Abstract
The crassispirids are a large branch of venomous marine gastropods whose venoms have not been investigated previously. We demonstrate that crassispirids comprise a major group of toxoglossate snails in a clade distinct from all turrids whose venoms have been analyzed. The isolation and biochemical definition of the first venom component from any crassispirid is described.
Crassipeptide cce9a from Crassispira cerithina (Anton, 1838) was purified from crude venom by following biological activity elicited in young mice, lethargy and a lack of responsiveness to external stimuli. Using Edman sequencing and mass spectrometry, the purified peptide was shown to be 29 amino acid residues long, with the sequence: GSCGLPCHENRRCGWACYCDDGICKPLRV.
The sequence assignment was verified through the analysis of a cDNA clone encoding the peptide. The peptide was chemically synthesized and folded; the synthetic peptide was biologically active and coelution with the native venom peptide was demonstrated. When injected into mice of various ages, the peptide elicited a striking shift in behavioral phenotype between 14 and 16 days, from lethargy to hyperactivity.
Keywords: Venom, Neuropharmacology, Developmental change, Crassipeptide, Mollusc
1. Introduction
The venom peptides from the cone snails (conotoxins, conopeptides) have become standard reagents in neuroscience, with significant biomedical applications (Terlau and Olivera, 2004). Many of the most important conopeptides were originally discovered through a simple assay: injection into the central nervous system of a mouse. This relatively unsophisticated assay was surprisingly effective in identifying different neuroactive peptides in Conus venoms, and proved to be key to identifying peptides that were later found to have important applications in neuroscience and biomedicine (Olivera et al., 1990). The peptide, ε-conotoxin MVIIA (Olivera et al., 1987), originally characterized using this approach, was approved as a commercial drug, Prialt (Teichert and Olivera, 2010). The sleeper (Olivera et al., 1985) and sluggish peptides (Craig et al., 1998), which were purified using the behavioral phenotype they elicited, have reached human clinical trials for pain and epilepsy, respectively (Teichert and Olivera, 2010). Thus, for cone snail venoms, using an in vivo behavioral phenotype as an assay has clearly been productive for identifying novel neuroactive venom peptides.
However, cone snails in the family Conidae comprise only a minor component of the total biodiversity of venomous marine molluscs. Other groups include the auger snails in the family Terebridae and an enormous biodiversity of largely small, deep-water venomous gastropods traditionally included in a single family, Turridae (Powell, 1966); these three families (i.e., Conidae, Terebridae and Turridae) are generally grouped together in the superfamily Conoidea. While cone snails and auger snails each comprise several hundred species, it is now believed that the various “turrids” (broadly defined) comprise over 10,000 species (Bouchet et al., 2002). Recent phylogenetic data has suggested that “turrid” species fall into several major branches, each comprising well over 103 species.
Studies on the venom of some of the larger turrid species, which all fall within the classical subfamily Turrinae (Bouchet and Rocroi, 2005; Powell, 1967), have been reported (Aguilar et al., 2009; Heralde et al., 2008; López-Vera et al., 2004). It is now clear that the other major branches of “turrids” are not closely related to the Turrinae, and probably should be regarded as separate families. In this report, we characterize for the first time a peptide from the venom of a “turrid”, Crassispira cerithina (Fig. 1), a species that does not belong to the turrine branch. Instead, it belongs to a highly biodiverse group that has variously been referred to as the family Crassispiridae or subfamily Crassispirinae and more rarely as Pseudomelatomidae or Drillidae. In this article, we will refer to this major clade of venomous gastropods as the “crassispirids.”
Fig. 1.

Crassisipira cerithina shell specimens. Variations in shell color of samples collected in Olango Island, Cebu, Philippines. Live specimens of mature snails (average length: 1.3 cm) were sorted into light brown, brown, and dark brown variants (left to right in the photo). The dark brown variant, which was most abundant, was used in this work.
We adopted the general approach used previously for characterizing venom peptides from Conus, i.e., injecting venom components into the central nervous system of a mouse. One venom peptide from Crassispira cerithina proved to have an unusual activity when assayed; this peptide has been purified and synthesized. The characterization of this peptide elucidates for the first time a venom component from the highly biodiverse crassispirids, a major group of venomous animal species that is likely to exceed both the cone snails and the auger snails in number of species.
2. Materials and Methods
2.1. Specimens
Live C. cerithina snails were collected by hookah divers at a depth of 10–20 m off Olango Island, Cebu, Philippines. These divers also provided most of the other species used in the molecular phylogeny analysis. Live specimens were dissected on ice to get the venom ducts. Ducts for peptide analysis were stored in liquid nitrogen on-site and at −70 °C in the laboratory prior to use. For RNA work, snails were dissected in RNAlater® (Ambion, Inc., USA) and 50 ducts were pooled per tube with RNAlater®. Foot samples were preserved in 95% ethanol for molecular analysis. Voucher specimens were preserved in 95% ethanol and deposited at The Marine Science Institute, University of the Philippines, Quezon City, Philippines.
2.2. DNA extraction, amplification, and sequencing
Genomic DNA was extracted from 10 mg of foot tissue using the Gentra® Puregene® DNA Isolation Kit (Qiagen, Inc., CA, USA) following the manufacturer’s standard protocol.
