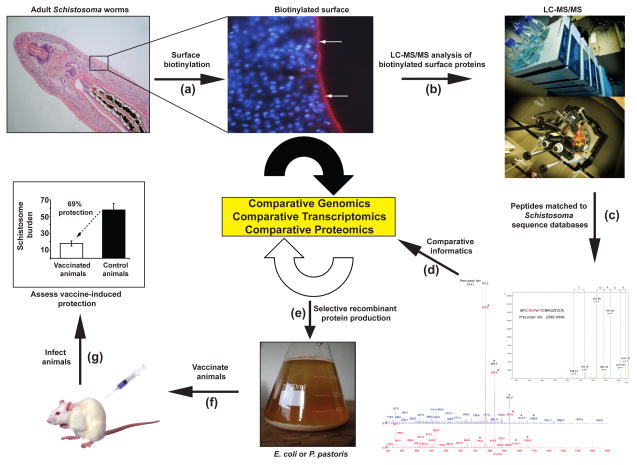
Figure 4. Comparative informatics approaches for identifying vaccine candidates from the schistosome tegument.
Due to advances in ‘omics’ technologies, next-generation schistosome vaccine discovery has been revitalised. Outlined is an example of how complementary strategies are currently being used to identify next-generation, anti-schistosomal vaccine targets. (a) To identify tegument proteins exposed at the host/parasite interface, schistosome worms (adults represented here) are labelled with biotin [60] (tegument visualised here by incubating adult worms with Cy3-labelled anti-biotin Abs (red sinuous band indicated by white arrows) and co-stained with DAPI to distinguish sub-tegumental nuclei (blue)). (b) Subsequent processing of these biotin-labelled tegumental preparations generates a collection of putative, surface-exposed proteins that can be analysed by LC-MS/MS. (c) Peptide spectra generated from MS/MS analysis are matched to gene/cDNA sequences from Schistosoma databases, allowing for the identification of parasite proteins. (d) Protein sequences are filtered through a cyclic round of informatics involving comparative genomics (protein sequences found only in the Platyhelminthes or also present in other genomes), comparative transcriptomics (intra-Schistosoma gene expression similarities throughout the parasite lifecycle [40, 41]) and comparative proteomics (proteins found in tegumental preparations across the Schistosoma[59–61]). (e) Selected recombinant proteins that display desirable features (i.e. expressed in schistosomula as well as adults and are surface exposed in the Schistosoma teguments) are produced in a heterologous expression system (e.g. Escherichia coli or Pichia pastoris), purified and used to (f) vaccinate laboratory animals to (g) assess protective efficacy.
Integration of ‘omics’ technologies has accelerated the development of new post-genomic tools to aid vaccine discovery. For example, high throughput protein expression techniques (e.g. in vitro transcription/translation systems), coupled to high-throughput immunoscreening with sera from resistant humans and animals, have thrust schistosomiasis research into the immunomics era [64]. Recently, the first Schistosoma immunomics protein microarray was described as a vaccine discovery tool[65]. Antigens included on this microarray were chosen from those identified in previous proteomic investigations in addition to those sequences selected from comparative in silico screening of both S. mansoni and S. japonicum genomes and transcriptomes. The goal here was to identify target antigens based on sub-cellular location with a particular emphasis on those proteins expressed in the tegument. Proteins were expressed in a cell-free in vitro transcription/translation system and contact-printed onto nitrocellulose-coated slides to form protein microarrays. The arrays are currently being probed with IgG and IgE antibody subclasses from resistant and chronically infected human and animal populations (S. Gaze and A. Loukas, unpublished). This approach is revealing antigens that are targets of both protective IgG responses and potentially harmful IgE responses [66]. This innovative technology of immunomics or, essentially, reverse vaccinology that relies on the outputs of comparative genomics, transcriptomics, and proteomics analyses has the potential to transform vaccine research for schistosomiasis and other parasitic diseases.
