Abstract
STAT1 mediates response to interferons and regulates immunity, cell proliferation, apoptosis, and sensitivity of Fanconi Anemia cells to apoptosis after interferon signaling; the roles of STAT1 in embryos, however, are not understood. To explore embryonic functions of STAT1, we investigated stat1b, an unstudied zebrafish co-ortholog of human STAT1. Zebrafish stat1a encodes all five domains of the human STAT1-alpha splice form but, like the human STAT1-beta splice variant, stat1b lacks a complete transactivation domain; thus, two unlinked zebrafish paralogs encode protein forms translated from two splice variants of a single human gene, as expected by subfunctionalization after genome duplication. Phylogenetic and conserved synteny studies showed that stat1b and stat1a arose as duplicates in the teleost genome duplication (TGD) and clarified the evolutionary origin of STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6 by tandem and genome duplication. RT-PCR revealed maternal expression of stat1a and stat1b. In situ hybridization detected stat1b but not stat1a expression in embryonic hematopoietic tissues. Morpholino knockdown of stat1b, but not stat1a, decreased expression of the myeloid and granulocyte markers spi and mpo and increased expression of the hematopoietic progenitor marker scl, the erythrocyte marker gata1, and hemoglobin. These results suggest that zebrafish Stat1b promotes myeloid development at the expense of erythroid development.
Keywords: Stat1α, Stat1β, hematopoiesis, myeloid, granulocyte, zebrafish, genome duplication, subfunctionalization, Fanconi anemia
1. Introduction
Blood cell lineages arise by lineage commitment and cell maturation. The helix-loop-helix transcription factor SCL/TAL1 causes hemangioblasts to become hematopoietic stem cells (HSC) and mice lacking SCL activity fail in embryonic erythropoiesis (Forrai and Robb, 2003; Shivdasani et al., 1995; Xiong, 2008). HSCs develop into erythroid, myeloid, and lymphoid lineages marked by lineage-specific transcription factors, including GATA1 for erythroid differentiation (Crispino, 2005; Fujiwara et al., 1996), SPI1 (PU.1) for the myeloid lineage (Kastner and Chan, 2008; Oikawa et al., 1999), and IKAROS for lymphoid differentiation (Georgopoulos et al., 1997; Schmitt et al., 2002). Although much is known, we lack details concerning the networks of factors that regulate these cell fate-marking genes.
Proinflammatory cytokines, such as IFN-gamma, can impact hematopoietic development (Baldridge et al., 2010). IFN-gamma binds its receptor, thus activating JAK proteins, which phosphorylate STAT (signal transducer and activator of transcription) proteins that then translocate to the nucleus where they bind DNA and activate expression of IFN-stimulated genes (Schindler et al., 1992; Stark et al., 1998). IFN-gamma, for example, promotes the proliferation of repopulating HSCs and this response requires STAT1 (Baldridge et al., 2010; Zhao et al., 2010). Furthermore, about half of AML (acute myeloid leukemia) patients have constitutive STAT1 activity (Aronica et al., 1996; Gouilleux-Gruart et al., 1997; Weber-Nordt et al., 1996). Mammals have seven STAT genes (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6) but their historical origins have not yet been fully explained.
STAT1 transduces signals in response to a variety of cytokines and growth factors (Najjar et al., 2010; Najjar and Fagard, 2010) and regulates a number of cellular functions, mainly in the immune system. In addition to type I and type II IFNs, which use STAT1 to fight against opportunistic virus infections (Durbin et al., 1996; Meraz et al., 1996), growth factors including EGF (Chin et al., 1996) and PDGF (Vignais et al., 1996) can activate STAT1. Moreover, STAT1 is activated by numerous interleukins (ILs) that regulate growth of various white blood cells (Najjar and Fagard, 2010). cytokines [69]. In mouse and human, Fanconi Anemia mutant hematopoietic progenitor cells are hypersensitive to IFN signaling due to increased apoptosis, and this is mediated by STAT signaling (Haneline et al., 1998; Li et al., 2004; Otsuki et al., 1999; Pang et al., 2000; Rathbun et al., 1997; Si et al., 2006; Whitney et al., 1996; Zhang et al., 2004).
Human STAT1 produces two splice variants that differ at their carboxy terminus. STAT1-alpha contains a TAZ2-binding, trans-activation domain (TAD) that is missing from the shorter, STAT1-beta isoform, which acts as a dominant negative regulator of STAT1 function (Bromberg et al., 1996). STAT1-beta inhibits the apoptotic effect of STAT1-alpha in B lymphocytes (Baran-Marszak et al., 2004) but induces cell death in B-cells by a p53-independent mechanism (Najjar et al., 2008). Other functions of these two splice forms have yet to be identified.
Zebrafish provides a good model for the investigation of the conserved functions of STAT1 and vertebrate hematopoiesis (Bertrand and Traver, 2009; Ciau-Uitz et al., 2010; Ellett and Lieschke, 2010; Paik and Zon, 2010). Transcriptional mechanisms of both primitive (embryonic) and definitive (adult) hematopoiesis are evolutionarily conserved among vertebrates but the anatomical location varies among species (de Jong and Zon, 2005). In zebrafish, primitive hematopoiesis begins at 10 hours post-fertilization (hpf) when the ventral lateral plate mesoderm gives rise to embryonic HSCs, which occupy the intermediate cell mass (ICM) and differentiate mainly into pro-erythroblasts, and the rostral blood island (RBI), which produces mostly macrophages (Davidson et al., 2003; Long et al., 1997). At 24 hpf, about 300 erythrocytes enter the blood stream from the ICM (Patterson et al., 2005), and several hours later, definitive HSCs begin to appear in the ventral wall of the dorsal aorta (the aorta-gonad-mesonephros, AGM) (Burns et al., 2005). Hematopoiesis continues in the AGM until a few hours before hatching, when the kidney marrow initiates lifelong definitive hematopoiesis (Traver, 2004)).
While STAT1 functions have been well studied in mammalian tissue culture cells, comparatively little work has been directed toward the roles of STAT1 in intact developing vertebrate embryos. Zebrafish has several advantages for the investigation of STAT1 in living embryos. First, as in mammals, zebrafish responds to LPS (the bacterial cell wall component lipopolysaccharide) by increased expression of IL1 and TNF (tumor necrosis factor) (Sullivan et al., 2009). Second, zebrafish stat1a can rescue interferon signaling-mediated cell growth inhibition in a STAT1-deficient human cell line (Oates et al., 1999b). Third, zebrafish conserves a role of jak2a in hematopoiesis (Oates et al., 1999a), raising the hypothesis that it may act upstream of zebrafish stat1. Fourth, due to the teleost genome duplication (TGD) at the base of the teleost radiation, many single copy genes in human have two orthologs in zebrafish (Amores et al., 1998; Amores et al., 2004; Jaillon et al., 2004; Postlethwait et al., 1998; Taylor et al., 2003). Such duplicates often share between them ancestral gene functions that are conserved in a single orthologous gene in mammals (Force et al., 1999; Postlethwait et al., 2004). Thus, knockdown of just one of the two duplicates can reveal gene functions that might be difficult to identify in mammals due to pleiotropy. We capitalized on this approach to investigate the functions of two zebrafish STAT1 co-orthologs and thereby provide new insights into the roles of STAT1 in hematopoietic development. Here we show that stat1b arose in the teleost genome duplication event, that it is expressed in the embryonic hematopoietic compartment, and that it promotes development of the myeloid lineage at the expense of the erythroid lineage.
2. Results
2.1. Zebrafish has two genes related to human STAT1
Because about a quarter of human genes have two co-orthologs in zebrafish due to the teleost genome duplication (Postlethwait et al., 1998), we searched for a zebrafish duplicate of the previously described stat1a gene (Oates et al., 1999b). A BLAST search (Altschul and Koonin, 1998) of stat1a (GenBank: M97936) against the zebrafish EST database identified EST RK103A3G02.T3 (GenBank: CD596714), which was isolated from adult zebrafish kidneys, the site of definitive hematopoiesis (Song et al., 2004), and had just 78% identity at the amino acid level with stat1a; this low level of identity suggested that these ESTs were from two different genes. A search of CD596714 against the Ensembl zebrafish genome hit a genomic sequence (ENSDARG00000076182, GenBank: XR_029517) that we now call stat1b. We designed primers to amplify overlapping fragments of the gene model and used them to amplify gene fragments from cDNA extracted from 24 – 48 hpf wild-type zebrafish embryos. We sequenced these fragments and assembled them to reconstruct the sequence of a complete cDNA (GenBank: FJ986224).
Comparing the structure of our 3,569 bp-long stat1b cDNA with genomic DNA at Ensembl revealed 23 exons. Both zebrafish stat1 genes appeared to lack the first of two untranslated exons in human STAT1 (Fig. 1A), consistent either with the loss of the first untranslated exon in the zebrafish lineage or the gain of this exon in the human lineage. A BLASTX search of the human genome with either our new stat1b gene or the previously known stat1a gene returned STAT1 as the most similar human gene, and reciprocally, a BLASTP search of zebrafish sequences with the human STAT1 protein hit zebrafish Stat1a and Stat1b as the two most similar sequences, as would be expected if stat1a and stat1b were co-orthologs of STAT1.