Approximately 10 ng of genomic DNA was used as template for the succeeding polymerase chain reaction (PCR) with oligonucleotide primers targeting segments of mitochondrial 12S rRNA: 12S-I (5′ TCG CAG CG YCG GGG TTA), 12S-III (5′ AGA GYG RCG GGC GAT GTG T) (Simon et al, 1991); cytochrome oxidase I (COI): LCO1490 (5′ GGT CAA CAA ATC ATA AAG AYA TGY G 3′), HCO2198 (5′ TAA ACT TCA GGG TGA CCA AAR AAY CA 3′) (Folmer et al, 1994); and 16S rRNA: 16SH (5′ CCG GTC TGA ACT CAG ATC ACT G 3′), 16LC (5′ GTT TAC CAA AAA CT GGC TTC 3′) (Palumbi, 1996). Amplification with Advantage® 2 Polymerase Mix (Clontech Laboratories, Inc., CA, USA) was done using an MJ Research PTC-100 thermal cycler (Bio-Rad Laboratories, Inc., USA). The PCR profile was as follows: initial denaturation (94 °C, 5 min); followed by 40 cycles of denaturation (94 °C, 20 s), annealing (55 °C for 12S rRNA and CO1 and 58 °C for 16S rRNA, 20 s), and extension (72 °C, 30 s); and final elongation (72 °C, 7 min).
PCR products were purified by gel electrophoresis in 1.5% agarose gel and recovered using a QIAquick Gel Extraction Kit (Qiagen, Inc., CA, USA). The PCR fragments were ligated to pGEM®-T Easy using T4 DNA ligase (Promega Corp., USA) according to the manufacturer’s suggested protocol and consequently transformed into DH10α competent cells. Nucleic acid sequences from 8 – 10 clones containing 12S rRNA, 16S rRNA, and COI fragments were determined by automated DNA sequencing (Core Sequencing Facility, University of Utah, USA). This methodology was also employed for generating sequences from the other species used in the molecular phylogeny analysis.
2.3. Phylogenetic analysis
For phylogenetic analyses, COI, 12SrRNA and 16SrRNA sequences were obtained from the genomic DNA of each species and aligned using Muscle (Edgar, 2004). Gene alignments were concatenated using MacClade (Maddison and Maddison, 2005).
Maximum likelihood parameters describing sequence evolution were optimized with a generalized linear model (GTR, Tavaré, 1986) with invariant sites and across-site rate heterogeneity parameters (GTR+I+G). Trees were inferred using maximum likelihood methods (PhyML, Guindon and Gascuel, 2003), and for partitioned maximum-likelihood inference of the concatenated sequences, the GTR+G model parameters were estimated independently for each gene (RaxML, Stamatakis et al., 2008).
Bayesian analyses (Huelsenbeck et al., 2001; Ronquist and Huelsenbeck, 2003) comprised 1,000,000 generations with the first 25% of the sampled generations discarded as burn-in trees. Two MCMCMC runs (Metropolis-Coupled Markov Chain Monte Carlo) of four chains each were used to thoroughly explore tree space. Convergence of the likelihoods was determined by comparing the average standard error of the difference (ASED) in split frequencies between the two runs and by comparing plots of the log-likelihood scores. Optimality was also judged adequate when the potential scale reduction factor (PSRF) for the total tree length and for each model parameter reached 1.00.
2.4. RNA isolation and sequencing of cDNA clones containing cce9a nucleic acid sequence
RNA from pooled venom ducts was isolated using TRIzol® reagent (Invitrogen, USA) following the manufacturer’s standard protocol. Subsequently, polyA+ RNA was purified from 25 ng of total RNA using Oligotex mRNA Mini Kit (Qiagen Inc., USA) and cDNA was synthesized using Super SMART PCR cDNA Synthesis Kit (Clontech, USA) according to the manufacturer’s standard protocols. First strand cDNA was used as template to amplify the gene encoding the toxin cce9a. Degenerate primers were designed based on the least degenerate region (-CYCDDG-) of the cce9a peptide sequence: CceF (5′ TGY TAY TGY GAY GAY GGC 3′), CceR (5′ RTC RTC RCA RTA RCA CGC 3′). PCR products were purified from 1.5% NuSieve GTG agarose gel (Cambrex Bio Science Rockland, Inc., USA) using High Pure PCR Product Purification Kit (Roche, USA) and annealed to pNEB206A vector using the USER Friendly Cloning Kit (New England Biolabs, Inc., USA) based on the manufacturer’s standard protocol. The resulting product was transformed into competent E. coli DH5α cells. Nucleotide sequences of several clones were determined by standard automated sequencing procedure.
2.5. Extraction of crude venom and peptide purification and characterization
Venom ducts (~400) in an Eppendorf tube were homogenized with a disposable teflon pestle. A series of extractions was made using 1ml each of 10%, 40%, and 60% aqueous acetonitrile (CH3CN) in 0.1% trifluoroacetic acid (TFA). Each mixture was incubated at 4 °C for 1 h and centrifuged at 10,000 rpm for 15 min at 4 °C. The supernatants were combined (crude venom extract) and stored at −20 C until further purification (Teichert et al., 2007).
The crude venom extract was fractionated at 27 °C by reversed-phase high pressure liquid chromatography (RP-HPLC) using a C18 analytical column (Zorbax Eclipse XDB; 4.6 × 250 mm, 5μm particle diameter, 300 Å pore size) equipped with a C18 guard column (Phenomenex; 4.6 × 10 mm, 5 μm particle diameter, 300 Å pore size) and a 1 ml sample-loading loop. Elution was conducted at 1 ml/min employing solutions A (0.1% (v/v) aqueous TFA) and B (0.1% (v/v) TFA in 90% (v/v) aqueous CH3CN). A linear gradient from 6% to 60% solution B was applied over 60 min followed by 60% to 100% solution B over 20 min (McIntosh et al., 1994). The absorbance of the eluate was monitored at 220 nm, 254 nm, and 280 nm. The bioactive fractions that were identified by the intracranial mouse bioassay were further purified using shallower linear gradients (0.2 % – 0.5% solution B per min).
