Abstract
The fields of mass spectrometry (MS) and stem cell biology have expanded greatly in the past twenty years. Taken alone, these fields occupy entirely different branches of science; however, the points where they overlap provide valuable insight, both in the biological and technical arenas. From a biological perspective, MS-based proteomics offers the capacity to follow post-transcriptional regulation and signaling that are 1) fundamental to pluripotency and differentiation, 2) largely beyond the reach of genomic technologies, and 3) otherwise difficult or impossible to examine on a large-scale. At the same time, addressing questions fundamental to stem cell biology has compelled proteomic researchers to pursue more sensitive and creative ways to probe the proteome, both in a targeted and high-throughput manner. Here, we highlight experiments that straddle proteomics and stem cell biology, with an emphasis on studies that apply mass spectrometry to dissect pluripotency and differentiation.
Key Terms: Mass spectrometry, Embryonic stem cell, Differentiation, Large-scale analysis, Quantitative proteomics
Introduction
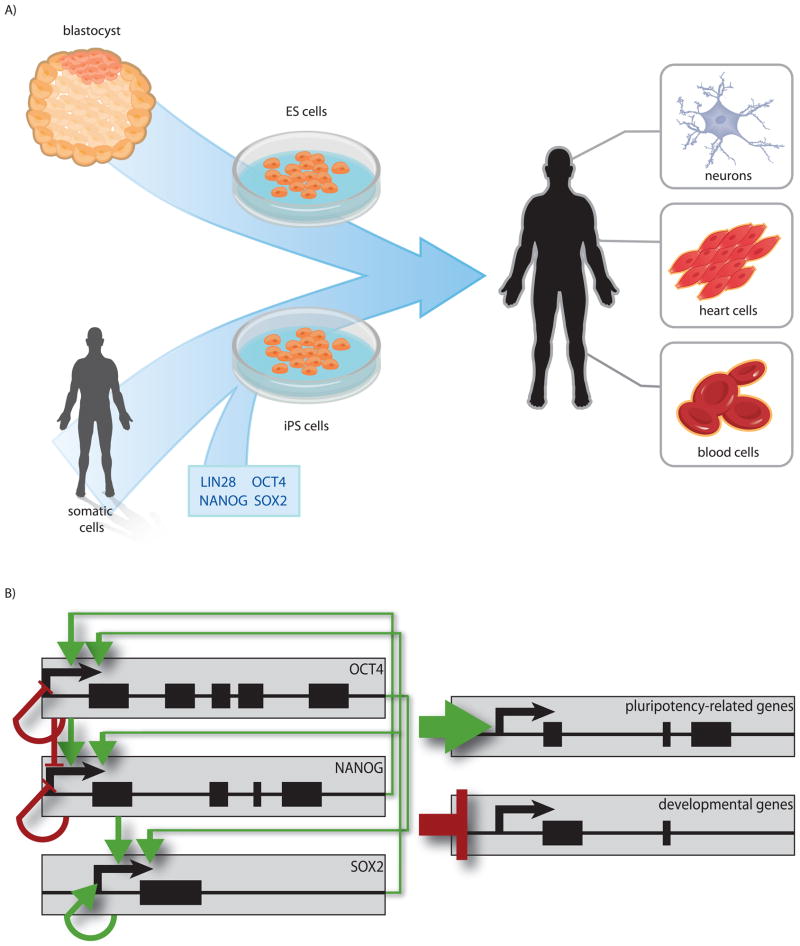
Pluripotent stem cells are endowed with the extraordinary capacity to differentiate into all three germ layers that constitute the adult body (Figure 1A) (Bradley et al., 1984; Nagy et al., 1990; Thomson et al., 1998; Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). This characteristic, together with the ability to self-renew indefinitely, makes pluripotent cells an ideal model for studying early development. Further, as we learn more about how pluripotent cells mature into various specialized tissues, we move closer to making regenerative therapies a reality. Hanna et al. demonstrated this potential using induced pluripotent stem (iPS) cells from a humanized sickle cell anemia mouse model (Hanna et al., 2007). In this study, iPS cells bearing the disease-causing mutation were genetically corrected, differentiated in vitro into hematopoietic progenitors, and reintroduced into irradiated mice. Remarkably, these mice exhibited virtually no pathological remnants of the disease. Significant efforts have also been made toward regenerative therapies for spinal cord injuries and retinal degenerative disease, illustrating the great promise that pluripotent cells hold for both fundamental science and medicine (McDonald et al., 1999; Liu et al., 2000; Keirstead et al., 2005; Howden et al., 2011).
Figure 1.
Pluripotency. (A) Two examples of pluripotent cell types are depicted. Human ES cells are derived from the inner cell mass of pre-implantation embryos whereas iPS cells are directly reprogrammed from somatic cells. Both of these pluripotent cell types are capable of differentiating into cells that constitute all three germ layers. (B) Pluripotency is maintained by a complex transcriptional network anchored by OCT4, SOX2, and NANOG. These genes activate transcription from the promoters of genes responsible for sustaining pluripotency, while repressing genes involved in development. This network is densely interconnected and forms numerous feedback loops, illustrating the importance of tight regulation in this system. Green arrows represent transcriptional activation while red symbols represent transcriptional repression. Color figures are available online.
Given this promise, enormous effort has been devoted to understanding cell potency, self-renewal, and differentiation. In pluripotent cells these processes are anchored by core transcription factors, OCT4, SOX2, and NANOG (Scholer et al., 1990; Chambers et al., 2003; Mitsui et al., 2003; Masui et al., 2007; Pan and Thomson, 2007; Takahashi et al., 2007; Yu et al., 2007; Kim et al., 2008; Yu et al., 2009). Expression of OCT4 and NANOG is primarily restricted to pluripotent cell types, while SOX2 is also expressed in some neural lineages (Rosner et al., 1990; Kamachi et al., 2000; Chambers et al., 2003; Mitsui et al., 2003). Knockdown of any of these factors in pluripotent cells promotes differentiation (Ivanova et al., 2006); however, it was recently shown that Nanog is not essential for maintaining the pluripotent state in mouse embryonic stem (ES) cells (Mullin et al., 2008). Conversely, overexpression of Sox2 or Oct4 by as little as 1.5- or 2-fold is sufficient to induce differentiation in pluripotent mouse cells (Niwa et al., 2000; Chew et al., 2005), yet overexpression of NANOG supports pluripotency, even in the absence of otherwise requisite growth factors (Chambers et al., 2003; Mitsui et al., 2003; Xu et al., 2008). Furthermore, forced expression of OCT4 and SOX2, either alone or in combination with NANOG and a handful of other genes, is sufficient to reprogram somatic cells to the pluripotent state (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007; Yu et al., 2009). The proportion of expression for OCT4 and SOX2 during reprogramming is one determining factor in the efficiency of this process (Yu et al., 2007). Thus, the function of these proteins is complex and dose dependent.