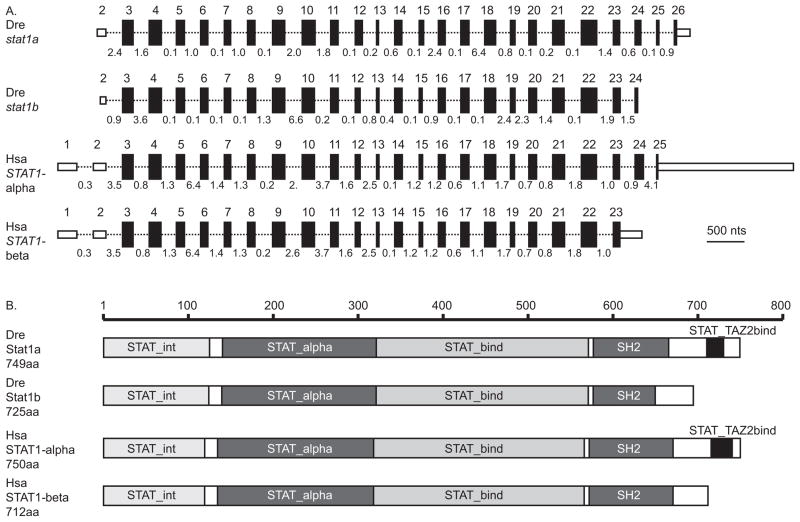
Figure 1.
A comparison of zebrafish and human STAT1-related genes and proteins. A. The structure of STAT1-related genes. Exons are represented as boxes; small, unfilled boxes represent 3′ and 5′ UTRs and filled boxes represent translated regions. The length of each box is proportional to the length of each exon. Introns are represented as thin dotted lines connecting boxes; the length of each intron is indicated in kilobases but introns are not drawn to scale. B. The structure of STAT1-related proteins. Domains that characterize STAT proteins are marked on diagrams representing lengths in amino acid residues according to the scale bar for proteins encoded by the two zebrafish genes stat1a and stat1b and the two human STAT1 splicing isoforms STAT1-alpha and STAT1-beta. White boxes represent linker regions between domains. The human STAT1-alpha splice form and the zebrafish Stat1a protein possess a trans-activation domain lacking from the human STAT1-beta splice form and the zebrafish Stat1b protein. This trans-activation domain is approximately 20 amino acids long and it binds selectively to the TAZ2 domain of CREB-binding protein to act as a transcriptional activator. Abbreviations: Dre, Danio rerio; Hsa, Homo sapiens; SH2, Src homology-2 domain; STAT_alpha, alpha helical, coiled-coil domain; STAT_bind, DNA binding domain; STAT_int, STAT dimer interaction stabilization domain; STAT_TAZ2bind, trans-activation domain.
2.2. Conserved domains in zebrafish Stat1 proteins
The protein predicted from our stat1b cDNA sequence contains 725 amino acids (GenBank: ACR83062). The Conserved Domain Database (Marchler-Bauer et al., 2009) showed that the translated Stat1b peptide has all but one of the five domains that are required for STAT function (Brierley and Fish, 2005). Conserved domains (Fig. 1B) include (1) the N-terminal 130 amino acids that constitute the STAT dimer interaction stabilization domain (STAT_int) (Vinkemeier et al., 1998); (2) a coiled-coil domain (STAT_alpha, at about residues 135 – 315) that contains four alpha helices with hydrophilic surfaces that mediate interactions with non-STAT1 alpha-helical proteins (Becker et al., 1998); (3) the DNA binding domain (DBD or STAT_bind, at about residues 320 – 480) consisting of a beta-barrel that determines DNA binding specificity (Chen et al., 1998); (4) the phosphotyrosine-binding SH2 domain (SH2, at about residues 580–680) that recognizes phosphorylated tyrosine motifs in cytokine receptors, JAKs, and other STAT proteins (Kisseleva et al., 2002). Stat1b, however, lacks (5) the C-terminal trans-activation domain (TAD or STAT1_TAZ2bind, at about residues 723–747 in zebrafish Stat1a) that binds TAZ2 domains and mediates interactions with cofactors that facilitate transcriptional activation (Decker and Kovarik, 2000) (Fig. 1B and Supplementary Fig. S1). This domain analysis showed that zebrafish Stat1b has the domain structure of the human STAT1-beta splice variant and that zebrafish Stat1a has the domain structure of the human STAT1-alpha splice variant. This domain distribution would be expected if the ancestral STAT1 protein had all five domains and that in the zebrafish lineage, the TAZ2bind domain was preserved by the stat1a gene but was lost by the stat1b gene, while in the human lineage, the TAZ2bind domain was preserved by the single STAT1 gene and that differential splicing provides the two isoforms, one with and one without the TAZ2bind domain; thus, two different molecular genetic mechanisms provide a pair of structurally similar isoforms in zebrafish and human.
2.3. Functional amino acid residues in Stat1b
Protein alignments showed that zebrafish Stat1b and Stat1a proteins generally conserve amino acids required for STAT1 function (Brierley and Fish, 2005) (Supplementary Fig. S1). In human STAT1, an arginine at position 31 becomes methylated (Mowen et al., 2001), and this site is conserved in zebrafish Stat1a and Stat1b proteins. The human DBD requires K366, V426, and T427, which correspond to K372, V432, and T433 in zebrafish Stat1b, and these residues are also conserved in Stat1a. Crystal structure of human STAT1 shows that H431, R378, and V457 are in positions critical for DNA binding (Chen et al., 1998), and these are all conserved in both zebrafish Stat1a and Stat1b proteins. Although STAT1 bound to DNA shows N460 located in the major groove, its mutation did not block IFN-gamma-induced binding of STAT1 to DNA (Yang et al., 2002) and, correspondingly, this residue is conserved in Stat1a but not Stat1b. R602 and P633, which are required for activity of the SH2 domain (Brierley and Fish, 2005; Improta et al., 1994), are conserved in both Stat1b and Stat1a. In the C-terminal region, the phosphorylation site Y701 in the human STAT1 protein is conserved in zebrafish Stat1a but not in Stat1b (Supplementary Fig. S1). Finally, the phosphorylated serine S727 in the TAZ2bind transactivation domain of human STAT1alpha is conserved in Stat1a and a serine is also present in a similar position in Stat1b despite its greatly disrupted TAZ2bind domain. We conclude that both zebrafish Stat1 proteins generally conserve protein domains shown to be functional in human STAT1, except for the C-terminal transactivation domain that is greatly disturbed in zebrafish Stat1b and the human STAT1beta splice form.
2.4. Zebrafish STAT1 co-orthologs originated in the teleost genome duplication
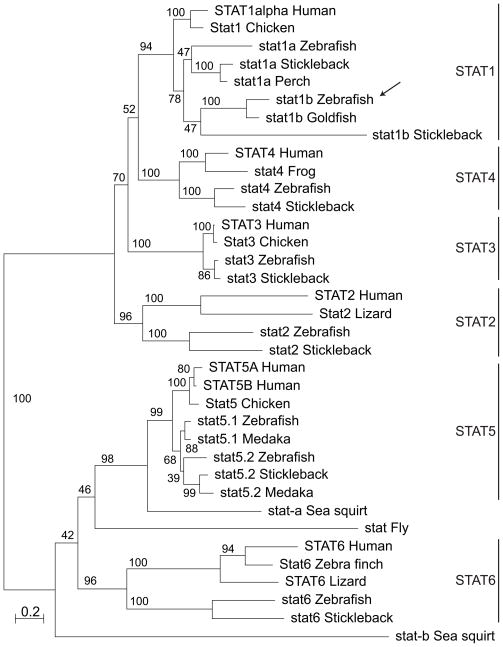
To help understand the origin of stat1b, we conducted a phylogenetic analysis of full-length STAT proteins using maximum likelihood (Guindon and Gascuel, 2003). The seven human STAT proteins occupied six well-supported clades (Fig. 2). The zebrafish Stat1b protein fell snugly into the STAT1 clade with good bootstrap support. The sister clade to zebrafish stat1b was represented by a highly similar sequence in goldfish, which is a cyprinid like zebrafish. Stickleback, a percomorph fish, contained a rapidly evolving sequence related to stat1b, although other sequenced percomorphs, including medaka and two pufferfish, appeared to lack a stat1b ortholog. The stat1b clade branched as a sister to the stat1a clade, which included sequences from several percomorphs. This tree morphology is predicted by the hypothesis that stat1a and stat1b arose in a gene duplication event after the divergence of ray-fin and lobe-fin fish but before the teleost radiation, a situation consistent with the hypothesis that these two paralogs arose in the teleost genome duplication event.
Figure 2.