Prior to sequencing, the peptide was reduced and alkylated using dithiothreitol and 4-vinylpyridine (Imperial et al., 2008). Native and linearized peptides were analyzed by MALDI-MS using a Voyager GE STR mass spectrometer (Salk Institute Peptide Biology Lab) and the linear peptides were sequenced using an automated Edman degradation method on an Applied Biosystem model 492 sequenator (University of Utah Health Sciences Center Core Research Facilities).
2.6. Synthesis, folding and co-elution of native and synthetic cce9a
Crassipeptide cce9a was synthesized in an Apex 396 automated peptide synthesizer (AAPPTec, Louisville, KY, USA) using a standard solid-phase Fmoc (9-fluorenylmethyloxycarbonyl) protocol. The peptide was constructed on preloaded Fmoc-L-Val-Wang resin (substitution: 0.53 mmolg-1, Peptides International Inc, KY, USA). All amino acids were purchased from AAPPTec and the side-chain protection for each amino acid was: Glu and Asp: O-tert-butyl; Arg: 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl; Lys and Trp: tert-butyloxycarbonyl; Tyr and Ser: tert-butyl; Asn, Cys and His: trityl. The peptide was synthesized at a 50-μmol scale. Coupling activation was achieved with 1 equivalent of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate and 2 equivalents of 2 M N, N-diisopropylethyl amine in N-methyl-2-pyrrolidone. A 10-fold excess of each amino acid was used. Each coupling reaction was conducted for 60 min. Fmoc deprotection reaction was carried out for 20 min with 20% piperidine in DMF. The peptide was removed from the resin by treatment with reagent K (TFA/water/phenol/thioanisole/ethanedithiol 82.5/5/5/5/2.5 by volume) for 4 h, subsequently filtered, precipitated and washed twice with cold methyl-tert-butyl ether. The crude peptide was dissolved in 10% of solution B and purified by RP-HPLC in a semi-preparative C18 Vydac column (218TP510, 250 mm × 10 mm, 5 μm particle size) with a flow rate of 4 ml/min using a linear gradient from 15% to 45% of solution B in 30 min. Solutions A and B were 0.1% (v/v) TFA in water and 0.1% TFA (v/v) in 90% aqueous acetonitrile, respectively. The absorbance of the eluate was monitored at 220 nm. The purity of peptide was assessed using an analytical C18 Vydac reversed-phase HPLC column (218TP54, 250 mm × 4.6 mm, 5 μm particle size) with a flow rate of 1 ml/min and a linear gradient ranging from 15% to 45% of solution B in 30 min. The peptide was further characterized by ESI-MS confirming the correct mass of: 3211.386 [M+1] (calculated: 3211.375).
Oxidative folding of cce9a was performed by resuspending the linear cce9a in a 0.01% TFA solution and adding it to a solution containing: 0.1 M Tris-HCl (pH 7.5), 0.1 mM EDTA, 1 mM GSH and 1 mM GSSG. The final peptide concentration in the folding mixture was 20 μM. The folding reaction was allowed to proceed for 30 min then quenched by adding formic acid to a final concentration of 8%. The reaction mixture was separated by RP-HPLC on C18 columns with a linear gradient of 15% to 45% of solution B in 30 min. Flow rates of 1 ml/min and 4 ml/min were used for analytical and semi-preparative columns, respectively. All HPLC runs were monitored using absorbance at 220 nm. The identity of the final product was confirmed by ESI-MS with the correct mass of: 3205.338 [MH+] (calculated: 3205.375).
To determine whether the native and the synthetic cce9a are identical, both native and synthetic peptides were separately loaded onto a C18 analytical column and profiled by HPLC using a linear gradient of 15% to 45% of solvent B in 60 min. Native cce9a and synthetic cce9a were mixed in a 40:60 ratio (796 pmol of native: 1194 pmol of synthetic peptide), applied on the C18 analytical column and eluted using the gradient described above.
2.7. Intracranial mouse bioassay
Swiss Webster mice (12 to 16 days old) were injected intracranially with 1 to 50 nmol synthetic cce9a dissolved in 20 μl of 0.9 % NaCl (NSS) (McIntosh et al., 1994). Negative control mice were injected with 20 μl NSS. Mouse behavior in response to stimuli, which included pushing the mouse, pinning the tail, dropping the mouse from a height of 7 inches and creating a loud acoustic input by dropping the cage lid, was observed for 2 hours to determine differences between treated and control groups for both young and older mice.