Mechanistically, OCT4, SOX2, and NANOG function as part of an intricate regulatory network (Boyer et al., 2005). Detailing this network and its dynamics during differentiation has been the aim of numerous studies and to this end, transcriptomic and genomic methods have been applied with great effect (Boyer et al., 2005; Kim et al., 2008; Chin et al., 2009; Yu et al., 2011)(reviewed in (Brien and Bracken, 2009; Do and Scholer, 2009; Hochedlinger and Plath, 2009; Young, 2011)). It is now clear that OCT4, SOX2, and NANOG bind and activate transcription from the promoters of genes important for pluripotency, while repressing developmental genes (Figure 1B) (Boyer et al., 2005; Pan and Thomson, 2007). They also bind to each other’s promoters, and their own, forming a complex regulatory loop (Figure 1B). To date, the majority of regulatory information regarding these key genes has come from transcriptional or epigenetic studies; however, an equally important aspect of stem cell biology occurs post-transcriptionally (Van Hoof et al., 2008; Phanstiel et al., 2011). For example, it is well established that mRNA levels do not necessarily reflect protein abundances and a number of mechanisms that function post-transcriptionally are key to pluripotency (Houbaviy et al., 2003; Lee et al., 2007; Judson et al., 2009; Xu et al., 2009). In fact, forced expression of only a small set of miRNAs, together with histone deacetylase inhibitors, is sufficient to initiate reprogramming of somatic cells to the pluripotent state (Miyoshi et al., 2011). Further, post-translational modification (PTM) of proteins provides a dynamic and rapid means to modulate protein activity and transduce signals, both of which play key roles in maintaining pluripotency and inducing differentiation (Seuntjens et al., 2009; Chng et al., 2011). Thus, although genomic and transcriptomic techniques are staple methods for the developmental biologist, the proteome represents an equally rich and informative experimental plane (Heck et al., 2007).
Mass spectrometry (MS) is well suited to proteomic studies because it is highly sensitive, it can be quantitative, and, unlike many antibody-based methods, it does not require a priori knowledge of protein targets. In a typical MS discovery experiment, proteins extracted from tissues or cell cultures are digested with one or more enzymes (e.g. Trypsin, Lys-C, etc.) to produce peptides. These peptides are separated to reduce sample complexity and interfaced to a mass spectrometer, often through an electrospray ionization source. Ionized peptides enter the mass spectrometer and are analyzed based on both mass and charge (output as a mass to charge ratio (m/z)). Peptides of a particular m/z are isolated and fragmented to produce distinctive fragment ions from which their primary sequence, complete with PTMs, is inferred.
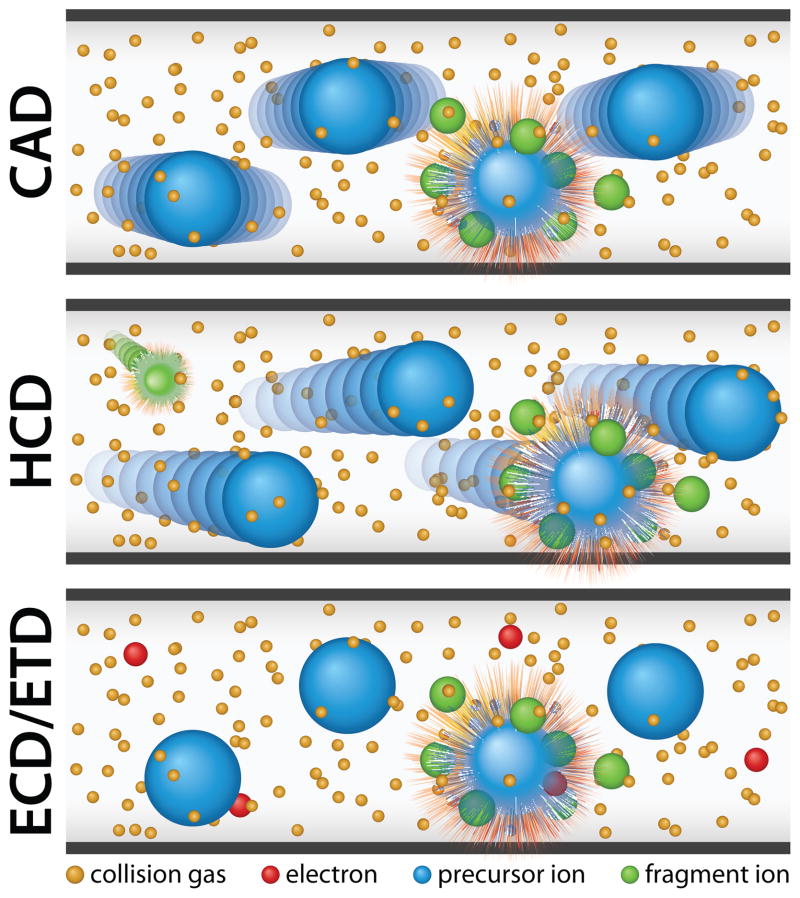
Many mass spectrometers offer a variety of peptide fragmentation methods for this purpose –each with benefits and drawbacks. Resonant excitation collision activated dissociation (CAD) is commonly used to dissociate peptides by inducing collisions with a bath gas (e.g. helium) (Figure 2). CAD is common in global proteomics experiments due to its short activation time and effectiveness in fragmenting a wide variety of peptides. However, CAD fragmentation is often biased toward more labile bonds and the major dissociation product of many PTM-containing peptides is therefore the loss of a PTM, rather than sequence informative ion fragments. In contrast, electron-based dissociation methods (electron transfer dissociation (ETD) or electron capture dissociation (ECD)) are well suited for the characterization of PTMs and intact proteins as these methods use either radical anions (ETD) or free electrons (ECD) to induce random “soft” fragmentation (Figure 2). This form of dissociation often leaves PTMs attached to the specific amino acid (Zubarev et al., 2000; Coon et al., 2004; Syka et al., 2004; Coon et al., 2005). The advantages of soft fragmentation methods are accentuated when studying heavily modified peptides. For example, ETD and ECD have been used to characterize combinations of PTMs on histones in human embryonic stem (ES) and cancer cells (Garcia et al., 2007; Phanstiel et al., 2008).
Figure 2.
Commonly used dissociation techniques. Fragmentation with resonant excitation CAD results from collisions between analyte ions and a bath gas. HCD is a higher energy form of CAD that retains many labile PTMs lost during resonant excitation CAD. ETD and ECD use anions or electrons to induce “soft fragmentation” that also retains many labile PTMs. Color figures are available online.
More recently, higher energy beam-type CAD, termed HCD, led to increased phosphopeptide identifications in both human ES and induced pluripotent stem (iPS) cells (Olsen et al., 2007; Phanstiel et al., 2011). Unlike resonant excitation, HCD fragmentation is accomplished by accelerating analyte peptides into a higher pressure collision cell, usually filled with nitrogen (Figure 2). Like CAD, HCD works for a wide variety of peptides, but is less hampered by the dominance of non-random PTM fragmentation, increasing the number of sequence-informative fragment ions. Also, HCD is not constrained by low mass cutoff and is therefore compatible with isobaric tag-based quantitation techniques. An increasingly common practice is to use multiple dissociation methods, in combination, to achieve increases in identification and/or quantitative accuracy (Guo et al., 2008; Swaney et al., 2008; Phanstiel et al., 2009; Frese et al., 2011). Thus, a number of options are available for tandem MS analysis and fragmentation can be tailored to address particular biological questions of interest.