Phylogenetic analysis of vertebrate STAT proteins: a maximum likelihood tree. The arrow indicates the zebrafish Stat1b protein, which groups as expected if the Stat1b and Stat1a clades arose in the teleost genome duplication event. Bootstrap values as a percent of 100 repetitions. Abbreviations and gene designators (those beginning with ENS are from Ensembl): Chicken (Gallus gallus) Stat1 (ENSGALG00000007651), Stat3 (ENSGALG00000003267), Stat5 (ENSGALT00000039892); Fly (Drosophila melanogaster) Stat92E-RG; Frog (Xenopus tropicalis): stat4 (ENSXETT00000010127); Goldfish (Carassius auratus) stat1b (GenBank: AAO88245); Human (Homo sapiens) STAT1alpha (ENST00000361099), STAT2 (ENST00000314128), STAT3 (ENST00000264657), STAT4 (ENST00000358470), STAT5A (ENST00000345506), STAT5B (ENST00000293328), STAT6 (ENST00000300134); Lizard (Anolis carolinensis): STAT2 (ENSACAG00000016385); STAT6 (ENSACAG00000009680); Medaka (Oryzias latipes) stat5.1 (ENSORLT00000004951), stat5.2 (ENSORLT00000017980); Perch (Siniperca chuatsi) stat1a (GenBank: ACU12484) Sea squirt (Ciona intestinalis) stat-a (ENSCINT00000020503), stat5 (ENSCING00000004044); Stickleback (Gasterosteus aculeatus) stat1a (ENSGACT00000020606), stat1b (ENSGACT00000003511), stat2 (ENSGACG00000000707), stat3 (ENSGACT00000011411), stat4 (ENSGACG00000002684), stat5 (ENSGACG00000015405), stat6 (ENSGACG00000008477); Zebra Finch (Taeniopygia guttata) Stat6 (ENSTGUG00000015716); Zebrafish (Danio rerio) stat1a (ENSDARG00000006266), stat1b (ENSDART00000059638), stat2 (ENSDART00000050682), stat3 (ENSDART00000022006), stat4 (ENSDART00000105764), stat5.1 (ENSDARG00000019392), stat5.2 (ENSDARG00000055588), stat6 (ENSDART00000059346).
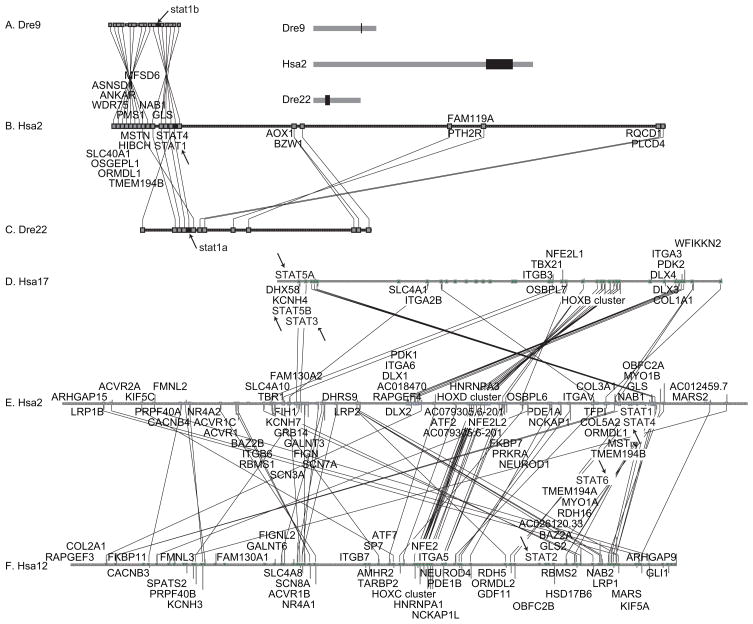
To confirm the affinities of the stat1b clade (78% bootstrap support), we explored conserved syntenies around stat1b using the Synteny Database (Catchen et al., 2009) and the zebrafish genome at Ensembl. The hypothesis that stat1a and stat1b arose in the TGD predicts that zebrafish should have two chromosomal segments co-orthologous with the region on human chromosome 2 (Hsa2) surrounding STAT1. Figure 3 shows that orthologs of genes surrounding STAT1 (Fig. 3B) appear in two locations in the zebrafish genome, one on Danio rerio linkage group 9 (Dre9, Fig. 3A) and the other on Dre22 (Fig. 3C). STAT1 lies adjacent to STAT4 on Hsa2 and the two genes are transcribed in the same direction. Likewise, on Dre9, stat1b resides next to stat4 in the same orientation. Except for an inversion, the human orthologs of an additional 15 genes surrounding stat1b are located in the same order and direction surrounding STAT1 on Hsa2 (compare Fig. 3A and 3B). Zebrafish stat1a lies on Dre22 near ten other genes whose human orthologs are located near STAT1. This double syntenic conservation, coupled with the phylogenetic analysis showing that stat1b and stat1a occupy sister clades, strongly supports the conclusion that stat1b and stat1a arose in the TGD. Furthermore, the conserved synteny of eight of eleven genes in a row (including stat4) in the stickleback and zebrafish genomes confirms the orthology of stat1b in these two species despite the rapid evolution of the stickleback gene (Supplemental Fig. S3).
Figure 3.
Conserved syntenies verify the history of the stat1 gene family. A. Conserved syntenies around zebrafish stat1b on zebrafish chromosome Dre9 were searched in the human genome using the Synteny Database (Catchen et al., 2009). B. Extensive conserved syntenies were found around STAT1 on human chromosome Hsa2. Lines between large boxes connect orthologs; small boxes represent genes without orthologs in the diagramed regions. C. Conserved syntenies around human STAT1 also appeared around zebrafish stat1a on Dre22. Chromosomal regions shown in black rectangles in the insert to the right of part A are blown up in parts A, B, and C. Arrows indicate the positions of stat genes. Note that on Hsa2 and Dre9 STAT1/stat1 and STAT4/stat4 reside adjacent to each other. Genes (indicated in the order: HUMAN gene, dre9 gene, dre22 gene): STAT4, stat4; STAT1, stat1b, stat1a; GLS, gls, gls; NAB1, NP_001116745.1, si:dkeyp-84a8.1; TMEM194B, B0UYT4; MFSD6, zgc:92925, si:dkey-188p4.2; HIBCH, hibch; MSTN, mstn, gdf8; PMS1, pms1; ORMDL1, ormdl1; OSGEPL1, osgepl1; ANKAR, ankar; slc40a1, slc40a1; WDR75, wdr75. The following genes are HUMAN, dre22gene: RQCD1, rqcd1; PLCD4, plcd4a; PTH2R, si:dkeyp-4h4.1; FAM119A, zgc:110528; FAM119A; AOX1, aox1; BZW1, bzw1a. D–F. Human STAT genes occupy paralogy groups that appear to have originated in the R1 and R2 rounds of genome duplication that preceded the vertebrate radiation. Human paralogs of genes residing in the 20 Mb region of Hsa2 surrounding STAT1 between 180 and 200 Mb were plotted on the rest of the human chromosomes with gene order according to the location on human chromosome 2 (Hsa2) using the Dotplot feature of the Synteny database (Catchen et al., 2009). Results showed that Hsa2 genes, including STAT1 and STAT4, showed extensive paralogy with Hsa12, which contains STAT2 and STAT6, with Hsa17, which contains STAT3, STAT5A and STAT5B (arrows), and with Hsa7, which lacks any STAT genes and is thus omitted from the figure, but has many other paralogs closely linked to STAT genes, e.g., potassium channel family H (KCNH2, 7q36.1; KCNH7, 2q24.2; KCNH3 12q13; KCNH4, 17q21.2), the HOX clusters (HOXA4, 7p15.2; HOXD4, 2q31.1; HOXC4, 12q13.3; HOXB4, 17q21.32), erythroid-derived nuclear factor-2 (NFE2L3, 7p15.2; NFE2L2, 2q31; NFE2, 12q13; NFE2L1, 17q21.3), and many other gene families.
The second copy of stat4 that would have been produced by the TGD and should lie next to stat1a appears to be missing from the zebrafish genome. Relative to Dre9 and the human STAT1 region, an inversion breakpoint is in the location expected for the duplicate copy of stat4 on Dre22, suggesting that a chromosome break occurred in this location after the TGD. This break could have initiated the destruction of this stat4 copy. The loss of the stat4 duplicate on the ancestral chromosome segment of Dre22 would be expected to have had no phenotypic penalty if its functions were fully redundant with the stat4 copy retained on Dre9. Because gene orientation in both zebrafish Dre9 and human Hsa2 is 5′-stat4-stat1–3′, an inversion between the genes could have disrupted cis-acting regulatory elements located in the intergenic region, thus resulting in the loss of ancestral regulatory elements originally associated with the ancestral stat1a and its originally adjacent stat4 gene.