3. Results
3.1. Phylogenetic analysis
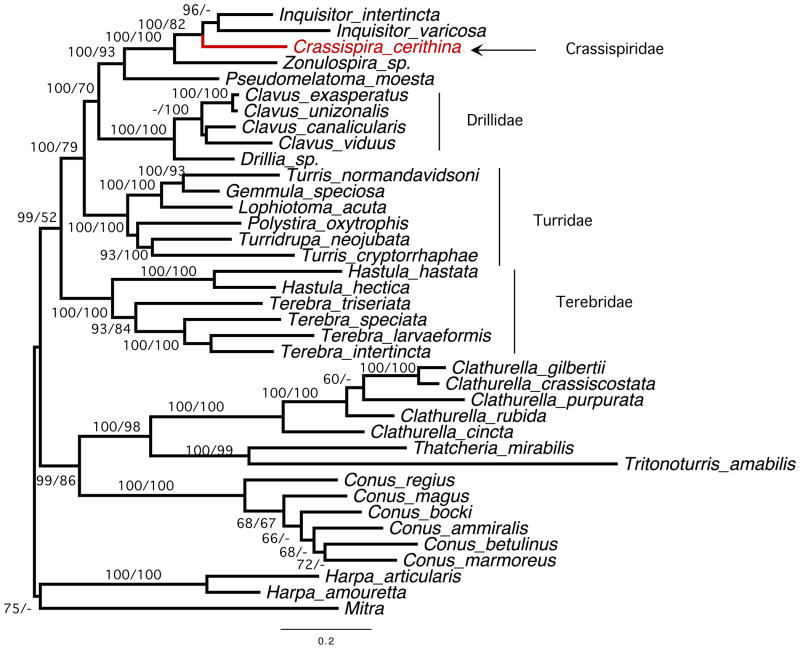
The molecular phylogeny of Crassispira cerithina was evaluated using standard methods. Maximum likelihood and Bayesian analyses were carried out of a diverse set of Conoideans using three standard mitochondrial markers (12S rRNA, 16S rRNA and COI) and for all three concatenated genes included. All of the analyses were consistent in separating Crassispira cerithina from the genera assigned to the subfamily Turrinae (e.g., Turris, Lophiotoma, Gemmula, Turridrupa, Polystira). Crassispira cerithina was originally assigned to the Turrinae in a number of earlier taxonomic treatments (in most books, the species is referred to as Turridrupa cerithina). However, the phylogenetic analysis presented in Fig. 2, which shows a tree generated from the concatenated sequences from all three genetic markers, clearly supports the conclusion that this is a non-turrine species and that assignment to Turridrupa is untenable.
Fig. 2.
Maximum likelihood phylogenetic tree inferred from concatenated 16S rRNA, 12S rRNA, and COI gene sequences. C.cerithina appears to belong to a separate clade well separated from the Turridae comprising Lophiotoma, Gemmula, Turris, Turridrupa, and Polystira. Instead it appears to be closely related to Inquisitor species and is provisionally placed in the family Crassispiridae. Branches are labeled with Bayesian posterior probabilities (left) and maximum-likelihood bootstrap values (right).
The phylogenetic analysis reveals that C. cerithina belongs to a major clade distinct from the turrines that also includes species assigned to the genus Inquisitor; we provisionally refer to this branch as the “crassispirines”, and refer to their venom peptides as “crassipeptides” (to differentiate them from “conopeptides” and “turripeptides”).
3.2. Isolation of bioactive peptide cce9a from crude venom
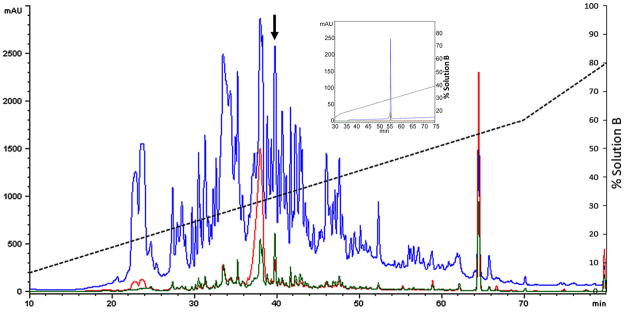
When the crude venom of C. cerithina was analyzed by RP-HPLC, an elution profile (Fig. 3A) comparable in complexity to that observed for Conus venoms was revealed. Bioassay-guided identification of neuroactive HPLC-separated fractions allowed us to purify a fraction that induced lethargy and a delayed response to stimuli at a dose of approximately 1 nmol (100 mAU = 1 nmol for peptidic components ~2000–4000 Da) per mouse (5 – 6 g; 12 – 14 d old). The major component of this fraction was purified to homogeneity; the purified biologically active component was further characterized as described under experimental procedures (Fig. 3B–3D). The peptide was designated Crassipeptide cce9a.
Fig. 3.
Purification of Crassipeptide cce9a. Samples were fractionated on an analytical C18 HPLC column and eluted with gradients of acetonitrile in 0.1% TFA indicated by the dashed lines. The elution profiles at 220nm (blue), 254 nm (red) and 280 nm (green) are shown. The neuroactive fraction is marked with an arrow. The pure peptide (inset) was isolated by subfractionation of the active fraction in A using a shallower linear gradient. The peptide was named cce9a following turripeptide nomenclature (Heralde et al., 2008).
The measured monoisotopic mass of the native peptide was 3204.11 Da (MALDI, reflector mode) and the reduced-alkylated form gave an average mass (MALDI, linear mode) that was approximately 630 Da greater than that of the native form indicating a reaction with 6 molecules of 4-vinylpyridine. This suggested that cce9a contains 6 cysteine residues forming 3 disulfide bonds. The reduced and alkylated peptide was sequenced using standard Edman methods, which confirmed that cce9a has 6 cysteines. The peptide has 29 amino acid residues and its sequence is: GSCGLPCHENRRCGWACYCDDGICKPLRV.