One of the most critical, recent movements in modern proteomics has been the development and rapid advancement of hybrid mass spectrometers. A key, defining feature of these mass spectrometers is the inclusion of two or more mass analyzers, which are the physical components that separate analytes based on mass and charge. Whereas some mass analyzers excel at isolation and fragmentation of peptide precursors (i.e., quadrupole mass filters and ion traps), others offer superior mass accuracy and resolution (i.e., time-of-flight, fourier transform ion cyclotron resonance, and orbitrap mass analyzers) (Glish and Burinsky, 2008). Hybrid mass spectrometers combine different types of mass analyzers to harness the unique benefits of each, which has drastically enhanced the ability to detect and quantify proteins and post-translational modifications in complex biological mixtures (Shevchenko et al., 2000; Syka et al., 2004; Makarov et al., 2006; Glish and Burinsky, 2008). For example, in 2001 Yates and co-workers identified approximately 22% of known and predicted yeast ORFs (1,484 proteins) in an early demonstration of a large-scale proteomics workflow (Washburn et al., 2001). However, using hybrid instruments just seven years later, Mann and co-workers identified nearly all yeast proteins known to be expressed (66% of known or predicted ORFs corresponding to 4,399 proteins) (de Godoy et al., 2008). This illustrates the power of MS instrumentation-advances in proteomics.
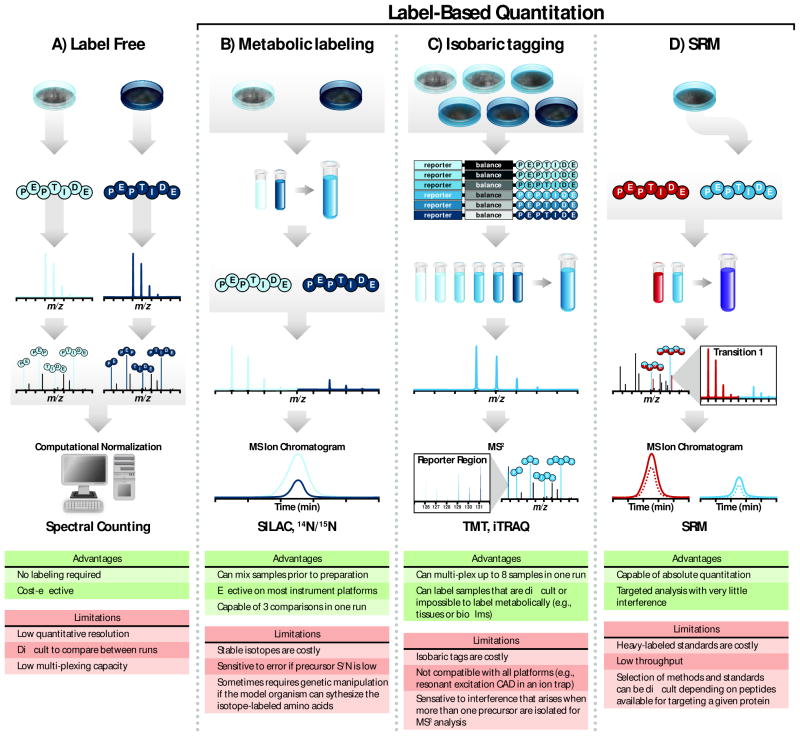
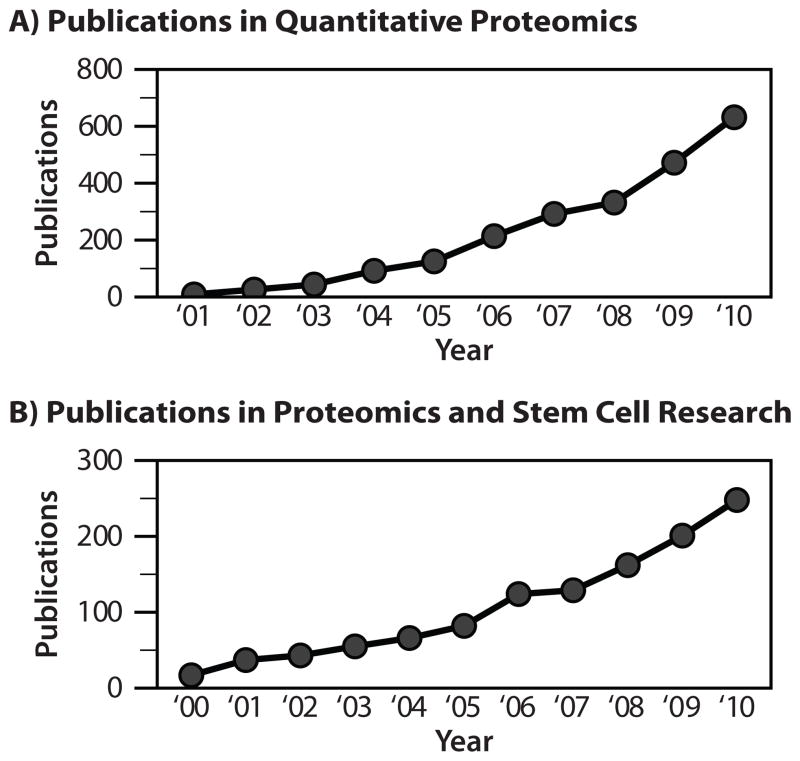
In addition to improved instrumentation, several quantitation methods are available, providing flexible solutions for a wide range of experimental conditions (Figure 3). Label-free methods allow quantitation across many samples and do not require additional steps during sample preparation; however, care must be taken when comparing results between samples since it is difficult to perfectly replicate chromatographic conditions, ionization efficiency, and sampling between runs (Wang et al., 2003; Liu et al., 2004). Metabolic labeling, in which stable isotopes are introduced via media or diet, can be used to quantify differences between two or three different samples in a single run (Jiang and English, 2002; Ong et al., 2002). For example, stable isotope labeling with amino acids in cell culture (SILAC) is a well-established form of this technique (reviewed (Ong et al., 2003; Pimienta et al., 2009)). A key advantage of metabolic labeling is that samples can be mixed prior to preparation, which reduces variation from sample processing and decreases instrument analysis time. On the other hand, chemical labeling strategies such as isobaric tags for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMT) allow for up to eight sample comparisons, further increasing throughput (Thompson et al., 2003; Ross et al., 2004; Choe et al., 2007). Like metabolic labeling, this approach has been used to quantify thousands of proteins and PTMs (Phanstiel et al., 2011); however isobaric labeling is compatible with a number of model systems that are difficult or impossible to metabolically label (i.e., tissue and biofilms). A different approach altogether, selected reaction monitoring (SRM), targets specific proteins and fragment ions to achieve highly accurate quantitation (Gerber et al., 2003; Gerber et al., 2007). While the aforementioned tactics are largely used for relative quantitation, SRM can be used to determine absolute protein amount. Still, this targeted approach currently has much lower throughput than other quantitation methods. Each quantitative method has advantages and limitations (Figure 3); however, quantitative MS is an enabling approach that is garnering increasing attention in the literature (Figure 4A).
Figure 3.
Quantitative proteomics strategies. (A) During label-free quantitation, peptides extracted from samples are identified through tandem MS. Spectral counting measures the number of peptide spectral matches (i.e., the number of times the peptide is observed) followed by normalization that accounts for protein size. This technique allows for relative quantitation without the use of isotopes. (B) Metabolic labeling incorporates heavy isotopes into proteins through amino acids or nutrients (i.e., 15N or 13C). Differently-labeled samples are combined during protein preparation and their corresponding stable isotopes generate a shift in m/z values observed in MS1 scans. The difference between extracted ion chromatograms for peaks corresponding to heavy and light samples is proportional to the relative abundance of each. (C) Isobaric labeling strategies, such as TMT and iTRAQ, are capable of comparing up to six or eight samples in a single run depending on the method. Like metabolic labeling, samples are combined during protein preparation. Labeled peptides from different samples within the mixture have the same nominal mass and co-elute with reversed-phase chromatography. Peptide fragmentation during tandem mass spectrometry produces both sequence ions for peptide identification and reporter tags for quantitation. (D) Quantitation via single reaction monitoring (SRM) is capable of absolute quantitation and is often performed on triple quadruple mass spectrometers. Peptides from samples are mixed with a chemically synthesized heavy peptide that serves as a quantitation standard. Because the exact amount of this standard is known, absolute quantitation is possible through this method. Figure panels adapted from (Gerber et al., 2007) and (Ross et al., 2004). Color figures are available online.