Functionally exploring comparative genomic outputs: revisiting the tegument
Identifying the precise role(s) of short-listed chemotherapeutic or immunoprophylactic targets is often problematic, especially for those genes without orthology outside of the genus or that encode proteins of unknown function [1, 2]. For schistosomes, RNAi has emerged as a useful technique to experimentally manipulate the expression of specific schistosome genes and, possibly, gain insight into gene function. Newer protocols employing shRNAs may further extend its utility [31, 32]. Comparative schistosome genomics has revealed that genes encoding known RNAi pathway effectors are shared between S. mansoni and S. japonicum [67, 68]. Furthermore, targeted gene suppression (mediated by RNAi) has been demonstrated in both species [69, 70]. Though RNAi can be both rapid and long -lasting [71, 72] as well as functional in several schistosome life cycle stages [71, 73], it is not always definitive. For reasons unknown, some genes appear recalcitrant to robust suppression [72, 74], and striking phenotypes are seldom seen [72, 73]. In other cases, RNAi has helped identify drug targets and gain insight into parasite biochemistry, physiology and host-parasite interactions [34, 70, 75, 76].
Perhaps due to the voluminous genome data, schistosome functional genomics has most often focused on a conserved biochemical pathway, tissue type or employed a gene-by-gene approach. As an example, we review how RNAi has contributed to the functional characterisation of conserved (within the Schistosoma) proteins in one such tissue, the aforementioned tegument. Part of its functioning at the host-parasite interface is to import nutrients from the host bloodstream. Within the S. mansoni tegument, two facilitated diffusion, glucose-importing proteins have been identified; schistosome glucose transporter (SGTP) 1 and 4 [77]. SGTP4 is present in the host interactive, apical tegumental membranes, whereas SGTP1 is found in the tegumental basal membrane and other tissues. Using schistosomula and adult worms, RNAi of SGTP1 or SGTP4 impaired the parasite’s ability to import glucose, and this effect was compounded by suppression of both transporter genes simultaneously [78]. Though RNAi-treated parasites cultured in rich medium were not phenotypically different from controls as assessed microscopically, their survival in vivo was significantly impaired [78]. Thus, RNAi has provided direct evidence for the importance of SGTP1 and SGTP4 in glucose transport and survival in the mammalian host.
Proteomic analysis of schistosome tegumental membranes also revealed the presence of an aquaporin homolog (SmAQP)[56]. Aquaporins (found within HSBs, Figure 1 and Table S2) are membrane proteins that form pores to selectively conduct water molecules into and out of cells. RNAi-mediated SmAQP suppression impaired the ability of schistosomula to osmoregulate and revealed the previously unrecognized role of the tegument in controlling water movement into, and out of, the parasites [79]. Unexpectedly, SmAQP-suppressed adult parasites in vitro failed to rapidly acidify their culture medium and were found to excrete less lactate compared to controls [80]. Heterologous expression of SmAQP in Xenopus oocytes demonstrated that this protein, in addition to transporting water, could also transport lactate [80]. These findings provide a molecular understanding of how schistosomes cope with the significant quantities of lactate created from the largely anaerobic glucose catabolism that is a hallmark of their intravascular lives[81]. Collectively, the comparative- and functional- genomic data suggest that the syntenic position of aquaporins is an important feature of schistosome/host relationships driven by the need to extend tegumental functions in metabolic waste excretion.
The contribution of the tegument to immune evasion by the worms is less well understood, although many investigations have provided functional explanations in this regard[55]. The presence of nucleotide-metabolizing enzymes associated with the parasite surface[56, 57] has led to the suggestion that these can catabolize host pro-inflammatory metabolites such as ATP and generate anti-inflammatory mediators such as adenosine [82]. Accordingly, schistosomes may prevent their hosts from focusing immunological mediators in their vicinity [82]. In support of this notion, RNAi of adult tegumental alkaline phosphatase (SmAP) prevented production of the anti-inflammatory adenosine from an exogenously added precursor (AMP) [83].
These examples illustrate the power of RNAi to provide insight into schistosome gene function. Combined with schistosome transcriptomics and proteomics, RNAi use can be extended to systematically identify the function of many comparatively-identified genome products in parasite biology, development and host interactions.
Concluding remarks and future perspectives
As outlined, comparative genomics offers an established set of principles for understanding gene and genome biology as well as exploiting that information to identify targets for chemo- and immunotherapy. The intra- and intergenus comparative genomic analyses discussed here are an encouraging first step, but will be improved (Box 1) once other schistosome genomes are sequenced (e.g. S. haematobium) and existing schistosome genomes are refined, reassembled and mapped (e.g. S. japonicum). When additional platyhelminths are included, a robust framework for comparative flatworm genomics will be realised. This information will lead to (i) detection of genes affected by the natural selection processes operating on each platyhelminth species; (ii) identification of the species- and class- specific gene network changes; and (iii) characterisation of gene birth/death rates due to lineage-specific genome rearrangements, duplications and mobile genetic element activity. This greater knowledge of genome biology (experimentally verified by post-genomic technologies) will aid the discovery of novel schistosomiasis control strategies and allow for a better understanding of lifestyle diversity and evolution within the Platyhelminthes.