The sequence is consistent with the monoisotopic mass of the peptide; there are no post-translational modifications. The sequence assignment was verified by isolating a cDNA clone. The nucleic acid sequence encoding the mature toxin region, the partial propeptide region and the 3″ untranslated region is shown in Fig. 4. The cDNA sequence confirms the amino acid sequence determined directly and verifies that the C-terminus is a free carboxyl group.
Fig. 4.
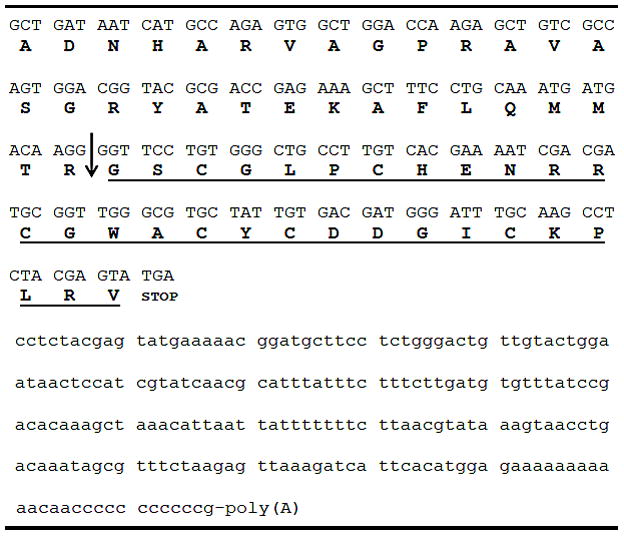
cDNA sequence of the gene encoding cce9a. The mature peptide region is underlined; amino acid residues prior to the mature peptide region are part of the prepropeptide region. Nucleotides in the 3′ untranslated region are in small letters.
3.3. Chemical synthesis and coelution
The predicted peptide cce9a was chemically synthesized using the standard Fmoc solid phase protocol. The purified linear form was folded in the presence of oxidized and reduced glutathione using methods described previously for μ-conotoxins (Fuller et al. 2005) and explained in detail in the Materials and Methods section. The time course of the folding reaction was monitored using analytical HPLC analysis of aliquots withdrawn after 2, 10, 30 and 60 min (data not shown for the last time point) and quenched with formic acid (Fig. 5). Extending the folding time beyond 30 min did not result in any significant change in distribution and accumulation of folded products. To determine whether the accumulated product represented the fully oxidized form, the peak from the 30-min folding reaction was collected, dried and analyzed by ESI-MS. The peptide had the expected molecular weight [MH+] 3205.338 (calculated: 3205.375). The identity of the folded cce9a was confirmed using HPLC coelution experiments with the native peptide, which was purified from the venom of C. cerithina (Fig. 3). Furthermore, the biological activity of the synthetic peptide was similar to that of the native cce9a.
Fig. 5.
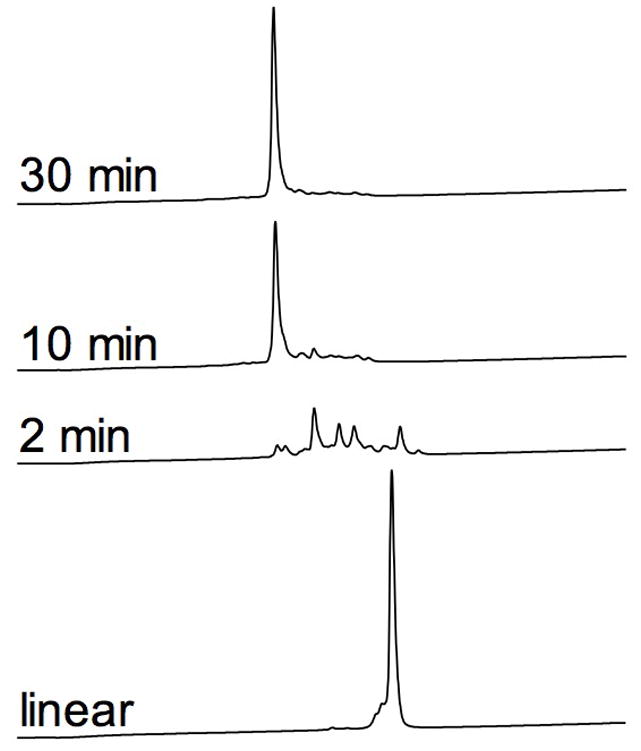
Oxidative folding of conotoxin cce9a. The folding reaction was carried out in 0.1 M Tris-HCl (pH 7.5), 0.1 mM EDTA, in the presence of 1 mM GSSG and 1 mM GSH at the ambient temperature (23–25 °C). At each time point, an aliquot was collected, quenched and subjected to HPLC analysis as described under Materials and Methods. The folded peptide was shown to be identical to the native sample by mass spectrometry and HPLC coelution.
3.4. Developmentally dependent biological activity
Intracranial bioassay using synthetic cce9a revealed that the behavioral phenotype elicited in the mice was age-dependent (Table 1). In younger mice (12 and 14 days old), cce9a induced lethargy and a delayed response to stimuli at doses of 5 to 40 nmol per mouse (6 – 7 g of body weight). Recovery to normal behavior took a longer period as the administered dose was increased. Control mice exhibited non-lethargic and responsive behavior. In older mice (16 days old), the peptide elicited hyperactivity at doses of 10 to 50 nmol per mouse. The onset of hyperactivity, which was characterized by climbing, running, and jumping, was more immediate at the higher injected doses.
TABLE 1.