Figure 4.
Publication trends for proteomics and stem cell research. (A) Publications relating to quantitative proteomics since 2000. (B) Publications relating to both stem cell research and proteomics since 2000.
To accommodate the complex data generated by these technological improvements, a number of software packages have emerged, many of which are freely available. The Trans-Proteomic Pipeline is a flexible software resource that can process data from a number of instrument formats (Keller and Shteynberg, 2011). For more specific needs, Cox et al. developed the MaxQuant platform for high resolution MS data analysis (Cox and Mann, 2008). Incorporated into the program are tools for analyzing quantitative SILAC data and more recently, Andromeda, a peptide search engine that is compatible with MaxQuant (Cox et al., 2011). Our own lab recently unveiled COMPASS, a modular software suite developed around the Open Mass Spectrometry Search Algorithm. COMPASS facilitates database searching, spectral reduction, estimation of false discovery rates, and large-scale quantitation through isobaric tagging (Wenger et al., 2011). Together, these tools and many others have greatly facilitated data processing and proteomic analysis. Advances in peptide fragmentation, quantitation, instrumentation, and software have propelled MS-based proteomic research over the past ten years.
Stem cell research is an exciting and exceedingly relevant field, both for medicine and basic science. Mass spectrometry is a highly technical and equally powerful analytical tool. Combining these disciplines has become increasingly popular and, as shown in Figure 4B, publication trends that span both fields continue to rise. Here we review recent literature that lies at the intersection of pluripotent stem cell research and mass spectrometry to provide perspective on these multi-disciplinary efforts.
The proteome of pluripotent cells
Although key functional properties of pluripotent cells, such as developmental potential and self-renewal, are readily appreciated, the mechanisms that establish and maintain these properties are considerably more complex. As a result, a number of large-scale studies aimed to define the pluripotent landscape, including the ES cell proteome (Nagano et al., 2005; Van Hoof et al., 2006; Schulz et al., 2007). Early MS work by Nagano and colleagues benchmarked nearly 1,800 proteins in mouse ES cells that were representative of a wide range of cellular compartments, functional categories, and expression levels (Nagano et al., 2005). Van Hoof et al. similarly identified 1,871 proteins in mouse ES cells but also expanded their study to cover the human ES cell-proteome at a depth of 1,775 proteins (Van Hoof et al., 2006). After subtracting proteins identified in differentiated cells, the authors intersected mouse and human datasets to compile a list of 191 orthologs that were ES cell-specific in both species. This set of proteins included many transcription factors with well-established roles in pluripotency but also contained many proteins of unknown function, which, the authors argue, are good candidates for further study in the context of ES cell biology.
In more recent years, the advent of hybrid mass spectrometers has improved shotgun proteomics analyses by coupling, for example, highly sensitive linear ion traps to Fourier transform mass analyzers with superior resolution and mass accuracy. The result is higher confidence peptide identification that provides far deeper proteomic coverage on a similar time scale. Recently, Graumann et al. achieved sub-ppm mass accuracy using a linear ion trap-orbitrap hybrid instrument, resulting in over 5,100 protein identifications in mouse ES cells (Graumann et al., 2008). Gene enrichment analysis suggested an over-representation of proteins associated with proliferation, which is consistent with the rapid cycling times typical of pluripotent cells (Becker et al., 2007). This study also adapts SILAC for ES cell culture, permitting quantitative analysis through metabolic labeling; however, it is used here distinguish peptide peaks from noise peaks, rather than comparing different cell states. Phanstiel and colleagues used similar instrumentation with multiple dissociation techniques to identify nearly 7,000 proteins in human ES and iPS cells (Phanstiel et al., 2011). To date, this is the largest compendium of pluripotent proteins available and points to the remarkable similarity of these cell types (vide infra).
A major advantage of mass spectrometry-based proteomics is the capacity to identify post-translational modifications (Grimsrud et al., 2010). For instance, tools for enriching and detecting phosphorylation are now well developed and several studies have explored this PTM in both mouse and human pluripotent cells (Brill et al., 2009; Swaney et al., 2009; Van Hoof et al., 2009; Li et al., 2011; Phanstiel et al., 2011). In particular, Swaney et al. presented data of interest to both the stem cell and proteomics community. Almost 11,000 unique phosphorylation sites were identified using a combination of resonant excitation CAD and ETD (Swaney et al., 2009). One noteworthy observation was the relationship between dissociation technique and phosphopeptide localization. ETD cleaved more frequently in the six amino acids surrounding phospho-serine and phospho-threonine, improving the chances of localizing that phosphorylation to a single amino acid. CAD, on the other hand, performed slightly better than ETD for tyrosine phosphorylation. Consistent with previous work, this suggests that ETD and CAD are complementary for phosphorylation analysis (Good et al., 2007), although overall, ETD identified many more phosphopeptides in partial tryptic digests (8,087 for ETD compared to 3,868 for CAD). Finally, 16 previously unknown phosphorylation motifs were reported in this work based on MotifX, a de novo motif-generating program (Schwartz and Gygi, 2005). Still other experiments have focused on key portions of the proteome (e.g. plasma membrane proteins and the secretome) and have been thoroughly reviewed elsewhere (Ahn et al., 2008; Dormeyer et al., 2008; Van Hoof et al., 2008; Skalnikova et al., 2011). Discovery experiments like these have identified key components of the pluripotent proteome and have laid the groundwork for future, directed research.
Comparative analyses
The development of quantitative proteomics has vastly increased the experimental repertoire available through mass spectrometry. Comparing between cell states, treatments, and differentiation time points is particularly informative in stem cell biology and a number of studies have applied MS technology to this end. Histone analysis is one highly relevant area where mass spectrometry has a distinct advantage since it can distinguish multiple post-translational modifications on a single molecule. One study identified and tracked 74 different histone H4 isoforms as human ES cells differentiate (Phanstiel et al., 2008). Using label-free quantitation, the authors show that global H4 acetylation rapidly decreases as ES cells differentiate, suggesting a move to a less transcriptionally permissive chromatin structure. Consistent with this is a concordant increase in H4K20 methylation, another mark characteristic of repressive chromatin. Together, these observations provide evidence at the protein level to support a concept suggested through genomic technologies: ES cells maintain more open, accessible chromatin relative to differentiated cells (Meshorer and Misteli, 2006; Brumbaugh et al., 2008). Interestingly, methylation of H4 appears to be sequential and concerted since methylation of other residues was never observed in the absence of di-methylated K20. This work establishes an epigenetic profile for human ES cells and shows the utility of quantitative MS in tracking changes during differentiation.