Box 1. Outstanding questions: the future of schistosome comparative genomics.
How important is genome reassembly?
In order to maximise the power of comparative genomics, Schistosoma genomes need to be continually reassembled into the smallest set of overlapping scaffolds as possible. This allows for large stretches of multiple genomes to be systematically compared. While the S. mansoni genome has undergone several rounds of reassembly (genome version 5.0 has 885 scaffolds; http://www.sanger.ac.uk/resources/downloads/helminths/schistosoma-mansoni.html), there has been very little progress in reassembling the S. japonicum genome since publication (SJR2 has 25 048 scaffolds; http://www.chgc.sh.cn/japonicum/Resources.html). Therefore, our preliminary intragenus comparative genome analysis is limited by the fragmented nature of the S. japonicum genome. Future analyses will be improved upon refined and reassembled Schistosoma genomes facilitated by application of next generation sequencing technologies and sophisticated informatics. Assigning the reassembled S. japonicum genome to chromosomes (similar to that performed for S. mansoni [13]) would also aid future intragenus analyses.
Is the S. haematobium genome important for future comparative analyses?
The preliminarySchistosoma comparative genomic analysis provided in this review would significantly benefit from the elucidation of the S. haematobium genome. A variety of evolutionary and control questions could be more readily addressed upon the inclusion of this most derived (amongst S. japonicum, S. mansoni and S. haematobium) schistosome species. Efforts are currently underway to sequence the genome of this species (e.g. http://www.cebio.org/projetos/schistosoma-haematobium-genome). Also, comprehensive genome information for S. haematobium would be essential given the operational simplicity demanded for a single drug or vaccine to control both S. mansoni and S. haematobium in sub-Saharan Africa.
How important are additional platyhelminth genomes?
Comparative genomics of only two species in one class within the phylum (illustrated in this review) is obviously providing a very limited snapshot of platyhelminth biology. Therefore, the elucidation of new flatworm genomes is necessary to understand the evolution of this important group of animals. Genomes of Echinoccocus granulosus (http://www.sanger.ac.uk/resources/downloads/helminths/echinococcus-granulosus.html), E. multilocularis (http://www.sanger.ac.uk/resources/downloads/helminths/echinococcus-multilocularis.html), Hymenolepsis microstoma (http://www.sanger.ac.uk/resources/downloads/helminths/hymenolepis-microstoma.html), Schmidtea mediterranea (http://smedgd.neuro.utah.edu/) and Taenia solium are all at different states of assembly, but upon completion, will advance our ability to perform comparative flatworm genomics. Information contained within new platyhelminth transcriptomes [84–87] will also complement certain aspects of comparative genome investigations (i.e. identification of conserved drug or vaccine targets).
How can post-genomic technologies effectively complement platyhelminth comparative genomics investigations?
A large variety of post-genomics technologies (e.g. [88, 89]) have been developed over the last decade to study platyhelminth biology. However, the optimism that these technologies would quickly deliver new platyhelminth drugs or vaccines has yet to be fully realised. Perhaps, in light of the enormous energy put forth in sequencing new platyhelminth genomes, equal emphasis should be dedicated towards the meta-analysis of existing post-genomics datasets to identify those conserved targets suitable for functional validation. Further refinement of post-genomic technologies applied to conserved targets identified by meta-analysis of available datasets and new targets identified by platyhelminth comparative genomics investigations will transform the state of anthelmintic discovery. Indeed, it is this type of approach that is beginning to make an impact on Plasmodium biology and the identification of novel anti-malarials[90].
Supplementary Material
Acknowledgments
We thank the many individuals within and outside the global worm community who have supported and encouraged us throughout the years. These interactions have stimulated our inspection of the topics addressed in this review. We also acknowledge the Wellcome Trust, the U.S. National Institutes of Health (AI-056273), the Australian National Health and Medical Research Council and the Sandler Foundation for financially supporting our research activities.