Age-dependent Effects of cce9a on Mice
| Age of Mice (days) | cce9a Dose (nmol) | Observed Behavior (time = min post injection) |
|---|---|---|
| 12 (Mice wt.: 6–7 g) | 0 | Normal: moving, responsive when poked/pinned/touched/dropped (stimuli) |
| 5 | Lethargic; less responsive to non-responsive to stimuli 15 min to 35 min | |
| 10 | Lethargic; less responsive to non-responsive to stimuli 15 min to 50 min | |
| 20 | Lethargic; less responsive to non-responsive to stimuli 10 – 15 min to > 60 min | |
| 30 | Lethargic; less responsive to non-responsive to stimuli 10 – 15 min to > 60 min | |
| 14 (Mice wt.: 7.5–8 g) | 0 | Normal: moving, responsive to stimuli |
| 5 | Similar to control | |
| 10 | Lethargic; less responsive to stimuli; hunched back 15 min to 50 min | |
| 20 | Lethargic; less responsive to stimuli; hunched back 10 min to 60 min | |
| 30 | Lethargic; less responsive to stimuli; hunched back 5 min to > 60 min | |
| 40 | Lethargic; less responsive to stimuli; hunched back 5 min to > 60 min | |
| 16 (Mice wt.: 8–9 g) | 0 | Normal: moving, responsive to stimuli |
| 5 | Similar to control | |
| 10 | Hyperactive (constantly moving around cage, climbing walls, jumping) 10 min to 45 min | |
| 20 | Hyperactive 10 min to > 60 min | |
| 30 | Hyperactive 5 – 10 min to > 60 min | |
| 40 | Hyperactive 5 – 10 min to > 60 min | |
| 50 | Hyperactive 3 – 5 min to > 60 min |
4. Discussion
The present study describes the purification and characterization of the first venom component from a member of a biodiverse group of toxoglossate molluscs, the crassispirids. It is likely that the crassispirids will be comparable in their biodiversity, and perhaps even exceed the ~700 species of cone snails and ~400 species of terebrids. Thus, this group of venomous animals has the potential to yield as many bioactive venom components as the cone snails.
The species that we investigated, Crassispira cerithina, is one of >10,000 species that were loosely referred to as “turrids”; the phylogeny and taxonomy of the species has had a confusing history. The species was known earlier as Turridrupa cerithina (Powell, 1966); despite the earlier assignment to the genus Turridrupa (a genus in the subfamily Turrinae), our phylogenetic analysis definitively establishes that Crassispira cerithina does not belong to this subfamily. Instead, as shown in Fig. 2, Crassispira cerithina, together with species in the genus Inquisitor, as well as the new world genus Zonulospira, comprise a different major branch. These genera were assigned to the subfamily Crassispirinae in most prior taxonomic work. Another new world species, Pseudomelatoma moesta, is more distantly related, but appears to be allied to these genera.
It has recently been suggested that the various families conventionally included in the superfamily Conoidea fall into two major groups, and that the families less closely related to Conus be moved to a new superfamily, Turroidea (Tucker and Tenorio, 2009). This suggestion is consistent with the molecular phylogeny shown in Fig. 2; the proposed superfamily Turroidea would encompass 4 major branches of the phylogenetic tree shown in Fig. 2: the auger snails or terebrids, (family Terebridae), the turrids (family Turridae, narrowly defined), the drillids (family Drillidae) and the crassispirids.
The “crassispirids”, as defined above, include several (but not all) genera assigned to the subfamily Crassispirinae (i.e., Crassispira, Inquisitor and Zonulospira); this corresponds well to “Clade 2” of Puillandre et al., (2008). It should be noted that the molecular phylogeny in Fig. 2 is somewhat divergent from the recent literature on the conventional phylogeny of the group (see Bouchet and Rocroi, 2005; Poppe, 2008; Taylor et al., 1993; Tucker and Tenorio, 2009) that generally treat crassispirids as more closely related to the turrines (subfamily Turrinae) than to the drillids (Dillinae/Drillidae). The phylogenetic analysis that we carried out and prior molecular phylogeny indicate that the converse is true: the sister group of the crassispirids are the drillids (e.g., Drillia, Clavus, etc.). It should be noted that the taxonomy and phylogeny of the crassispirids need further elucidation, since there are many species that may be crassispirids, but in the absence of molecular data, their assignment to this group is uncertain. These two major branches of venomous molluscs, the crassispirids and the drillids, are unexplored territory for toxinologists, with the present work providing the very first characterization of any venom component from these species-rich lineages of venomous gastropods. We propose to call venom peptides from crassispirids and drillids “crassipeptides” and “drillipeptides”, respectively, to distinguish them from “conopeptides”, “augerpeptides” and “turripeptides” from cone snails, terebrids and turrids, respectively.
The approach that we used to purify and characterize the first crassipeptide was one that has been extraordinarily productive for conopeptides—assessing the behavioral phenotype of a mouse after injection of a venom component into the central nervous system. The peptide we purified caused lethargy in younger mice, with a muted response to external stimuli. This behavioral phenotype allowed the purification and characterization of crassipeptide cce9a, a peptide with 29 amino acid residues and three disulfide bonds.
As shown in Table 2, the mature crassipeptide cce9a, whose sequence was determined using standard Edman methods on the purified native peptide and confirmed by sequencing a cDNA clone (Fig. 4), has a striking similarity to conopeptides in the Conus P-superfamily with Framework IX cysteine pattern (Lirazan et al., 2000) with respect to the arrangement and spacing of cysteine residues. However, apart from the pattern of cysteine residues, there is no other sequence similarity between cce9a and P-superfamily Conus peptides. Furthermore, the symptomatology induced by the spasmodic peptides, tx9a from Conus textile and gm9a from Conus gloriamaris, and cce9a from Crassispira cerithina are strikingly divergent.