2.5. History of the vertebrate STAT gene family
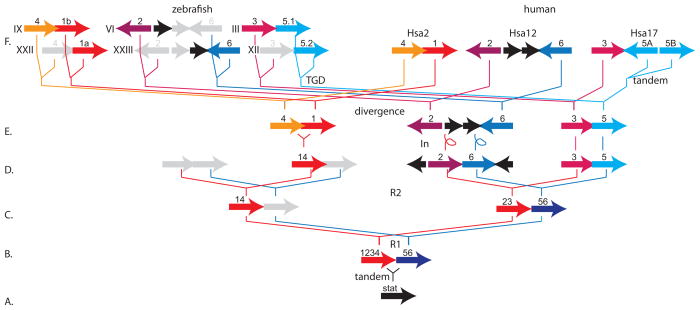
Phylogenetic and conserved synteny analysis help infer the origins of the entire STAT family. In a maximum likelihood tree, the seven human STAT genes (STAT1, 2, 3, 4, 5A, 5B, and 6) fell into six well-supported clades reflected by their names (Fig. 2). Two stat genes from the sea squirt Ciona intestinalis, a Urochordate representing the sister group to the vertebrates, and a stat-related gene from fruit fly fell in the phylogeny unexpectedly in the STAT5 and STAT6 clades rather than as outgroups. These locations may be artifacts due to the long branches of these taxa. The six vertebrate STAT clades further subdivided into two strongly supported subclades, one containing STAT1, 2, 3, and 4, and the other containing STAT5 and 6 (Fig. 2). This evidence suggests that an ancestral STAT gene experienced an early gene duplication event that produced a STAT1234 gene and a STAT56 gene (Fig. 4A, B). Because the seven human STAT genes occur in three gene clusters (Fig. 3E–F), this ancient event was probably a tandem duplication. Use of the Synteny Database (Catchen et al., 2009) showed that the three STAT clusters appear on Hsa2 (STAT1 and STAT4), Hsa12 (STAT2 and STAT6), and Hsa17 (STAT3, STAT5A, and STAT5B) in genome neighborhoods that contain many paralogous genes, including three HOX clusters (HOXD, HOXC, and HOXB, respectively). These genome neighborhoods, along with the HOXA region on Hsa7, which lacks any STAT family genes, arose in two rounds of whole genome duplication at the base of the vertebrate radiation (Dehal and Boore, 2005; Holland et al., 1994; Ohno, 1970)(Fig. 4C, D). STAT4 and STAT1 are adjacent and transcribed in the same direction in human and stat4 and stat1b in zebrafish share this genomic arrangement (Fig. 4F). The fact that the STAT1 and STAT4 clades are both in the STAT1234 clade with strong bootstrap support and are sister groups (although with low bootstrap support, Fig. 2) suggests that STAT1 and STAT4 arose by tandem duplication after the second round of vertebrate genome duplication R2 and that this chromosome’s STAT56 paralog was lost (Fig. 4E). Because STAT2 and STAT6 are spaced 18 genes apart in the GRCCh37 version of the human genome and are transcribed in opposite orientations with respect to stat3 and stat5.1 in zebrafish, inversions must have separated them after the R2 event (Fig. 4D, E). After the divergence of the zebrafish and human lineages (Fig. 4E, F), the TGD event would have duplicated the ancestral gene content in the zebrafish lineage. After the TGD, gene loss continued, as duplicate copies of stat4, stat2, stat6, and stat3 were lost (Fig. 4F). Finally, after the human lineage diverged from the chicken lineage, a tandem duplication event produced STAT5A and STAT5B (Fig. 4F). Note that stat5.1 and stat5.2 in zebrafish are both co-orthologous to both STAT5A and STAT5B in human due to independent duplications in each lineage (Lewis and Ward, 2004).
Figure 4.
A model for the origin of vertebrate STAT genes. A. The single STAT gene in the last common ancestor of all vertebrates. B–E. Sequential steps of genome duplication and gene loss (gray arrows). F. Current arrangements of STAT genes in today’s zebrafish and human genomes. See text for more explanation. Abbreviations: 1234, the ancestor of STAT1, STAT2, STAT3, and STAT4; 14, the ancestor of STAT1 and STAT4; 56, the ancestor of STAT5 and STAT6; divergence, the speciation event that separated the human and zebrafish lineages; In, chromosomal inversions; R1, R2, two sequential rounds of whole genome duplication at the base of vertebrate origins; roman numerals, zebrafish chromosomes; tandem, a tandem duplication event; TGD, teleost genome duplication event.
2.6. stat1a and stat1b are both expressed in early zebrafish development
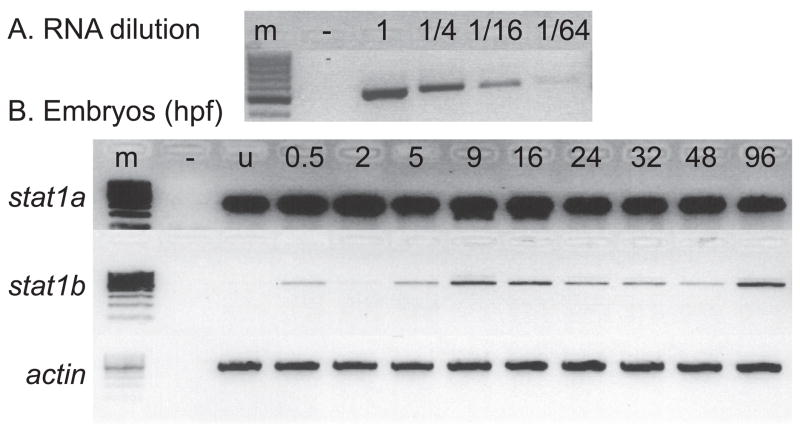
Having established the historical relationships of the vertebrate STAT gene family, we turned to the investigation of stat1 functions. First, we followed the developmental time course of stat1a and stat1b expression by RT-PCR using total RNA samples from zebrafish embryos of various ages. To calibrate this semi-quantitative technique, we made a dilution series of the RNA template and amplified with stat1a or stat1b primers or with actin primers for loading control and amplified for just 21 cycles of PCR to avoid saturation. Results showed that under these conditions, band intensity depended on template concentration (Fig. 5A). Use of the same conditions on embryo RNAs revealed strong and steady expression of stat1a from 0.5 hpf (two-cell stage) until 96 hpf (= 2 day post hatching) (Fig. 5B). We conclude that stat1a is expressed maternally followed by persistence of the transcript and that the zygotic stat1a gene is expressed later, verifying previous results (Oates et al., 1999b). For stat1b, transcript levels were low in two-cell embryos (0.5hpf) and then disappeared during cleavage (2 hpf); but then by 5 hpf, about two hours after the mid-blastula transition when transcription commences from zygotic genes (Kane and Kimmel, 1993), quantities of stat1b mRNA began to rise, dipped somewhat after 24 hpf, and then rose again at 96 hpf. We conclude that stat1a is expressed maternally and that both stat1a and stat1b are expressed zygotically through embryogenesis.
Figure 5.
stat1 gene expression assayed by RT-PCR. A. Calibration series. A gradient of RNA dilutions and 21 amplification cycles with stat1b primers verified a semi-quantitative analysis of target mRNAs. B. Detection of stat1a and stat1b expression using RT-PCR at various stages of development using beta actin expression as loading control. RNA samples were extracted from zebrafish embryos at the indicated ages. Abbreviations: –, negative control with no RNA added; u, unfertilized eggs; 0.5, 2, etc., ages at hours post fertilization (hpf).
2.7. Hematopoietic tissues express stat1b
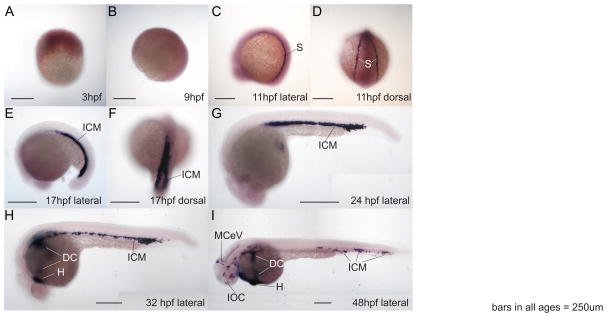
To verify the RT-PCR experiments and to learn which tissues express stat1 transcripts, we explored gene expression by whole mount in situ hybridization. In situ hybridization experiments revealed that stat1a is expressed ubiquitously at a low level throughout the embryo (data not shown), confirming the previous report (Oates et al., 1999b). In contrast to stat1a, stat1b showed strong and restricted expression by in situ hybridization. Beginning at the 2-somite stage (about 11 hpf), signal for stat1b appeared strongly and specifically in bilateral stripes in the lateral plate mesoderm (Fig. 6A–D). These stripes eventually migrated medially and fused to form the intermediate cell mass (ICM) by the 18-somite stage (17hpf, Fig. 6E, F). This pattern is similar to that shown by gata1, where at 16 hpf, gata1-expressing cells mark stripes of erythroid progenitors (de Jong and Zon, 2005). At 24 hpf, stat1b expression appeared in the ICM (Fig. 6G), the location of erythropoiesis at this stage (de Jong and Zon, 2005). Later, at 32 hpf and 48 hpf, stat1b expression broke up into islands in the ventral vasculature and advanced to the heart and the ducts of Cuvier, which form after 25 hpf and contain both erythrocytes and myeloid cells (Fig. 6H, I). The expression pattern of stat1b mimics those of hematopoietic marker genes, including gata1, gata2 and hbbe (Detrich et al., 1995).
Figure 6.
Whole mount RNA in situ hybridization shows specific expression pattern of stat1b in zebrafish hematopoietic regions. A, B. 3 hpf and 9 hpf embryos, respectively, did not show specific expression of stat1b. C, D. At 11 hpf (2 somites), stat1b was expressed in stripes flanking the paraxial mesoderm, shown in lateral and dorsal views, respectively. E, F. At 17 hpf (18 somites), stat1b was expressed in the intermediate cell mass, shown in lateral and dorsal views, respectively. G. By 24 hpf, stat1b expression stripes had merged centrally. H. At 32 hpf, the stat1b expression compartment had spread from the intermediate cell mass to the heart and ducts of Cuvier on the yolk. I. At 48 hpf, the expression of stat1b continued in hematopoietic regions and veins. Abbreviations: dc, ducts of Cuvier; h, heart; icm, intermediate cell mass; IOC, inner optic circle; MCeV, middle cerebral vein (Isogai et al., 2001); s, stripes flanking the paraxial mesoderm.
2.8. stat1b functions in hematopoietic cell lineage specification
The early and specific expression of stat1b in the ICM suggested that stat1b might play a role in primitive hematopoietic development. To test this hypothesis, we knocked down activity of stat1a and stat1b in developing zebrafish embryos by injecting early cleavage embryos with either stat1a or stat1b antisense morpholino oligonucleotide (MO) directed against intron/exon borders and then queried various blood cell lineages by investigating the expression of lineage-specific gene markers. We first verified the efficacy of the splice-inhibiting MOs by RT-PCR of cDNA extracted from 24–48 hpf embryos that were injected with MO at the 1–2 cell stage. Results showed that the stat1a and stat1b MOs were both effective inhibitors of transcript splicing (Supplementary Fig. S2). Translation of the unspliced transcript into intron-3 of stat1a predicted a premature in-frame stop codon at 127 residues of the 749 amino acid protein and translation reading into intron-5 of stat1b predicted a premature stop codon at 201 amino acids of the full length 725 residue protein.