Glossary
- Acoelomate
Schistosomes lack a coelom (an internal fluid filled body cavity) and are comprised of a solid, triploblastic (ectoderm, mesoderm and endoderm), bilaterally symmetrical, body plan. Due to the acoelomate (lacking a coelom) nature, schistosome organs develop in direct contact with these triploblastic tissues and not in fluid filled cavities
- BAC (bacterial artificial chromosome)
A DNA construct used in whole-genome, shotgun sequencing projects that can be propagated through Escherichia coli. Genomic DNA insert sizes of 150–350 kb are typically found within BACs. BAC DNA constructs (found in Sm1 and CHORI 103 BAC libraries) were used to map genome scaffolds to S. mansoni chromosomes via FISH
- Comparative genomics
An informatics based technique used to compare the DNA sequences that comprise draft or finished genomes of organisms within the same phylum or across different phyla. By identifying similarities (homologous synteny blocks, HSBs) and differences (evolutionary break point regions, EBRs) across genomes, schistosome comparative genomics attempts to: (i) provide insight into the processes that shape species evolution, (ii) identify essential genes suitable for chemotherapy development and (iii) predict potential immunoprophylactic gene products. For this review, intragenus (comparing draft S. mansoni and S. japonicum genomes) and intergenus (contrasting Schistosoma genomes to comparator genomes) comparative genomics analyses are discussed
- Contig
A contiguous sequence of DNA assembled from overlapping cloned DNA fragments. The S. mansoni genome was assembled from 50 376 contigs and the S. japonicum genome was assembled from 95 265 contigs
- Evolutionary breakpoint regions (EBRs)
A non-aligning, genomic interval found between two adjacent HSB boundaries. These can comprise interchromosomal or intrachromosomal rearrangements. At 100 kb resolution, 52 EBRs were found whereas at 10 kb resolution, 477 EBRs were uncovered
- FISH (Fluorescence in situ hybridisation)
An experimental technique by which genome scaffolds are physically mapped to chromosomes and visualised by fluorescence microscopy. Upon publication, 43% of the S. mansoni genome assembly was unambiguously assigned to the 7 autosomes and the ZW sex-determining pairs. A total of 81% of the S. mansoni genome assembly (version 5.0) has now been physically mapped to the karyotype
- Gap
A portion in the scaffold that is linked to two sequence verified contigs but has not been sequenced
- Gene Ontology (GO)
A standardised vocabulary of gene product attributes that is species neutral and applicable to both prokaryotes and eukaryotes. See http://www.geneontology.org/ for more information
- Homologous synteny blocks (HSBs)
A minimum of two adjacent markers in the same chromosome/scaffold between S. mansoni and S. japonicum genomes that share the same order in both species without interruption by an HSB from a different region of the same chromosome/scaffold or from a different chromosome/scaffold. In this preliminary intra-Schistosoma comparative genome analysis, HSBs of 100 kb and 10 kb were considered
- Interchromosomal rearrangements
An intraspecies genomic rearrangement that takes place between different Schistosoma chromosomes. At 100 kb resolution, 10 interchromosomal rearrangements were found whereas at 10 kb resolution, 67 EBRs were uncovered
- Intrachromosomal rearrangements
An intraspecies genomic rearrangement that occurs within the same Schistosoma chromosome. At 100 kb resolution, 42 intrachromosomal rearrangements were found whereas at 10 kb resolution, 410 intrachromosomal rearrangements were uncovered
- Resolution
The length threshold (in bps) that defines a HSB. For 100 kb resolution, all HSBs shorter than 100 kb are excluded from further analyses. Likewise, for 10 kb resolution, all HSBs shorter than 10 kb are excluded from further analyses
- Scaffold
A reconstructed portion of the genome made by assembling overlapping contigs and gaps. The 50 376 S. mansoni contigs were assembled into 5 745 scaffolds and the 95 265 S. japonicum contigs were assembled into 25 048 scaffolds. These scaffolds comprised the draft genome sequences of each schistosome species
- Schistosomulum
A schistosome lifecycle stage that develops immediately after cercarial penetration of the definitive vertebrate host. Characteristic features of this developmental form include a tegumental heptalaminate (seven membranes comprised of two opposing trilaminte lipid bilayers) covering and an ability to initiate haematophagy (blood feeding). The schistosomulum is believed to be a major target of protective host immune responses[51]
- Tegument