TABLE 2.
Comparison of Venom Peptides with cysteine Framework IX
| Venom Peptide Class | Name | Sequence | Reference |
|---|---|---|---|
| Crassipeptides: | cce9a | GSCGLPCHENRRCGWACYCDDGICKPLRV | This work |
| Conopeptides: | gm9a | -SCNNSCQSHSDCASHCICTFRGCGAVN* | Lirazan et al. (2000) |
| tx9a | GCNNSCQγHSDCγ SHCICTFRGCGAVN* | Lirazan et al. (2000) | |
| Turripeptides: | gsp9a | IDOORYCNHIICYγDSγ CSQWCTAGCNSITSKCDT | Heralde et al. (2008) |
| gsp9b | GDOORFCRDKLCSGDGDCSV WCTAGCNHDMGKCDTL | Heralde et al. (2008) | |
| Augerpeptides: | hhe9a | YEENCGTEYCTSKIGCPGRCVCKEYNYNGEITRRCRA | Imperial et al. (2007) |
Other peptides with the Framework IX cysteine pattern have been purified from Gemmula speciosa, which belongs to the family Turridae (ss) (Heralde et al., 2008), and Hastula hectica, which belongs to the family Terebridae (Imperial et al., 2007). The sequences of the peptides purified from the venom of G. speciosa and H. hectica are compared to the conopeptides and to cce9a in Table 2; it is notable that although in the phylogenetic tree, the crassispirids are more closely related to turrids and terebrids than to Conus, the peptides from the turrid and terebrid appear to be more divergent from cce9a than are the corresponding conopeptides, considering the size of the intercysteine regions.
Crassipeptide cce9a was further characterized after the successful chemical synthesis of the biologically active peptide. As shown in Table 1, the behavioral phenotype elicited by the peptide is dependent upon the age of the mice injected; the peptide causes a lethargic state in young mice (12 and 14 days old), which is refractory to external stimuli; however, in older mice (16 days old) the peptide elicited hyperactivity, which is characterized by climbing, running and jumping. The switch in behavioral phenotype as a function of age is reminiscent of conopeptides that belong to the conantokin family. The first conantokin characterized, Conantokin G from Conus geographus (Olivera et al., 1985), caused a sleep-like state in young mice, but at approximately the same age interval, there was a transition to hyperactivity. Conantokin G is a well-established NMDA receptor antagonist; given the parallel developmental switch in behavioral phenotype, we tested cce9a for activity on NMDA receptors—these assays were negative (results not shown). Thus, the basis for the change in behavioral phenotype as a function of age is apparently due to a molecular mechanism different from the one for the conantokins; however, it remains possible that the behavioral shift indicates that cce9a interacts with at least 2 types of molecular target with the effects of one type predominating over the other differentially with age (Rivier et al., 1987).
Thus, the characterization of the venom component described in this work, Crassipeptide cce9a, is noteworthy in several respects: it is the first biochemical characterization of any toxin from the venom of a crassispirid species, and it has a novel biological activity in mammals: the behavioral symptomatology elicited by intracranial injections into mice is developmentally dependent. Thus, it is reasonable to expect that many crassipeptides with novel pharmacology will be discovered and characterized that should prove useful for investigating molecular mechanisms in nervous systems.
Supplementary Material
Highlights.
Crassispira cerithina belongs to a new major clade, the “crassispirines”.
Crassipeptide cce9a is 29 amino acids long and has the Framework IX cysteine pattern.
Crassipeptide cce9a is active by intracranial injection in mice.
Acknowledgments
This work was carried out through the Philippine PharmaSeas Drug Discovery Program, supported by the Philippine Council for Aquatic and Marine Research and Development, Department of Science and Technology (DOST), Philippines. This work was supported in part by a Program Project grant from the National Institutes of Health, GM 48677. The field collection was done in collaboration with a team supported by the Philippine Mollusk Symbiont International Cooperative Biodiversity Grant (NIH 1U01TW008163-01) from the Fogarty International Center, National Institutes of Health. JSI thanks the DOST for visits to the University of the Philippines Marine Science Institute through the Balik Scientist Program. We thank Vernon Twede for carrying out the NMDA assays, and Greg Bulaj and Pradip Bandyopadhyay for advice.