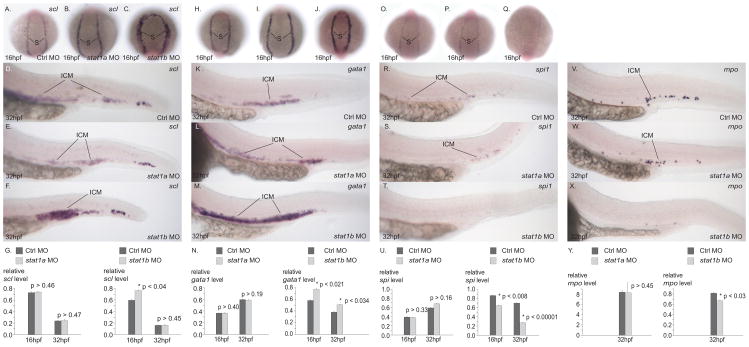
To test whether stat1b plays a role in the development of hematopoietic stem cells (HSC), we investigated expression of scl (tal1) (Liao et al., 1998) in stat1a and stat1b knockdown animals. Results showed that 16 hpf and 32 hpf embryos injected with stat1a MO showed the same expression compartment of scl as those injected with control MO (Fig. 7A, B). In contrast, 16 hpf and 32 hpf embryos injected with stat1b MO contained a substantially broader band of scl expression than control-MO injected animals (Fig. 7C). To quantitate levels of scl expression, we performed quantitative PCR on RNA extracted from control and treated embryos. Results confirmed that knockdown of stat1a had no effect on scl transcript levels, but that stat1b knockdown resulted in significantly higher levels of scl expression than in controls, although the effect had diminished by 32 hpf (Fig. 7G). We conclude that stat1b, but not stat1a, normally limits the width of the striped compartment of scl-expressing hematopoietic progenitor cells and the amount of scl expression in 16 hpf embryos.
Figure 7.
Gene knockdowns investigate the roles of stat1a and stat1b in hematopoiesis. In situ hybridization with probes for scl (A–G), gata1 (H–N), spi1 (O–U), and mpo (V–Y) in 16 hpf embryos (A–C, H–J, O–Q) and 32 hpf embryos (D–F, K-M, R–T, V–X) injected with control sequence MO (A, D, H, K, O, R, V), stat1a MO (B, E, I, L, W), and stat1b MO (C, F, J, M, X). Quantitative PCR experiments (G, N, U, Y). Knockdown of stat1a did not alter the expression of the genes tested, but knockdown of stat1b led to increased expression of scl and gata1, markers of HSCs and erythrocytes, and to depressed expression of spi1 and mpo, markers of the myelocyte lineage. These results would be expected if normal stat1b activity helped promote a lineage switch from the erythroid to the myeloid lineage.
After the specification of hematopoietic stem cells, the primitive erythroid lineage expresses gata1 (Detrich et al., 1995). Knockdown of stat1a had little effect on gata1 expression at 16 hpf or 32 hpf (Fig. 7H, I, K, L), but knockdown of stat1b activity resulted in broader and denser gata1 expression compartments at both stages (Fig. 7J, M). Q-PCR experiments confirmed these results (Fig. 7N). These experiments suggest that stat1b normally suppresses the extent of the gata1 expression compartment. If stat1b activity inhibits the formation of gata1 expressing cells, then it should affect erythrocyte development. To test whether stat1b activity is involved in the development of hemoglobin-producing cells, we stained control and stat1b knockdown individuals at 2dpf with o-dianisidiine, a stain for hemoglobin. Results showed a substantial increase in hemoglobin-producing cells after stat1b knockdown relative to uninjected controls, control MO injected animals, and stat1a treated embryos (Supplementary Figure s4). We conclude that stat1b activity inhibits erythrocyte development.
To investigate the role of stat1b on myeloid cell development, we studied spi1 (pu.1) (Lieschke et al., 2002) and to assay heterophil granulocytes, we used myeloperoxidase (mpo) (Bennett et al., 2001). Both markers were depressed after knockdown of stat1b in both RNA in situ hybridization assays and in Q-PCR assays, while stat1a knockdown had no effect (Fig. 7). The decrease in expression of myeloid cells and granulocytes after stat1b knockdown shows that Stat1b normally promotes myeloid cell development. Markers for definitive HSC, macrophages, and lymphoid cells – as marked by c-myb and runx1, l-plastin, and rag1, respectively (Herbomel et al., 1999; Kalev-Zylinska et al., 2002; Thompson et al., 1998; Willett et al., 1997) -- showed no expression differences between control animals and embryos injected with either stat1a or stat1b MOs (data not shown). To test whether cell death plays a prominent role in the effect of stat1b knockdown, we stained uninjected controls, and animals injected with control-MO, stat1a MO, or stat1b MO with acridine orange, which stains apoptotic cells, and then counted stained cells after spinning disc confocal microscopy. Results showed no differences among treatment groups (data not shown), suggesting that cell death does not play a large role in stat1b-mediated effects. Taken together, these analyses show that the knockdown of stat1b results in an expansion of the erythroid territory and a depression of the myeloid territory, while other hematopoietic lineages remain intact.
3. Discussion
Experiments reported here cloned stat1b cDNA, showed that its sequence is similar to that of the human STAT1beta splice form, defined stat1b history in the context of the evolution of the full vertebrate STAT family through tandem duplication and three rounds of whole genome duplication, identified stat1b expression in primitive hematopoietic cells, and showed that stat1b functions to promote the myeloid lineage at the expense of the erythroid lineage.
Mice lacking both STAT1alpha and STAT1beta develop normally but do not respond normally to interferons (Durbin et al., 1996; Meraz et al., 1996). In mammals, STAT1alpha contains a C-terminal transcription activation domain and is a fully functional transcriptional activator, while STAT1beta is a splice variant that lacks the TAD and fails to stimulate transcription in vitro from a chromatin template (although it can stimulate transcription from naked DNA) (Schindler et al., 1992; Zakharova et al., 2003). In a human lymphoblastoid cell line, forced expression of STAT1beta inhibited the phosphorylation, DNA-binding activity, and transcription activating function of STAT1alpha, and the expression of STAT1alpha target genes, while also increasing cell proliferation and resistance to fludarabine-induced cell cycle arrest and apoptosis, suggesting that STAT1beta is a dominant negative regulator of STAT1alpha (Baran-Marszak et al., 2004; Bromberg et al., 1996; Muller et al., 1993). Expression of a STAT1 form that, like STAT1beta, lacks critical features of transcription activating domain acts as a dominant negative inhibitor of STAT1 action (Bromberg et al., 1998).
Comparative analysis showed that, like human STAT1beta, zebrafish Stat1b protein lacks a complete TAD, but zebrafish Stat1a maintains a full C-terminal TAD like human STAT1alpha. Thus, zebrafish Stat1b has a structure homologous to the human dominant negative form, suggesting that Stat1b may act in zebrafish as a dominant regulator of Stat1a. Because the zebrafish Stat1a protein rescues interferon-signaling in a STAT1-deficient human cell line (Oates et al., 1999b), the zebrafish and human Stat1 systems appear to conserve basic mechanisms in cytokine signaling.
This raises the hypothesis that some of the STAT functions that partition between splice forms in mammals divide between duplicated genes in zebrafish. This type of subfunctionalization of protein functional domains is predicted by the duplication-degeneration-complementation hypothesis (Force et al., 1999; Postlethwait et al., 2004).
For vertebrate genes in the STAT3, STAT4, and STAT5 clades, teleosts and tetrapods have evolutionary rates (branch lengths on the phylogenetic tree) that are approximately equivalent, whether they are duplicated (like stat5.1 and stat5.2) or not (like stat3 and stat4). In contrast, in the STAT1 clade, teleost genes are evolving more rapidly than their tetrapod orthologs: stat1b is evolving especially rapidly in stickleback and appears to have been lost by other sequenced perciform fish. The sharing of functions between gene duplicates is thought to relax pleiotropy and thus may provide opportunities to contribute to more rapid protein sequence evolution in some situations. In contrast to the STAT3, 4, and 5 clades, the STAT2 and 6 clades are rapidly evolving in both in tetrapods and in teleosts. Curiously, STAT2 and STAT6 genes appear to have experienced an inversion event that separated these paralogs, which initially arose from a tandem duplication event, with 20 genes between them now in the human genome. By inserting break points in intergenic regions, this inversion may have altered gene regulation and expression compartments in ways that relaxed selection on the protein-coding regions of STAT2 and STAT6 genes.
Functionally, zebrafish stat1a and stat1b genes differ in several key respects. First, stat1b is expressed early and strongly in hematopoietic tissues in zebrafish embryos, while in situ hybridization studies showed that stat1a is expressed at low levels ubiquitously, confirming previous results (Oates et al., 1999b). These findings are consistent with our RT-PCR studies on whole embryos that revealed transcript for both stat1a and stat1b in embryos. According to the duplication-degeneration-complementation hypothesis, the partitioning of ancestral subfunctions, including tissue-specific regulatory elements, between duplicated genes as we observed for stat1 genes is predicted to occur frequently by neutral evolution due to the reciprocal extinction of regulatory elements in the two duplicates (Force et al., 1999; Postlethwait et al., 2004).