A protective syncytium layer sandwiched between the hepatolaminate surface covering and the acoelomate schistosome body plan. Nucleated cell bodies situated below the tegument produce the diverse biomolecules and vesicles that are transported throughout the syncytium. Due to the positional (host-parasite interface) and protective nature of this structure[55], proteins shuttled from sub-tegumental cell bodies to the hepatolaminate membrane surface are of interest for vaccinologists
- Whole genome shotgun sequencing
A genome sequencing strategy, by which fragmented genomic DNA (gDNA) is randomly sequenced, assembled into overlapping contiguous sequences (contigs) and built into large genome scaffolds. Scaffolds are subsequently assembled into whole draft genomes of the studied organism. Whole genome shotgun sequencing of the S. mansoni genome (363 Mb at 6X coverage) was initiated with mixed-sex cercariae gDNA whereas the S. japonicum genome (397 Mb at 6X coverage) was elucidated by gDNA isolated from mixed-sex adult schistosomes
- Z/W sex-determination pair
The diploid schistosome genome is maintained across seven autosomal- and one Z/W sex-determining- chromosomal pairs. The gender of individual schistosomes is dependent upon inheritance of these Z/W sex-defining chromosomes. A female schistosome will, therefore, contain a ZW combination of sex-determining chromosomes whereas a male schistosome will contain a ZZ combination. Due to challenges with physical mapping of the S. mansoni assembly (version 5.0) to individual Z or W chromosomes, a concatenated Z/W description is indicated (in this review as well as in the S. mansoni genome description[2]). Further studies undoubtedly will facilitate physical mapping of the most recent genome assembly to specific Z and W chromosomal regions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schistosoma japonicum sequencing consortium. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gryseels B, et al. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 4.King CH. Parasites and poverty: the case of schistosomiasis. Acta Trop. 2010;113:95–104. doi: 10.1016/j.actatropica.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han ZG, et al. Schistosoma genomics: new perspectives on schistosome biology and host-parasite interaction. Annu Rev Genomics Hum Genet. 2009;10:211–240. doi: 10.1146/annurev-genom-082908-150036. [DOI] [PubMed] [Google Scholar]
- 6.Webster JP, et al. Schistosome genomes: a wealth of information. Trends Parasitol. 2010;26:103–106. doi: 10.1016/j.pt.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Ghedin E, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitreva M, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H, et al. Core genome components and lineage specific expansions in malaria parasites Plasmodium. BMC Genomics. 2010;11(Suppl 3):S13. doi: 10.1186/1471-2164-11-S3-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Sayed NM, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 11.Larkin DM, et al. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19:770–777. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Attwood SW, et al. A DNA sequence-based study of the Schistosoma indicum (Trematoda: Digenea) group: population phylogeny, taxonomy and historical biogeography. Parasitology. 2007;134:2009–2020. doi: 10.1017/S0031182007003411. [DOI] [PubMed] [Google Scholar]
- 13.Criscione CD, et al. Genomic linkage map of the human blood fluke Schistosoma mansoni. Genome Biol. 2009;10:R71. doi: 10.1186/gb-2009-10-6-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardison RC. Comparative genomics. PLoS Biol. 2003;1:E58. doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sankoff D. The where and wherefore of evolutionary breakpoints. J Biol. 2009;8:66. doi: 10.1186/jbiol162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J, et al. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006;16:1557–1565. doi: 10.1101/gr.5383506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaitre C, et al. Analysis of fine-scale mammalian evolutionary breakpoints provides new insight into their relation to genome organisation. BMC Genomics. 2009;10:335. doi: 10.1186/1471-2164-10-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longhese MP, et al. The cellular response to chromosome breakage. Mol Microbiol. 2006;60:1099–1108. doi: 10.1111/j.1365-2958.2006.05186.x. [DOI] [PubMed] [Google Scholar]