Comparative MS approaches are equally valuable on the large-scale. Van Hoof et al. used multi-plexed SILAC samples to monitor protein and phosphorylation changes induced by BMP4-treatment in human ES cells (Van Hoof et al., 2009). This work suggests that differentiation works rapidly through signaling networks since approximately 50% of quantified phosphorylation sites were differentially regulated within one hour of BMP4 addition. Overall, phosphorylation levels increased as cells differentiated, suggesting that kinases are activated in response to bone morphogenetic protein 4 (BMP4). This effect appears to be transient, however, since no phosphorylation sites were differentially regulated after four hours of treatment and the largest changes were observed within the first hour. Early in the time course, developmental signaling molecules, SMAD5 and SMAD8, accumulated activating phosphorylation marks on residues S465 and S467, respectively. This is consistent with the differentiation regime inflicted by bone morphogenetic protein (Pera et al., 2004). Finally, the authors relate an interesting vignette that directly addresses the biological relevance of three SOX2 phosphorylation sites identified in the work. Mutational analysis of S249-S251 showed that phosphorylation of these sites is required for SUMOylation of SOX2. Although the direct functional relevance of the identified SUMOylation is not clear, this modification was previously shown to decrease the capacity for SOX2 to bind DNA and may point to a more general regulatory mechanism (Meshorer and Misteli, 2006). Given the relevance of SOX2 in pluripotency, this example illustrates one way that proteomics and biology combine to uncover specific regulatory events relevant to stem cell function.
Rigbolt and colleagues recently used a similar approach to compare and contrast differentiation induced by (1) phorbol myristate acetate (PMA) treatment and (2) culture in unconditioned media (Rigbolt et al., 2011). Provided in the analysis are novel statistical methods to group proteins and phosphorylation sites based on their dynamics during the treatment time course. Consistent with Van Hoof et al., roughly 50% of phosphorylation sites were differentially regulated. Notably, the vast majority of these changes were treatment specific, although phosphorylation on several known pluripotent regulators, for example UTF1, exhibited corresponding profiles in both treatments. Stoichiometry is another important consideration when interpreting phosphorylation changes. For example, if a given phosphorylation site increases fivefold during differentiation, it is tempting to presume that the event is significant; however, if the total protein abundance also increases five-fold, the increased phosphorylation abundance may simply correlate with increased protein. In this study, the authors note that the overall change in abundances was 45% and 19% greater for phosphorylation than protein after treatment with PMA and unconditioned media respectively.
In addition to providing valuable comparisons between cell types and treatments, quantitative analyses expand the coverage of the pluripotent proteome. The data represent a rich resource that can be further processed to explore biological relevance.
Protein interaction profiling: Oct4, Nanog, SOX2
A number of studies have applied mass spectrometry to resolve more specific biological questions. One well-established example is protein interaction profiling, which generally involves pulling down a protein of interest under native conditions, followed by MS to identify co-precipitated binding partners. These partners are validated, for example, through reciprocal pulldown and further studied to determine their biological relevance. This kind of targeted experiment is particularly well-suited to characterize key proteins, including OCT4, SOX2, and NANOG, that establish and maintain pluripotency and self-renewal (Boyer et al., 2005; Takahashi and Yamanaka, 2006; Pan and Thomson, 2007; Takahashi et al., 2007; Yu et al., 2007; Kim et al., 2008; Yu et al., 2009).
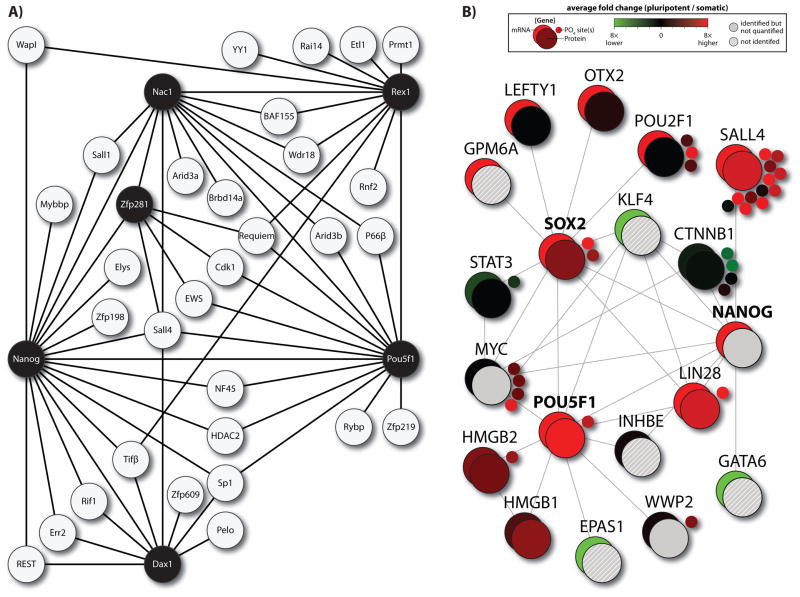
A Nanog-centered interaction network was first described by Wang et al. in mouse ES cells (Wang et al., 2006). The authors use both single- and dual-capture affinity purification, followed by tandem MS to isolate an initial set of 18 binding partners. Included in this set are numerous transcription factors and proteins with known roles in pluripotency, including Sall1, Sall4, and Oct4 (Pou5f1); however, a number of other proteins were identified that had not previously been explored in the context of pluripotency. Two of these, Zfp281 and Nac1, were selected for shRNA-mediated knockdown to determine whether they play a role in maintaining pluripotency. Quantitative RT-PCR showed that downregulation of Zfp281 led to a dramatic increase in primitive endoderm markers Gata6 and Gata4, while Nac1 downregulation correlated with an increase in neuroectoderm marker Isl1. Thus, protein interaction profiling identified factors with putative functional roles in early development. To extend the protein interaction network beyond Nanog, the authors iteratively tagged selected binding partners for further profiling. The resulting “interactome” consisted of nearly 40 proteins that were largely enriched for nuclear factors that are co-regulated during differentiation (Figure 5A). Also conspicuous in this analysis were epigenetic regulators including Hdac2 and several polycomb group proteins. In a separate publication, the same lab performed similar analysis to build a Myc-centered interaction network (Kim et al., 2010). Based on interaction profiling, the work separates three fundamental programs at work in pluripotent cells: a Myc-centered core, an epigenetic core, and a pluripotent core. The authors argue that these cores are modular since constituent proteins are co-expressed under various conditions and knockdown of proteins in one core does not necessarily impact proteins in another core. This information is pertinent to stem cell biology because it helps to categorize proteins into functional groups. Further, the protein cores can be used as an analytical tool to help determine cell state.
Figure 5.
Protein networks in pluripotent cells. (A) A protein interaction network constructed by Wang et al. Nodes established by iterative tagging of key proteins (indicated by black circles) are apparent as hubs and expand the network (Wang et al., 2006). (B) String database was used to collate proteins that interact with OCT4, SOX2, and NANOG. Large-scale proteomic and transcriptomic data was then applied to these proteins to compare gene expression profiles. Color figures are available online.
Recent back-to-back publications defined protein interactions associated with Oct4 (Pou5f1) (Pardo et al., 2010; van den Berg et al., 2010). Pardo et al. identified 92 Oct4 binding partners and importantly, performed experiments in biological triplicate. Many of these proteins shared expression profiles that matched Oct4 during differentiation. Closer inspection of the dataset revealed that five of the 92 partners were required for self-renewal, nine had a known role in pluripotency or self-renewal and 83% of the binding partners that had been knocked out during development were embryonic or peri-natal lethal. This shows the clear connection between Oct4 interacting proteins and development. Van den Berg et al. similarly identified 166 Oct4-interacting proteins, many of which overlapped with the analyses from Pardo et al. and Wang et al. (Wang et al., 2006; Pardo et al., 2010; van den Berg et al., 2010). The authors attribute increased identifications to higher sensitivity detection with improved methods for affinity purification. A major portion of Oct4 interacting proteins in this study was related to chromatin modification and epigenetic regulation. For example, every component of the NuRD complex was identified as Oct4-associated.