Second, the differential expression patterns of stat1a and stat1b may reflect different roles in development and physiology. Zebrafish Stat1a can rescue interferon signaling in human tissue culture cells that lack STAT1 activity, showing that Stat1a shares an ancestral virus-related cytokine-signaling mechanism with humans (Oates et al., 1999b). The virus-related function of Stat1 protein is expected to be required broadly in many cell types consistent with the broad expression pattern of stat1a. In contrast, stat1b expression and gene knockdown analysis reveal a role for stat1b in hematopoiesis and this gene is expressed specifically in the hematopoietic lineage.
Knockdown experiments confirmed a role for stat1b in zebrafish hematopoiesis. As assayed both by in situ hybridization and by quantitative PCR, the knockdown of stat1b activity led to the increased expression of scl and gata1 in primitive hematopoiesis and the increase in hemoglobin-producing cells but the decreased expression of spi and mpx. Because RNA in situ expression analyses showed broadened expression compartments, we conclude that the increased expression of scl and gata1 results from more cells expressing these markers after stat1b knockdown than in controls. Likewise, the smaller compartments of spi and mpx reflect fewer cells expressing these markers after stat1b knockdown. The scl gene marks primitive hematopoietic stem cells and gata1 marks erythroid cells, whereas spi1 and mpx mark myeloid cell lineages. The expression compartments of genes marking other lineages, including markers for definitive HSC (c-myb and runx1), macrophages (l-plastin), and lymphoid cells (rag1), were unaltered after stat1b knockdown. Thus, these results suggest that stat1b knockdown results in an increase in cells of the erythroid lineage at the expense of the myeloid lineage.
We interpret these results in the following framework. First, progenitor cells with high scl expression may be biased towards erythrocyte differentiation, whereas other markers of primitive HSCs, such as fli1a, gata2, and hhex, might be responsible for the differentiation of HSCs into other cell lineages, including myeloid and lymphoid cells (de Jong and Zon, 2005). Second, normal Stat1b function may be necessary to promote the myeloid lineage of spi1- and mpx-expressing cells and/or to suppress the erythroid lineage of gata1-expressing, o-dianisidine-positive cells. Note that the knockdown of stat1b reported here provides a result complementary to that derived from the knockdown of gata1, which leads to an increase of spi1 and mpx expression in zebrafish and suggests that gata1 suppresses the myeloid pathway (Galloway et al., 2005; Rhodes et al., 2005). Thus, stat1b and gata1 may play opposing roles in hematopoietic cell fate determination.
Whether mammalian STAT1 proteins play a role in hematopoiesis similar to that shown here for stat1b in zebrafish is as yet unclear. Recent studies showed that IFN treatments stimulated dormant HSCs in mice and that the effect depended on Stat1 (Baldridge et al., 2010; Essers et al., 2009; Sato et al., 2009), but the roles of STAT1-alpha and STAT1-beta forms were not tested independently. In addition, IFN--gamma produced by Th1 cells stimulates a significant expansion of LSK (lineage marker minus, Sca-1+, cKit+) hematopoietic stem cells that tend to differentiate into myeloid lineages through a STAT1 dependent pathway in mice (Zhao et al., 2010). In those experiments (Zhao et al., 2010), IFN--gamma led to the depletion of total progenitor cells but to the promotion of LSK cell proliferation into myeloid cells, which then compensated for the loss of total progenitors. IFN--gamma has long been known to regulate hematopoietic cell proliferation (Raefsky et al., 1985) but its effects are complicated. Depending on the stage of hematopoiesis and the hematopoietic lineage, IFN-gamma either inhibits hematopoiesis (Shimozato et al., 2002; Young et al., 1997) or promotes the expansion of specific stem cell populations and the differentiation of myeloid cells (Brugger et al., 1993; Caux et al., 1992), in each case mediated by STAT1. These results suggest the hypothesis that the basic functions we found for stat1b in myeloid development may be shared by the mammalian orthologs.
In our experiments, Stat1b could act in any of a number of ways. It might (1) promote the apoptosis of erythroid cells, which is unlikely given our apoptosis studies; (2) inhibit the differentiation of progenitor cells into erythrocytes; or (3) promote the differentiation of progenitor cells into myeloid cells. Further investigation of the regulatory effects of IFN and STAT1 in various hematopoietic cell lineages and at different developmental stages would shed light on the mechanisms by which those two important regulators act in normal development and perhaps in the bone marrow failure and myeloid cancers of Fanconi anemia patients. The partitioning of ancestral STAT1 functions between the zebrafish stat1a and stat1b gene duplicates provide a special opportunity to dissect the complicated hematopoietic roles of STAT1 genes in developing vertebrate embryos.
4. Experimental Procedures
4.1. Primers for cloning stat1b cDNA
Primers covering overlapping fragments of XR_029517 were designed for RT-PCR, with two primer pairs for each fragment. PCR products were cloned with the TOPO TA cloning kit into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, USA) and sequences were assembled to obtain a full-length cDNA sequence. Primers: stat1bF10 CGTTGAAAGATGACGCTCTG, stat1bR10 ACGGATCTGCTGGAGACACT; stat1bF11 GCTTATCCCGAGATACACTCC, stat1bR11 TCTTCAGCTGCTGACGGATCT; stat1bF20 GCTGAAGGTGTGCAAATGAA, stat1bR20 AGGCACTGGGTAACTGGTTG; stat1bF21 GCTGAAGGTGTGCAAATGAA, stat1bR21 TTCACCGCTGAGCATGTTGTA; stat1bF30 GACGCCTCTCATCGTTACAGA, stat1bR30 TTTTGGGATGTTCGGGTAAA; stat1bF31 GCAGCTGAATTTCGTCATTTG, stat1bR31 GCGACCAAAAGCTTCATCTT; stat1bF40 TGGTGAACCCAAGATCCATT, stat1bR40 GTGAAATGGCCTGTTCATCC; stat1bF41 TGCGCTTTAGTGAAAGCTGT, stat1bR41 TGAAATGGCCTGTTCATCCA; stat1bF50 CAGCAGATGGCTTGTATTGG, stat1bR50 TGCCAGCTTATGACCTTTGA; stat1bF51 TATCAGAGGAGTTACCCGAGT, stat1bR51 TGCTGATTGAAGAAAACTGCC.
4.2. Phylogenetic analysis
For phylogenetic analysis, STAT-related protein sequences were downloaded from NCBI and from Ensembl and were aligned by Muscle (MUltiple Sequence Comparison by Log-Expectation, http://www.ebi.ac.uk/Tools/muscle/index.html) using the JTT model (Abascal et al., 2005). PhyML (Guindon and Gascuel, 2003); http://www.atgc-montpellier.fr/phyml/) was used to construct maximum likelihood phylogenetic trees.
4.3. Zebrafish, morpholinos, and in situ hybridization
Zebrafish (ABC X TU strain) were maintained as described (Kimmel et al., 1995) with all protocols approved by the University of Oregon IACUC. The stat1b morpholino (AAAATGTAGCGGATGTTACTTCGAC, Gene Tools, LLC, Philomath, OR, USA) targets the splice donor site of exon-5-intron-5, leading to the inclusion of intron-5 and a premature translation stop codon. The stat1a MO (TCATGTGGTCAACAGGCACCTGCAA) targets the splice donor site of exon-3-intron-3, leading to the inclusion of intron-3 and a premature translation stop. The sequence of the control MO was CCTCTTACCTCAGTTACAATTTATA, which has no significant similarity in the zebrafish reference genome sequence database. A volume of 2nl of MO (5 mg/ml for stat1b MO and 10 mg/ml for stat1a MO and control MO) was injected into one-cell stage embryos.
For RNA in situ hybridization, embryos were collected at various ages and queried using as probe the 3′ untranslated regions of zebrafish stat1a and stat1b, and as negative control a cuttlefish sequence (GenBank: GU388435). RNA in situ hybridization with all three probes was carried out following methods previously described (Yan et al., 2002) except that stat1b probe was incubated at 50°C. RNA in situ hybridization with blood markers was carried out at 16 hpf and 32 hpf following methods previously described (Yan et al., 2002).