- 19.Stein LD, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geyer KK, et al. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat Commun. 2011;2:424. doi: 10.1038/ncomms1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohne A, et al. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- 22.Elsik CG, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergara IA, Chen N. Large synteny blocks revealed between Caenorhabditis elegans and Caenorhabditis briggsae genomes using OrthoCluster. BMC Genomics. 2010;11:516. doi: 10.1186/1471-2164-11-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robb SM, et al. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 2008;36:D599–606. doi: 10.1093/nar/gkm684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguilar-Diaz H, et al. The genome project of Taenia solium. Parasitol Int. 2006;55(Suppl):S127–130. doi: 10.1016/j.parint.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Tsai IJ, et al. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frearson JA, et al. Target assessment for antiparasitic drug discovery. Trends Parasitol. 2007;23:589–595. doi: 10.1016/j.pt.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caffrey CR, et al. A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosoma mansoni. PLoS ONE. 2009;4:e4413. doi: 10.1371/journal.pone.0004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowther GJ, et al. Identification of attractive drug targets in neglected-disease pathogens using an in silico approach. PLoS Negl Trop Dis. 2010;4:e804. doi: 10.1371/journal.pntd.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguero F, et al. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat Rev Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao ZR, et al. Schistosoma japonicum: inhibition of Mago nashi gene expression by shRNA-mediated RNA interference. Exp Parasitol. 2008;119:379–384. doi: 10.1016/j.exppara.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Ayuk MA, et al. Schistosoma mansoni U6 gene promoter-driven short hairpin RNA induces RNA interference in human fibrosarcoma cells and schistosomules. Int J Parasitol. 2011;41:783–789. doi: 10.1016/j.ijpara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdulla MH, et al. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 2007;4:e14. doi: 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuntz AN, et al. Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target. PLoS Med. 2007;4:e206. doi: 10.1371/journal.pmed.0040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade LF, et al. Eukaryotic protein kinases (ePKs) of the helminth parasite Schistosoma mansoni. BMC Genomics. 2011;12:215. doi: 10.1186/1471-2164-12-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerlotini A, et al. SchistoDB: a Schistosoma mansoni genome resource. Nucleic Acids Res. 2009;37:D579–582. doi: 10.1093/nar/gkn681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aurrecoechea C, et al. EuPathDB: a portal to eukaryotic pathogen databases. Nucleic Acids Res. 2010;38:D415–419. doi: 10.1093/nar/gkp941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarter JP. Genomic filtering: an approach to discovering novel antiparasitics. Trends Parasitol. 2004;20:462–468. doi: 10.1016/j.pt.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 39.Doyle MA, et al. Drug target prediction and prioritization: using orthology to predict essentiality in parasite genomes. BMC Genomics. 2010;11:222. doi: 10.1186/1471-2164-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitzpatrick JM, et al. Anti-schistosomal intervention targets identified by lifecycle transcriptomic analyses. PLoS Negl Trop Dis. 2009;3:e543. doi: 10.1371/journal.pntd.0000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gobert GN, et al. Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics. 2009;10:128. doi: 10.1186/1471-2164-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolly ER, et al. Gene expression patterns during adaptation of a helminth parasite to different environmental niches. Genome Biol. 2007;8:R65. doi: 10.1186/gb-2007-8-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doenhoff MJ, et al. Praziquantel: its use in control of schistosomiasis in sub-Saharan Africa and current research needs. Parasitology. 2009;136:1825–1835. doi: 10.1017/S0031182009000493. [DOI] [PubMed] [Google Scholar]
- 44.Rollinson D. A wake up call for urinary schistosomiasis: reconciling research effort with public health importance. Parasitology. 2009;136:1593–1610. doi: 10.1017/S0031182009990552. [DOI] [PubMed] [Google Scholar]
- 45.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 46.Carlson SM, White FM. Using small molecules and chemical genetics to interrogate signaling networks. ACS Chem Biol. 2011;6:75–85. doi: 10.1021/cb1002834. [DOI] [PubMed] [Google Scholar]