Sox2 is particularly difficult to analyze at the protein level because it is highly homologous and often co-expressed with a number of other Sox-family proteins. Mallanna and colleagues tackled this problem by establishing a stable mouse ES cell line harboring an ectopic, doxycycline-inducible version of Flag-tagged Sox2 (Mallanna et al., 2010). Overexpression of this construct served two purposes: 1) it was readily purified with high specificity and 2) forced expression of Sox2 induced differentiation, allowing the authors to track binding partners as ES cells exit the pluripotent state. Using MS analysis with spectral counting, over sixty proteins were categorized as Sox2 binding partners, including known chromatin modifiers and several developmentally related transcription factors. One protein, Sox21, increased greatly during differentiation and its overexpression was subsequently shown to induce development to mesoderm and neuroectoderm lineages. The authors conclude that Sox21 antagonizes Sox2 function to achieve this result.
Together, these data show the value of protein interaction profiling for ES cell-related proteins. Several common themes are apparent from Sox2, Nanog, and Oct4 studies. First, each of these factors interacts with known epigenetic regulators like polycomb group proteins (Wang et al., 2006; Mallanna et al., 2010; Pardo et al., 2010; van den Berg et al., 2010). The redundancy of these interactions reinforces the concept that DNA methylation and histone modification are tightly linked to stem cell maintenance and development. Also, there is significant overlap between proteins identified in the experiments described above. For example, roughly 20% of Sox2 binding partners were also present in either Nanog or Oct4 pulldown experiments (Mallanna et al., 2010). This confirms at the protein level a thesis that has garnered great attention at the transcriptional level: proteins important to pluripotency form a tightly-woven, interdependent network and function together to maintain the stem cell state.
Another way to apply MS analysis to study associated proteins is through protein interaction databases, such as the STRING database (Snel et al., 2000). Using existing empirical and computational data, proteomic data can be applied across entire networks to quantitatively monitor expression and modification of groups of proteins (Figure 5B). Here, large-scale data from different platforms can be layered onto protein networks to provide a systems-level view of protein interaction (Phanstiel et al., 2011).
Proteomics in systems-level analyses
The delicate balance between pluripotency and differentiation is determined by concerted and often inter-dependent regulation of transcripts, proteins, and PTMs. A full appreciation for this system requires multiple approaches. To this end, a handful of studies have paired proteomics with complementary genomic and transcriptomic techniques. Comparing between platforms presents a unique challenge since data outputs are rarely, if ever, directly comparable. As a result, most studies establish fold-differences between experimental conditions or normalize to a median quantitative value for comparisons at each regulatory level (Lu et al., 2009; Phanstiel et al., 2011).
Lu and colleagues used this approach, combining ChIP-chip, microarray technology, and quantitative mass spectrometry to study differentiation induced by Nanog depletion in mouse ES cells (Lu et al., 2009). On the protein level, the multiplexing capacity of isobaric labeling enabled the authors to examine four time points and identify hundreds of proteins that change in response to differentiation. Using these changes as a training set, the authors go on to categorize genes based on regulatory processes by comparing the protein data to epigenetic marks, Pol II occupancy on 3’ exons, and transcript abundances. For example, Nanog depletion did not appreciably alter Pol II occupancy on Esrrb; however, both mRNA and protein levels significantly decreased for this gene during differentiation. This suggests that Esrrb is actively transcribed under these conditions, but differentiation induces potent regulation at the transcript level and a subsequent decrease in protein abundance. Although more direct evidence is required to prove this hypothesis, these kinds of analyses show the power of large-scale proteomics in systems-analyses.
More recent work expanded to cover the transcriptome, proteome, and phosphoproteome in a large-scale comparison of human ES, iPS, and fibroblast cells (Phanstiel et al., 2011). In this study, Phanstiel and co-workers compiled 7,952 proteins and 10,499 phosphorylation sites, including quantitative data for key regulators of pluripotency, OCT4 (POU5F1) and SOX2. Leveraging eight-plex iTRAQ, four iPS cell and four ES cell lines were examined in biological triplicate, which provided the statistical power needed for a comparison of these highly similar cell types. A key point is that this study covered multiple different cell lines as a number of studies have noted variation between both ES and iPS lines (Bock et al., 2011; Lister et al., 2011). Although ES and iPS cells were remarkably similar at each tier of regulation, gene enrichment analysis identified statistically significant differences that were consistent at the transcript, protein, and PTM level. Further analysis showed that these differences are related to functional properties characteristic of the fibroblast cells from which the iPS cells are derived. This supports the notion that iPS cells bear residual regulation from their somatic origins (Kim et al., 2010).
Combining phosphorylation analysis with transcript and protein data enabled the authors to tease out further regulatory mechanisms at work in pluripotent cells. For example, phosphorylation of substrates associated with a number of kinases was higher in pluripotent cells relative to fibroblast cells, even though transcript and protein abundances for the kinases were constant (Phanstiel et al., 2011). This observation suggests that the activity of these kinases is controlled largely through post-transcriptional regulation rather than expression levels. Global phosphorylation analysis confirmed this model for CDK2 by showing that its activating mark, phosphothreonine-160, was increased nearly six fold in pluripotent lines. Conversely, phosphorylation by kinases like PKC and PKA may be more dependent upon expression levels since protein abundances correlate closely with substrate phosphorylation. In these ways, proteomics plays a key role in systems-level analyses, helping to clarify regulation in stem cells.
Data sharing
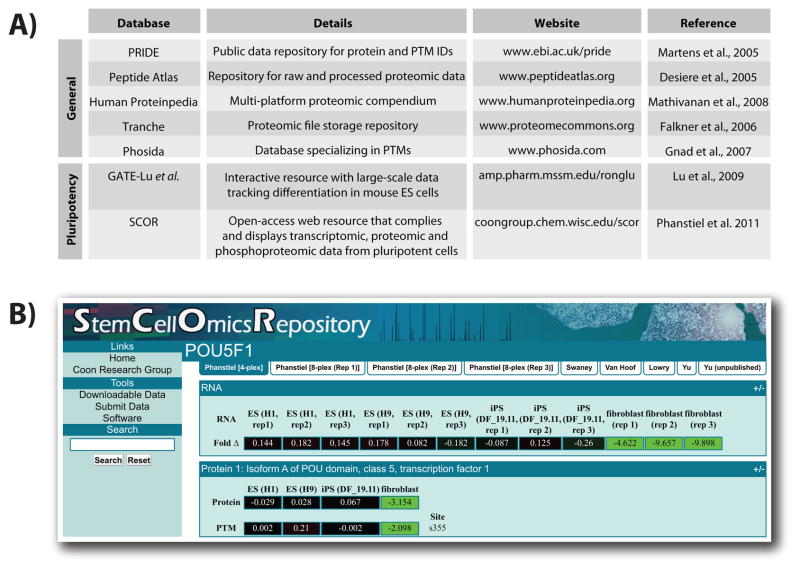
To truly make an impact in the field, researchers must share large-scale data for application to any biological question. A number of websites are commonly used to store proteomics data and Figure 6A lists information for several of the more established databases (Desiere et al., 2005; Martens et al., 2005; Falkner et al., 2006; Gnad et al., 2007; Mathivanan et al., 2008). In addition, several studies reviewed here have established proteomic resources devoted to stem cell biology. Lu et al. provide a website that displays their quantitative data as heatmaps (http://amp.pharm.mssm.edu/ronglu) (Lu et al., 2009). Time course data from these experiments can be viewed as a movie so that protein dynamics are easily visualized. Also included are transcriptomic and epigenetic data for comparison. Our own lab has created a dedicated repository for large-scale data, called the Stem Cell –Omics Repository (SCOR; Figure 6B) http://coongroup.chem.wisc.edu/scor) (Phanstiel et al., 2011). The website is open-access and allows the user to make queries based upon protein name, gene name, or any protein identifier present in the IPI protein description. A key feature of SCOR is the ability to visualize quantitative information for transcripts, proteins, and PTMs from numerous sources, including large-scale analyses from multiple labs. To ensure that SCOR remains relevant, it is now possible to submit published data for inclusion on the website. We expect these types of resources to enable and expand the field.