4.4. RT-PCR and Quantitative RT-PCR
Fertilized eggs were collected at indicated time points. Total RNA was extracted with TRI reagent (Molecular Research Center, Cincinnati, OH, USA) and genomic DNA was removed with DNA-free™ DNase Treatment and Removal Reagents (ABI, Foster City, CA, USA). Total RNA was quantified by spectrometry at 260 nm to ensure equal loading for RT-PCR. Product from each pair of primers was sequenced and verified to be the target mRNA fragment before the actual RT-PCR. RT-PCR was run with the OneStep RT-PCR kit (QIAGEN, Valencia, CA, USA). Primers used were: actinF GCCAACACAGTGCTGTCTGGAGG, actinR GGTCATGGACGCCCATTGTGAGG; stat1aMEF AGTCGCAGCAATGACTCAGTG, stat1aMER CTGCTGATGATCATCGCCATTG; stat1bMEF AGGTGACTCCATGCAGGGGAAC, stat1bMER GCGGGTCGTTGCTGTAGGTGA. cDNAs were prepared with the High Capacity RNA-to-cDNA kit (ABI, Foster City, CA, USA). Quantitative PCR was carried out using Power SYBR® Green PCR Master Mix (ABI, Foster City, CA, USA). Q-PCR was run with three biological repeats and three sampling repeats. The expression level of a house-keeping gene zgc:136952 (GenBank: NM_001040043), which encodes RPL32 (ribosomal protein L32) was run as an internal control. After collection, raw Ct data were normalized to that of this internal control. The expression level of each marker gene in control MO, stat1a MO, and stat1b MO treated embryos was then compared with a t-test assuming equal standard deviations with α = 0.05 (95% confidence interval). Primers used were: gata1qF GAATGCAGCTTCAGAGGTTTATCC, gata1qR TGGGTTCAGAGAATACGCTCCTA; mpxqF CCAAACCTCAGGGATGTTCTTG, mpxqR CCCAAACTACGAGTCCCTATGC; spiqF GGGTAGCCATCACATCCCTCTAG, spiqR TGGACGTTGTGAGGGTAACACA; sclqF GACTAATTTCTCGGGCTGACAACTA, sclqR GGGCGTTAACAGAAAGTCTTACGTA; RPL32qF CCCTCACCAAACCTAAGATCGT, RPL32qR CTCCAGTTTGCCCTGATCTTG.
4.5 Acridine orange and o-dianisidine staining
For analyses of cell death, 30 hpf embryos were dechorionated by hand and treated with 0.5 ug/ml acridine orange in embryo media for 30 minutes, washed 3x with embryo media, anesthetized with MS-222. For analyses of hemoglobin production, dechorionated embryos at 30 hpf and 48hpf were anesthetized, and treated with 0.06% o-dianisidine in 40% ethanol, 0.01M NaOAc, pH 4.5 and 0.6% of H2O2 for 15 minutes at room temperature in the dark. Embryos were washed with PBT to stop the reaction and fixed in 4% paraformaldehyde. After acridine orange and o-dianisidine treatments, embryos were imaged on a Leica spinning disc confocal microscope. Images were opened in ImageJ, max-projected, and red pixels in the ICM were counted.
Supplementary Material
Figure S1. Amino acid sequences for human STAT1alpha compared to zebrafish Stat1a and Stat1b proteins. STAT_int, STAT_alpha, STAT_bind, STAT_SH2 and STAT_TAZ2bind mark the locations of functional domains. Functional amino acids (e.g., R31) are also marked.
Figure S2. Validation of morpholino knockdown. A. The stat1a morpholino targets the exon-3/intron-3 border; it inhibits splicing, leads to a transcript that includes intron-3, and results in a premature stop codon that is truncated after 127 amino acid residues rather than the 749 amino acid residues of the normal protein. B. The stat1b morpholino targets the exon-5/intron-5 border; it inhibits splicing, leads to a transcript that includes intron-5 mRNA, and results in a premature stop codon that is truncated after 201 amino acid residues rather than the 725 amino acid residues of the normal protein.
Figure S3. Conserved syntenies of zebrafish and stickleback stat1b. A portion of zebrafish linkage group 9 (Dre LG9) surrounding stat1b has eight orthologs (connected by solid lines) with a small portion of stickleback linkage group 16 (Gac Group XVI), with the introduction of a small inversion.
Figure S4. Histochemical staining of hemoglobin with o-dianisidine. A. Embryos stained with o-dianisidine: uninjected controls (UIC, 1–3), control MO injected embryos (CMO, 4–6), stat1a MO treated embryos (7–9), and stat1b MO treated embryos (10–12). Knockdown of stat1b caused an increase in hemoglobin staining in the trunk compared to controls and stat1a MO injected embryos. B. Quantitative analysis of o-dianisidine-stained embryos, ten individuals scored for each of the four conditions. Pixels of o-dianisidine positive cells were counted and summed. Compared to the uninjected control using a t-test (single tailed), P values for significant only for stat1b MO (p = 0.0014), and were not significant for control MO (0.337) or stat1a MO (0.108).
Highlights Stat1b.
Zebrafish has co-orthologs of human STAT1: stat1a like STAT1α and stat1b like STAT1β.
STAT1α and STAT1β are splice isoforms but stat1a and stat1b are 2 duplicated genes.
stat1b but not stat1a expression was detected in embryonic hematopoietic tissues.
stat1b but not stat1a knockdown decreased myeloid and increased erythrocyte markers.
Conclusion: zebrafish Stat1b promotes myeloid at expense of erythroid development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Hao Song, Email: hsong@uoregon.edu.
Yi-lin Yan, Email: yan@uoneuro.uoregon.edu.
Tom Titus, Email: titus@uoneuro.uoregon.edu.
Xinjun He, Email: xhe@uoneuro.uoregon.edu.
John H. Postlethwait, Email: jpostle@uoneuro.uoregon.edu.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST--a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–7. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica MG, Brizzi MF, Dentelli P, Rosso A, Yarden Y, Pegoraro L. p91 STAT1 activation in interleukin-3-stimulated primary acute myeloid leukemia cells. Oncogene. 1996;13:1017–26. [PubMed] [Google Scholar]
- Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–7. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran-Marszak F, Feuillard J, Najjar I, Le Clorennec C, Bechet JM, Dusanter-Fourt I, Bornkamm GW, Raphael M, Fagard R. Differential roles of STAT1alpha and STAT1beta in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood. 2004;104:2475–83. doi: 10.1182/blood-2003-10-3508. [DOI] [PubMed] [Google Scholar]
- Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–51. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Traver D. Hematopoietic cell development in the zebrafish embryo. Curr Opin Hematol. 2009;16:243–8. doi: 10.1097/MOH.0b013e32832c05e4. [DOI] [PubMed] [Google Scholar]
- Brierley MM, Fish EN. Stats: multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733–44. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Fan Z, Brown C, Mendelsohn J, Darnell JE., Jr Epidermal growth factor-induced growth inhibition requires Stat1 activation. Cell Growth Differ. 1998;9:505–12. [PubMed] [Google Scholar]
- Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:7673–8. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger W, Mocklin W, Heimfeld S, Berenson RJ, Mertelsmann R, Kanz L. Ex vivo expansion of enriched peripheral blood CD34+ progenitor cells by stem cell factor, interleukin-1 beta (IL-1 beta), IL-6, IL-3, interferon-gamma, and erythropoietin. Blood. 1993;81:2579–84. [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–42. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caux C, Moreau I, Saeland S, Banchereau J. Interferon-gamma enhances factor-dependent myeloid proliferation of human CD34+ hematopoietic progenitor cells. Blood. 1992;79:2628–35. [PubMed] [Google Scholar]
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–39. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–22. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- Ciau-Uitz A, Liu F, Patient R. Genetic control of hematopoietic development in Xenopus and zebrafish. Int J Dev Biol. 2010;54:1139–49. doi: 10.1387/ijdb.093055ac. [DOI] [PubMed] [Google Scholar]
- Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16:137–47. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–6. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–37. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich HW, 3rd, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci U S A. 1995;92:10713–7. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–50. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Ellett F, Lieschke GJ. Zebrafish as a model for vertebrate hematopoiesis. Curr Opin Pharmacol. 2010;10:563–70. doi: 10.1016/j.coph.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrai A, Robb L. The hemangioblast--between blood and vessels. Cell Cycle. 2003;2:86–90. [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–8. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–16. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–76. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- Gouilleux-Gruart V, Debierre-Grockiego F, Gouilleux F, Capiod JC, Claisse JF, Delobel J, Prin L. Activated Stat related transcription factors in acute leukemia. Leuk Lymphoma. 1997;28:83–8. doi: 10.3109/10428199709058334. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haneline LS, Broxmeyer HE, Cooper S, Hangoc G, Carreau M, Buchwald M, Clapp DW. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac−/− mice. Blood. 1998;91:4092–8. [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–45. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994:125–133. [PubMed] [Google Scholar]