- 47.Durrant JD, et al. A multidimensional strategy to detect polypharmacological targets in the absence of structural and sequence homology. PLoS Comput Biol. 2010;6:e1000648. doi: 10.1371/journal.pcbi.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verjovski-Almeida S, et al. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat Genet. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- 49.Liu F, et al. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathog. 2006;2:e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeMarco R, Verjovski-Almeida S. Schistosomes--proteomics studies for potential novel vaccines and drug targets. Drug Discov Today. 2009;14:472–478. doi: 10.1016/j.drudis.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 51.McManus DP, Loukas A. Current status of vaccines for schistosomiasis. Clin Microbiol Rev. 2008;21:225–242. doi: 10.1128/CMR.00046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gobert GN, et al. Tissue specific profiling of females of Schistosoma japonicum by integrated laser microdissection microscopy and microarray analysis. PLoS Negl Trop Dis. 2009;3:e469. doi: 10.1371/journal.pntd.0000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nawaratna SS, et al. Gene atlasing of digestive and reproductive tissues in Schistosoma mansoni. PLoS Negl Trop Dis. 2011;5:e1043. doi: 10.1371/journal.pntd.0001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loukas A, et al. Schistosome membrane proteins as vaccines. Int J Parasitol. 2007;37:257–263. doi: 10.1016/j.ijpara.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Skelly PJ, Alan Wilson R. Making sense of the schistosome surface. Adv Parasitol. 2006;63:185–284. doi: 10.1016/S0065-308X(06)63003-0. [DOI] [PubMed] [Google Scholar]
- 56.Braschi S, et al. The tegument surface membranes of the human blood parasite Schistosoma mansoni: a proteomic analysis after differential extraction. Proteomics. 2006;6:1471–1482. doi: 10.1002/pmic.200500368. [DOI] [PubMed] [Google Scholar]
- 57.Castro-Borges W, et al. Enzymatic shaving of the tegument surface of live schistosomes for proteomic analysis: a rational approach to select vaccine candidates. PLoS Negl Trop Dis. 2011;5:e993. doi: 10.1371/journal.pntd.0000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Balkom BW, et al. Mass spectrometric analysis of the Schistosoma mansoni tegumental sub-proteome. J Proteome Res. 2005;4:958–966. doi: 10.1021/pr050036w. [DOI] [PubMed] [Google Scholar]
- 59.Perez-Sanchez R, et al. Proteomic analysis of the tegument and excretory-secretory products of adult Schistosoma bovis worms. Proteomics. 2006;6(Suppl 1):S226–236. doi: 10.1002/pmic.200500420. [DOI] [PubMed] [Google Scholar]
- 60.Mulvenna J, et al. Exposed proteins of the Schistosoma japonicum tegument. Int J Parasitol. 2010;40:543–554. doi: 10.1016/j.ijpara.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Braschi S, Wilson RA. Proteins exposed at the adult schistosome surface revealed by biotinylation. Mol Cell Proteomics. 2006;5:347–356. doi: 10.1074/mcp.M500287-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Tran MH, et al. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat Med. 2006;12:835–840. doi: 10.1038/nm1430. [DOI] [PubMed] [Google Scholar]
- 63.Da’dara AA, et al. DNA-based vaccines protect against zoonotic schistosomiasis in water buffalo. Vaccine. 2008;26:3617–3625. doi: 10.1016/j.vaccine.2008.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doolan DL. Plasmodium immunomics. Int J Parasitol. 2011;41:3–20. doi: 10.1016/j.ijpara.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Driguez P, et al. Schistosomiasis vaccine discovery using immunomics. Parasit Vectors. 2010;3:4. doi: 10.1186/1756-3305-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hotez PJ, et al. Developing vaccines to combat hookworm infection and intestinal schistosomiasis. Nat Rev Microbiol. 2010;8:814–826. doi: 10.1038/nrmicro2438. [DOI] [PubMed] [Google Scholar]
- 67.Luo R, et al. Analysis and characterization of the genes encoding the Dicer and Argonaute proteins of Schistosoma japonicum. Parasit Vectors. 2010;3:90. doi: 10.1186/1756-3305-3-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krautz-Peterson G, Skelly PJ. Schistosoma mansoni: the dicer gene and its expression. Exp Parasitol. 2008;118:122–128. doi: 10.1016/j.exppara.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zou X, et al. RNAi silencing of calcium-regulated heat-stable protein of 24 kDa in Schistosoma japonicum affects parasite growth. Parasitol Res. 2011;108:567–572. doi: 10.1007/s00436-010-2099-0. [DOI] [PubMed] [Google Scholar]