Figure 6.
Proteomics resources for data sharing. (A) A table showing various online resources for storing and viewing proteomic data. A short description and link to respective websites are provided. (B) A screen shot depicting the SCOR interface. SCOR integrates transcriptomic, proteomic, and phosphoproteomic data for ease of visualization (Phanstiel et al., 2011). Color figures are available online.
Conclusion and outlook
The fields of proteomics and stem cell biology have rapidly evolved over the past ten years. Initial studies benchmarked stem cell proteomes, identifying proteins enriched in pluripotent cells. Meanwhile, improvements to large-scale quantitation have enabled comparative analyses between cell types. More recent experiments have tackled more specific biological questions and incorporated genomic techniques for systems-level analysis.
The accumulating body of work that incorporates proteomic techniques and stem cell biology speaks to the power and potential of this interdisciplinary work (Figure 4B). Looking to the future, a key challenge will be integrating these rapidly evolving and technically demanding fields to answer relevant biological questions. The experiments outlined here and elsewhere provide important information; however, generating a list of proteins is not a sufficient endpoint for a project, and follow-up experiments are, of course, required. In addition, the interdisciplinary nature of this work can be daunting, forcing scientists out of traditional comfort zones and into other areas of expertise. Initiatives like the Proteome Biology of Stem Cells Consortium help greatly in this regard and provide a crucial forum for scientists interested in both topics (Heck et al., 2007). Still, it will ultimately fall to collaborating researchers to adapt both biological and analytical technology to the greatest effect.
Acknowledgments
The authors wish to thank AJ Bureta for graphic design and Pengzhi Yu, Ron Stewart, Alicia Williams, and Krista Eastman for critical reading of this manuscript.
Footnotes
Declaration of interest
J.A.T. is a founder, stockowner, consultant and board member of Cellular Dynamics International (CDI), and serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures. JJC is a consultant for Thermo Finnigan. JB was funded through the NSF Graduate Research Fellowship and NIH traineeship (5T32GM08349). CMR was funded through the NSF Graduate Research Fellowship and NIH traineeship (T32GM008505).
References
- Ahn SM, Goode RJ, Simpson RJ. Stem cell markers: insights from membrane proteomics? Proteomics. 2008;8(23–24):4946–4957. doi: 10.1002/pmic.200800312. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210(2):517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144(3):439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309(5965):255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Brien GL, Bracken AP. Transcriptomics: unravelling the biology of transcription factors and chromatin remodelers during development and differentiation. Semin Cell Dev Biol. 2009;20(7):835–841. doi: 10.1016/j.semcdb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5(2):204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Phanstiel D, Coon JJ. Unraveling the histone’s potential: a proteomics perspective. Epigenetics. 2008;3(5):254–257. doi: 10.4161/epi.3.5.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25(14):6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5(1):111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng Z, Vallier L, Pedersen R. Activin/nodal signaling and pluripotency. Vitam Horm. 2011;85:39–58. doi: 10.1016/B978-0-12-385961-7.00003-2. [DOI] [PubMed] [Google Scholar]
- Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7(20):3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon JJ, Syka JE, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. Biotechniques. 2005;38(4):519, 521, 523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. International Journal of Mass Spectrometry. 2004;236(1–3):33–42. [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, King NL, Eng JK, Aderem A, Boyle R, Brunner E, Donohoe S, Fausto N, Hafen E, Hood L, Katze MG, Kennedy KA, Kregenow F, Lee H, Lin B, Martin D, Ranish JA, Rawlings DJ, Samelson LE, Shiio Y, Watts JD, Wollscheid B, Wright ME, Yan W, Yang L, Yi EC, Zhang H, Aebersold R. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6(1):R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do JT, Scholer HR. Regulatory circuits underlying pluripotency and reprogramming. Trends Pharmacol Sci. 2009;30(6):296–302. doi: 10.1016/j.tips.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Dormeyer W, van Hoof D, Braam SR, Heck AJ, Mummery CL, Krijgsveld J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J Proteome Res. 2008;7(7):2936–2951. doi: 10.1021/pr800056j. [DOI] [PubMed] [Google Scholar]
- Falkner JA, Falkner JW, Andrews PC. ProteomeCommons.org JAF: reference information and tools for proteomics. Bioinformatics. 2006;22(5):632–633. doi: 10.1093/bioinformatics/btk015. [DOI] [PubMed] [Google Scholar]
- Frese CK, Altelaar AF, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, Heck AJ, Mohammed S. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10(5):2377–2388. doi: 10.1021/pr1011729. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nature Methods. 2007;4(6):487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Kettenbach AN, Rush J, Gygi SP. The absolute quantification strategy: application to phosphorylation profiling of human separase serine 1126. Methods Mol Biol. 2007;359:71–86. doi: 10.1007/978-1-59745-255-7_5. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glish GL, Burinsky DJ. Hybrid mass spectrometers for tandem mass spectrometry. J Am Soc Mass Spectrom. 2008;19(2):161–172. doi: 10.1016/j.jasms.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8(11):R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics. 2007;6(11):1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- Graumann J, Hubner NC, Kim JB, Ko K, Moser M, Kumar C, Cox J, Scholer H, Mann M. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7(4):672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol. 2010;5(1):105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Gan CS, Zhang H, Zhu Y, Kon OL, Sze SK. Hybridization of pulsed-Q dissociation and collision-activated dissociation in linear ion trap mass spectrometer for iTRAQ quantitation. J Proteome Res. 2008;7(11):4831–4840. doi: 10.1021/pr800403z. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, Jaenisch R. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318(5858):1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Heck AJ, Mummery C, Whetton A, Oh S, Lee B, Pera M, Lemischka I, Krijgsveld J. Proteome biology of stem cells. Stem Cell Res. 2007;1(1):7–8. doi: 10.1016/j.scr.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136(4):509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5(2):351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, Taapken SM, Vereide DT, Montgomery KD, Zhang K, Gamm DM, Thomson JA. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011;108(16):6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jiang H, English AM. Quantitative analysis of the yeast proteome by incorporation of isotopically labeled leucine. J Proteome Res. 2002;1(4):345–350. doi: 10.1021/pr025523f. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16(4):182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- Keirstead HS, Nistor G, Bernal G, Totoiu M, Cloutier F, Sharp K, Steward O. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25(19):4694–4705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Shteynberg D. Software pipeline and data analysis for MS/MS proteomics: the trans-proteomic pipeline. Methods Mol Biol. 2011;694:169–189. doi: 10.1007/978-1-60761-977-2_12. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3(12):e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QR, Xing XB, Chen TT, Li RX, Dai J, Sheng QH, Xin SM, Zhu LL, Jin Y, Pei G, Kang JH, Li YX, Zeng R. Large scale phosphoproteome profiles comprehensive features of mouse embryonic stem cells. Mol Cell Proteomics. 2011;10(4):M110 001750. doi: 10.1074/mcp.M110.001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Liu S, Qu Y, Stewart TJ, Howard MJ, Chakrabortty S, Holekamp TF, McDonald JW. Embryonic stem cells differentiate into oligodendrocytes and myelinate in culture and after spinal cord transplantation. Proc Natl Acad Sci U S A. 2000;97(11):6126–6131. doi: 10.1073/pnas.97.11.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, Troyanskaya OG, Whetton AD, Lemischka IR. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462(7271):358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78(7):2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28(10):1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C, States D, Gevaert K, Vandekerckhove J, Apweiler R. PRIDE: the proteomics identifications database. Proteomics. 2005;5(13):3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9(6):625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, Bjorling E, Blagoev B, Bose R, Brahmachari SK, Burlingame AS, Bustelo XR, Cagney G, Cantin GT, Cardasis HL, Celis JE, Chaerkady R, Chu F, Cole PA, Costello CE, Cotter RJ, Crockett D, DeLany JP, De Marzo AM, DeSouza LV, Deutsch EW, Dransfield E, Drewes G, Droit A, Dunn MJ, Elenitoba-Johnson K, Ewing RM, Van Eyk J, Faca V, Falkner J, Fang X, Fenselau C, Figeys D, Gagne P, Gelfi C, Gevaert K, Gimble JM, Gnad F, Goel R, Gromov P, Hanash SM, Hancock WS, Harsha HC, Hart G, Hays F, He F, Hebbar P, Helsens K, Hermeking H, Hide W, Hjerno K, Hochstrasser DF, Hofmann O, Horn DM, Hruban RH, Ibarrola N, James P, Jensen ON, Jensen PH, Jung P, Kandasamy K, Kheterpal I, Kikuno RF, Korf U, Korner R, Kuster B, Kwon MS, Lee HJ, Lee YJ, Lefevre M, Lehvaslaiho M, Lescuyer P, Levander F, Lim MS, Lobke C, Loo JA, Mann M, Martens L, Martinez-Heredia J, McComb M, McRedmond J, Mehrle A, Menon R, Miller CA, Mischak H, Mohan SS, Mohmood R, Molina H, Moran MF, Morgan JD, Moritz R, Morzel M, Muddiman DC, Nalli A, Navarro JD, Neubert TA, Ohara O, Oliva R, Omenn GS, Oyama M, Paik YK, Pennington K, Pepperkok R, Periaswamy B, Petricoin EF, Poirier GG, Prasad TS, Purvine SO, Rahiman BA, Ramachandran P, Ramachandra YL, Rice RH, Rick J, Ronnholm RH, Salonen J, Sanchez JC, Sayd T, Seshi B, Shankari K, Sheng SJ, Shetty V, Shivakumar K, Simpson RJ, Sirdeshmukh R, Siu KW, Smith JC, Smith RD, States DJ, Sugano S, Sullivan M, Superti-Furga G, Takatalo M, Thongboonkerd V, Trinidad JC, Uhlen M, Vandekerckhove J, Vasilescu J, Veenstra TD, Vidal-Taboada JM, Vihinen M, Wait R, Wang X, Wiemann S, Wu B, Xu T, Yates JR, Zhong J, Zhou M, Zhu Y, Zurbig P, Pandey A. Human Proteinpedia enables sharing of human protein data. Nat Biotechnol. 2008;26(2):164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- McDonald JW, Liu XZ, Qu Y, Liu S, Mickey SK, Turetsky D, Gottlieb DI, Choi DW. Transplanted embryonic stem cells survive, differentiate and promote recovery in injured rat spinal cord. Nat Med. 1999;5(12):1410–1412. doi: 10.1038/70986. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7(7):540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of Mouse and Human Cells to Pluripotency Using Mature MicroRNAs. Cell Stem Cell. 2011;8(6):633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Mullin NP, Yates A, Rowe AJ, Nijmeijer B, Colby D, Barlow PN, Walkinshaw MD, Chambers I. The pluripotency rheostat Nanog functions as a dimer. Biochem J. 2008;411(2):227–231. doi: 10.1042/BJ20080134. [DOI] [PubMed] [Google Scholar]
- Nagano K, Taoka M, Yamauchi Y, Itagaki C, Shinkawa T, Nunomura K, Okamura N, Takahashi N, Izumi T, Isobe T. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 2005;5(5):1346–1361. doi: 10.1002/pmic.200400990. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Diaz EM, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110(3):815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nature Methods. 2007;4(9):709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29(2):124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17(1):42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6(4):382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci. 2004;117(Pt 7):1269–1280. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- Phanstiel D, Brumbaugh J, Berggren WT, Conard K, Feng X, Levenstein ME, McAlister GC, Thomson JA, Coon JJ. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(11):4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel D, Unwin R, McAlister GC, Coon JJ. Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry. Anal Chem. 2009;81(4):1693–1698. doi: 10.1021/ac8019202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, Stewart R, Thomson JA, Coon JJ. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nature Methods. 2011;8(10) doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimienta G, Chaerkady R, Pandey A. SILAC for global phosphoproteomic analysis. Methods Mol Biol. 2009;527:107–116. x. doi: 10.1007/978-1-60327-834-8_9. [DOI] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4(164):rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345(6277):686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344(6265):435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Schulz TC, Swistowska AM, Liu Y, Swistowski A, Palmarini G, Brimble SN, Sherrer E, Robins AJ, Rao MS, Zeng X. A large-scale proteomic analysis of human embryonic stem cells. BMC Genomics. 2007;8:478. doi: 10.1186/1471-2164-8-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Seuntjens E, Umans L, Zwijsen A, Sampaolesi M, Verfaillie CM, Huylebroeck D. Transforming Growth Factor type beta and Smad family signaling in stem cell function. Cytokine Growth Factor Rev. 2009;20(5–6):449–458. doi: 10.1016/j.cytogfr.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Loboda A, Ens W, Standing KG. MALDI quadrupole time-of-flight mass spectrometry: a powerful tool for proteomic research. Anal Chem. 2000;72(9):2132–2141. doi: 10.1021/ac9913659. [DOI] [PubMed] [Google Scholar]
- Skalnikova H, Motlik J, Gadher SJ, Kovarova H. Mapping of the secretome of primary isolates of mammalian cells, stem cells and derived cell lines. Proteomics. 2011;11(4):691–708. doi: 10.1002/pmic.201000402. [DOI] [PubMed] [Google Scholar]
- Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nature Methods. 2008;5(11):959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(4):995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syka JE, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3(3):621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D, Heck AJ, Krijgsveld J, Mummery CL. Proteomics and human embryonic stem cells. Stem Cell Res. 2008;1(3):169–182. doi: 10.1016/j.scr.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5(2):214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, Heck AJ, Mummery CL, Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5(7):1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75(18):4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre-and post-search proteomics software tools for OMSSA. Proteomics. 2011;11(6):1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3(2):196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8(3):326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]