- Improta T, Schindler C, Horvath CM, Kerr IM, Stark GR, Darnell JE., Jr Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc Natl Acad Sci U S A. 1994;91:4776–80. doi: 10.1073/pnas.91.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol. 2001;230:278–301. doi: 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–57. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–30. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–56. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kastner P, Chan S. PU.1: a crucial and versatile player in hematopoiesis and leukemia. Int J Biochem Cell Biol. 2008;40:22–7. doi: 10.1016/j.biocel.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- Lewis RS, Ward AC. Conservation, duplication and divergence of the zebrafish stat5 genes. Gene. 2004;338:65–74. doi: 10.1016/j.gene.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Yuan J, Hong P, Freie B, Orazi A, Haneline LS, Clapp DW. Continuous in vivo infusion of interferon-gamma (IFN-gamma) preferentially reduces myeloid progenitor numbers and enhances engraftment of syngeneic wild-type cells in Fancc−/− mice. Blood. 2004;104:1204–9. doi: 10.1182/blood-2004-03-1094. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–6. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, Ho RK, Zon LI, Layton JE. Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–95. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–11. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–41. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell JE, Jr, Stark GR, Kerr IM. Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-alpha and -gamma signal transduction pathways. EMBO J. 1993;12:4221–8. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar I, Deglesne PA, Schischmanoff PO, Fabre EE, Boisson-Dupuis S, Nimmerjahn F, Bornkamm GW, Dusanter-Fourt I, Fagard R. STAT1-dependent IgG cell-surface expression in a human B cell line derived from a STAT1-deficient patient. J Leukoc Biol. 2010;87:1145–52. doi: 10.1189/jlb.1109714. [DOI] [PubMed] [Google Scholar]
- Najjar I, Fagard R. STAT1 and pathogens, not a friendly relationship. Biochimie. 2010;92:425–44. doi: 10.1016/j.biochi.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar I, Schischmanoff PO, Baran-Marszak F, Deglesne PA, Youlyouz-Marfak I, Pampin M, Feuillard J, Bornkamm GW, Chelbi-Alix MK, Fagard R. Novel function of STAT1beta in B cells: induction of cell death by a mechanism different from that of STAT1alpha. J Leukoc Biol. 2008;84:1604–12. doi: 10.1189/jlb.0508287. [DOI] [PubMed] [Google Scholar]
- Oates AC, Brownlie A, Pratt SJ, Irvine DV, Liao EC, Paw BH, Dorian KJ, Johnson SL, Postlethwait JH, Zon LI, Wilks AF. Gene duplication of zebrafish JAK2 homologs is accompanied by divergent embryonic expression patterns: only jak2a is expressed during erythropoiesis. Blood. 1999a;94:2622–36. [PubMed] [Google Scholar]
- Oates AC, Wollberg P, Pratt SJ, Paw BH, Johnson SL, Ho RK, Postlethwait JH, Zon LI, Wilks AF. Zebrafish stat3 is expressed in restricted tissues during embryogenesis and stat1 rescues cytokine signaling in a STAT1-deficient human cell line. Dev Dyn. 1999b;215:352–70. doi: 10.1002/(SICI)1097-0177(199908)215:4<352::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer-Verlag; New York: 1970. [Google Scholar]
- Oikawa T, Yamada T, Kihara-Negishi F, Yamamoto H, Kondoh N, Hitomi Y, Hashimoto Y. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 1999;6:599–608. doi: 10.1038/sj.cdd.4400534. [DOI] [PubMed] [Google Scholar]
- Otsuki T, Nagakura S, Wang J, Bloom M, Grompe M, Liu JM. Tumor necrosis factor-alpha and CD95 ligation suppress erythropoiesis in Fanconi anemia C gene knockout mice. J Cell Physiol. 1999;179:79–86. doi: 10.1002/(SICI)1097-4652(199904)179:1<79::AID-JCP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Paik EJ, Zon LI. Hematopoietic development in the zebrafish. Int J Dev Biol. 2010;54:1127–37. doi: 10.1387/ijdb.093042ep. [DOI] [PubMed] [Google Scholar]
- Pang Q, Fagerlie S, Christianson TA, Keeble W, Faulkner G, Diaz J, Rathbun RK, Bagby GC. The Fanconi anemia protein FANCC binds to and facilitates the activation of STAT1 by gamma interferon and hematopoietic growth factors. Mol Cell Biol. 2000;20:4724–35. doi: 10.1128/mcb.20.13.4724-4735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson LJ, Gering M, Patient R. Scl is required for dorsal aorta as well as blood formation in zebrafish embryos. Blood. 2005;105:3502–11. doi: 10.1182/blood-2004-09-3547. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–90. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, Westerfield M, Zon LI, Johnson SL, Talbot WS. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–9. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Raefsky EL, Platanias LC, Zoumbos NC, Young NS. Studies of interferon as a regulator of hematopoietic cell proliferation. J Immunol. 1985;135:2507–12. [PubMed] [Google Scholar]
- Rathbun RK, Faulkner GR, Ostroski MH, Christianson TA, Hughes G, Jones G, Cahn R, Maziarz R, Royle G, Keeble W, Heinrich MC, Grompe M, Tower PA, Bagby GC. Inactivation of the Fanconi anemia group C gene augments interferon-gamma-induced apoptotic responses in hematopoietic cells. Blood. 1997;90:974–85. [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- Schindler C, Fu XY, Improta T, Aebersold R, Darnell JE., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci U S A. 1992;89:7836–9. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, Tonnelle C, Dalloul A, Chabannon C, Debre P, Rebollo A. Aiolos and Ikaros: regulators of lymphocyte development, homeostasis and lymphoproliferation. Apoptosis. 2002;7:277–84. doi: 10.1023/a:1015372322419. [DOI] [PubMed] [Google Scholar]
- Shimozato O, Ortaldo JR, Komschlies KL, Young HA. Impaired NK cell development in an IFN-gamma transgenic mouse: aberrantly expressed IFN-gamma enhances hematopoietic stem cell apoptosis and affects NK cell differentiation. J Immunol. 2002;168:1746–52. doi: 10.4049/jimmunol.168.4.1746. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–4. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- Si Y, Ciccone S, Yang FC, Yuan J, Zeng D, Chen S, van de Vrugt HJ, Critser J, Arwert F, Haneline LS, Clapp DW. Continuous in vivo infusion of interferon-gamma (IFN-gamma) enhances engraftment of syngeneic wild-type cells in Fanca−/− and Fancg−/− mice. Blood. 2006;108:4283–7. doi: 10.1182/blood-2006-03-007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, Wu XY, Sheng Y, Chen Y, Ruan Z, Jiang CL, Fan HY, Zon LI, Kanki JP, Liu TX, Look AT, Chen Z. Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci U S A. 2004;101:16240–5. doi: 10.1073/pnas.0407241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH, Millard PJ, Kim CH. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol. 2009;183:5896–908. doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–90. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–69. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2004;76:127–49. doi: 10.1016/s0091-679x(04)76008-2. [DOI] [PubMed] [Google Scholar]
- Vignais ML, Sadowski HB, Watling D, Rogers NC, Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol. 1996;16:1759–69. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–52. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–16. [PubMed] [Google Scholar]
- Whitney MA, Royle G, Low MJ, Kelly MA, Axthelm MK, Reifsteck C, Olson S, Braun RE, Heinrich MC, Rathbun RK, Bagby GC, Grompe M. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- Willett CE, Cherry JJ, Steiner LA. Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics. 1997;45:394–404. doi: 10.1007/s002510050221. [DOI] [PubMed] [Google Scholar]
- Xiong JW. Molecular and developmental biology of the hemangioblast. Dev Dyn. 2008;237:1218–31. doi: 10.1002/dvdy.21542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–79. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- Yang E, Henriksen MA, Schaefer O, Zakharova N, Darnell JE., Jr Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J Biol Chem. 2002;277:13455–62. doi: 10.1074/jbc.M112038200. [DOI] [PubMed] [Google Scholar]
- Young HA, Klinman DM, Reynolds DA, Grzegorzewski KJ, Nii A, Ward JM, Winkler-Pickett RT, Ortaldo JR, Kenny JJ, Komschlies KL. Bone marrow and thymus expression of interferon-gamma results in severe B-cell lineage reduction, T-cell lineage alterations, and hematopoietic progenitor deficiencies. Blood. 1997;89:583–95. [PubMed] [Google Scholar]
- Zakharova N, Lymar ES, Yang E, Malik S, Zhang JJ, Roeder RG, Darnell JE., Jr Distinct transcriptional activation functions of STAT1alpha and STAT1beta on DNA and chromatin templates. J Biol Chem. 2003;278:43067–73. doi: 10.1074/jbc.M308166200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li J, Sejas DP, Rathbun KR, Bagby GC, Pang Q. The Fanconi anemia proteins functionally interact with the protein kinase regulated by RNA (PKR) J Biol Chem. 2004;279:43910–9. doi: 10.1074/jbc.M403884200. [DOI] [PubMed] [Google Scholar]
- Zhao X, Ren G, Liang L, Ai PZ, Zheng B, Tischfield JA, Shi Y, Shao C. Brief report: interferon-gamma induces expansion of Lin(−)Sca-1(+)C-Kit(+) Cells. Stem Cells. 2010;28:122–6. doi: 10.1002/stem.252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Amino acid sequences for human STAT1alpha compared to zebrafish Stat1a and Stat1b proteins. STAT_int, STAT_alpha, STAT_bind, STAT_SH2 and STAT_TAZ2bind mark the locations of functional domains. Functional amino acids (e.g., R31) are also marked.
Figure S2. Validation of morpholino knockdown. A. The stat1a morpholino targets the exon-3/intron-3 border; it inhibits splicing, leads to a transcript that includes intron-3, and results in a premature stop codon that is truncated after 127 amino acid residues rather than the 749 amino acid residues of the normal protein. B. The stat1b morpholino targets the exon-5/intron-5 border; it inhibits splicing, leads to a transcript that includes intron-5 mRNA, and results in a premature stop codon that is truncated after 201 amino acid residues rather than the 725 amino acid residues of the normal protein.
Figure S3. Conserved syntenies of zebrafish and stickleback stat1b. A portion of zebrafish linkage group 9 (Dre LG9) surrounding stat1b has eight orthologs (connected by solid lines) with a small portion of stickleback linkage group 16 (Gac Group XVI), with the introduction of a small inversion.
Figure S4. Histochemical staining of hemoglobin with o-dianisidine. A. Embryos stained with o-dianisidine: uninjected controls (UIC, 1–3), control MO injected embryos (CMO, 4–6), stat1a MO treated embryos (7–9), and stat1b MO treated embryos (10–12). Knockdown of stat1b caused an increase in hemoglobin staining in the trunk compared to controls and stat1a MO injected embryos. B. Quantitative analysis of o-dianisidine-stained embryos, ten individuals scored for each of the four conditions. Pixels of o-dianisidine positive cells were counted and summed. Compared to the uninjected control using a t-test (single tailed), P values for significant only for stat1b MO (p = 0.0014), and were not significant for control MO (0.337) or stat1a MO (0.108).