- 70.Rinaldi G, et al. RNA interference targeting leucine aminopeptidase blocks hatching of Schistosoma mansoni eggs. Mol Biochem Parasitol. 2009;167:118–126. doi: 10.1016/j.molbiopara.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krautz-Peterson G, et al. Optimizing gene suppression in schistosomes using RNA interference. Mol Biochem Parasitol. 2007;153:194–202. doi: 10.1016/j.molbiopara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 72.Stefanic S, et al. RNA interference in Schistosoma mansoni schistosomula: selectivity, sensitivity and operation for larger-scale screening. PLoS Negl Trop Dis. 2010;4:e850. doi: 10.1371/journal.pntd.0000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mourao MM, et al. Phenotypic screen of early-developing larvae of the blood fluke, Schistosoma mansoni, using RNA interference. PLoS Negl Trop Dis. 2009;3:e502. doi: 10.1371/journal.pntd.0000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krautz-Peterson G, et al. RNA interference in schistosomes: machinery and methodology. Parasitology. 2009;137:485–495. doi: 10.1017/S0031182009991168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swierczewski BE, Davies SJ. A schistosome cAMP-dependent protein kinase catalytic subunit is essential for parasite viability. PLoS Negl Trop Dis. 2009;3:e505. doi: 10.1371/journal.pntd.0000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tran MH, et al. Suppression of mRNAs encoding tegument tetraspanins from Schistosoma mansoni results in impaired tegument turnover. PLoS Pathog. 2010;6:e1000840. doi: 10.1371/journal.ppat.1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skelly PJ, et al. Cloning, characterization, and functional expression of cDNAs encoding glucose transporter proteins from the human parasite Schistosoma mansoni. J Biol Chem. 1994;269:4247–4253. [PubMed] [Google Scholar]
- 78.Krautz-Peterson G, et al. Suppressing glucose transporter gene expression in schistosomes impairs parasite feeding and decreases survival in the mammalian host. PLoS Pathog. 2010;6:e1000932. doi: 10.1371/journal.ppat.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faghiri Z, Skelly PJ. The role of tegumental aquaporin from the human parasitic worm, Schistosoma mansoni, in osmoregulation and drug uptake. FASEB J. 2009;23:2780–2789. doi: 10.1096/fj.09-130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faghiri Z, et al. The tegument of the human parasitic worm Schistosoma mansoni as an excretory organ: the surface aquaporin SmAQP is a lactate transporter. PLoS One. 2010;5:e10451. doi: 10.1371/journal.pone.0010451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tielens AG, et al. Schistosoma mansoni: rapid turnover of glycogen by adult worms in vivo. Exp Parasitol. 1989;68:235–237. doi: 10.1016/0014-4894(89)90103-3. [DOI] [PubMed] [Google Scholar]
- 82.Bhardwaj R, Skelly PJ. Purinergic signaling and immune modulation at the schistosome surface? Trends Parasitol. 2009;25:256–260. doi: 10.1016/j.pt.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 83.Bhardwaj R, Skelly PJ. Characterization of schistosome tegumental alkaline phosphatase (SmAP) PLoS Negl Trop Dis. 2011;5:e1011. doi: 10.1371/journal.pntd.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Young ND, et al. Elucidating the transcriptome of Fasciola hepatica - a key to fundamental and biotechnological discoveries for a neglected parasite. Biotechnol Adv. 2009;28:222–231. doi: 10.1016/j.biotechadv.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 85.Young ND, et al. Unlocking the transcriptomes of two carcinogenic parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl Trop Dis. 2010;4:e719. doi: 10.1371/journal.pntd.0000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Young ND, et al. A portrait of the transcriptome of the neglected trematode, Fasciola gigantica--biological and biotechnological implications. PLoS Negl Trop Dis. 2011;5:e1004. doi: 10.1371/journal.pntd.0001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adamidi C, et al. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 2011;21:1193–1200. doi: 10.1101/gr.113779.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tchoubrieva E, Kalinna B. Advances in mRNA silencing and transgene expression: a gateway to functional genomics in schistosomes. Biotechnol Genet Eng Rev. 2010;26:261–280. doi: 10.5661/bger-26-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hokke CH, et al. Integrating transcriptome, proteome and glycome analyses of Schistosoma biology. Trends Parasitol. 2007;23:165–174. doi: 10.1016/j.pt.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 90.Kooij TW, et al. Plasmodium post-genomics: better the bug you know? Nat Rev Microbiol. 2006;4:344–357. doi: 10.1038/nrmicro1392. [DOI] [PubMed] [Google Scholar]
- 91.Grabherr MG, et al. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics. 2010;26:1145–1151. doi: 10.1093/bioinformatics/btq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Donthu R, et al. SyntenyTracker: a tool for defining homologous synteny blocks using radiation hybrid maps and whole-genome sequence. BMC Res Notes. 2009;2:148. doi: 10.1186/1756-0500-2-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skovlund E, Fenstad GU. Should we always choose a nonparametric test when comparing two apparently nonnormal distributions? J Clin Epidemiol. 2001;54:86–92. doi: 10.1016/s0895-4356(00)00264-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.