Footnotes
Conflict of Interest Statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar MB, de la Rosa RA, Falcon A, Olivera BM, Heimer de la Cotera EP. Peptide pal9a from the venom of the turrid snail Polystira albida from the Gulf of Mexico: Purification, characterization, and comparison with P-conotoxin-like (framework IX) conoidean peptides. Peptides. 2009;30:467–476. doi: 10.1016/j.peptides.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet P, Lozouet P, Maestrati P, Heros V. Assessing the magnitude of species richness in tropical marine environments: high numbers of molluscs at a New Caledonia site. Biological Journal of the Linnean Society. 2002;75:421–436. [Google Scholar]
- Bouchet P, Rocroi JP. Malacologia: International Journal of Malacology, Classification and Nomenclator of Gastropod Families. Malacologia - International Journal of Malacology, ConchBooks 2005 [Google Scholar]
- Craig AG, Zafaralla G, Cruz LJ, Santos AD, Hillyard DR, Dykert J, Rivier JE, Gray WR, Imperial J, DelaCruz RG, Sporning A, Terlau H, West PJ, Yoshikami D, Olivera BM. An O-glycosylated neuroexcitatory Conus peptide. Biochemistry. 1998;37:16019–16025. doi: 10.1021/bi981690a. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Biology and Biotechnology. 1994;3:294 – 299. [PubMed] [Google Scholar]
- Fuller E, Green BR, Catlin P, Buczek O, Nielsen JS, Olivera BM, Bulaj G. Oxidative folding of conotoxins sharing an identical disulfide bridging framework. Federation of European Biochemical Societies Journal. 2005;272:1727–1738. doi: 10.1111/j.1742-4658.2005.04602.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Heralde FM, 3rd, Imperial J, Bandyopadhyay PK, Olivera BM, Concepcion GP, Santos AD. A rapidly diverging superfamily of peptide toxins in venomous Gemmula species. Toxicon. 2008;51:890–897. doi: 10.1016/j.toxicon.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Imperial JS, Chen P, Sporning A, Terlau H, Daly NL, Craik DJ, Alewood PF, Olivera BM. Tyrosine-rich conopeptides affect voltage-gated K+ channels. J Biol Chem. 2008;283:23026–23032. doi: 10.1074/jbc.M800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperial JS, Kantor Y, Watkins M, Heralde FM, 3rd, Stevenson B, Chen P, Hansson K, Stenflo J, Ownby JP, Bouchet P, Olivera BM. Venomous auger snail Hastula (Impages) hectica (Linnaeus, 1758): molecular phylogeny, foregut anatomy and comparative toxinology. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2007;308:744–756. doi: 10.1002/jez.b.21195. [DOI] [PubMed] [Google Scholar]
- Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, Olivera BM. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- López-Vera E, Heimer de la Cotera EP, Maillo M, Riesgo-Escovar JR, Olivera BM, Aguilar MB. A novel structural class of toxins: the methionine-rich peptides from the venoms of turrid marine snails (Mollusca, Conoidea) Toxicon. 2004;43:365–374. doi: 10.1016/j.toxicon.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4.08. 2005. [Google Scholar]
- McIntosh JM, Yoshikami D, Mahe E, Nielsen DB, Rivier JE, Gray WR, Olivera BM. A nicotinic acetylcholine receptor ligand of unique specificity, a-conotoxin ImI. Journal of Biological Chemistry. 1994;269:16733–16739. [PubMed] [Google Scholar]
- Olivera BM, Cruz LJ, de Santos V, LeCheminant G, Griffin D, Zeikus R, McIntosh JM, Galyean R, Varga J, Gray WR, Rivier J. Neuronal Ca channel antagonists. Discrimination between Ca channel subtypes using ε-conotoxin from Conus magus venom. Biochemistry. 1987;26:2086–2090. doi: 10.1021/bi00382a004. [DOI] [PubMed] [Google Scholar]
- Olivera BM, McIntosh JM, Clark C, Middlemas D, Gray WR, Cruz LJ. A sleep-inducing peptide from Conus geographus venom. Toxicon. 1985;23:277–282. doi: 10.1016/0041-0101(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Rivier J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, Mena EE, Woodward SR, Hillyard DR, Cruz LJ. Diversity of Conus neuropeptides. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. PCR. In: IM, et al., editors. Molecular Systematics. Sinauer Press; Sunderland, MA: 1996. [Google Scholar]
- Poppe GT. Philippine Marine Mollusks. In: Poppe GT, editor. Gastropoda. Part 2. ConchBooks; 2008. p. 848. [Google Scholar]
- Powell AWB. The molluscan families Speightiidae and Turridae. Bulletin of the Auckland Institute and Museum. 1966;5:1–184. + 23 plates. [Google Scholar]
- Powell AWB. The family Turridae in the Indo-Pacific. Part 1a. The subfamily Turrinae concluded. Indo-Pacific Moll. 1967;1:409–432. [Google Scholar]
- Puillandre N, Samadi S, Boisselier MC, Sysoev AV, Kantor YI, Cruaud C, Couloux A, Bouchet P. Starting to unravel the toxoglossan knot: molecular phylogeny of the “turrids” (Neogastropoda: Conoidea) Molecular Phylogenetic Evolution. 2008;47:1122–1134. doi: 10.1016/j.ympev.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Rivier J, Galyean R, Simon L, Cruz LJ, Olivera BM, Cruz LJ. Total synthesis and further characterization of the γ-carboxyglutamate-containing “sleeper” peptide from Conus geographus venom. Biochemistry. 1987;26:8508–8512. doi: 10.1021/bi00400a002. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Simon C. Molecular systematics at the species boundary: exploiting conserved and variable regions of the mitochondrial genome of animals via direct sequencing from amplified DNA. In: Hewitt GM, Johnston AWB, Young JPW, editors. Molecular techniques in taxonomy, NATO ASI Series. H57. 1991. pp. 33–71. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–71. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tavaré S. Some Probablsistic and Statistical Problems in the Analysis of DNA Sequences. American Mathematical Society: Lectures on Mathematics in the Lif Sciences. 1986;17:57–86. [Google Scholar]
- Taylor JD, Kantor Y, Sysoev AV. Foregut anatomy, feeding mechanisms, relationships and classification of the Conoidea (=Toxoglossa) (Gastropoda) Bulletin, Natural History Museum, London Zoology. 1993;59:125–170. [Google Scholar]
- Teichert RW, Jimenez EC, Twede V, Watkins M, Hollmann M, Bulaj G, Olivera BM. Novel conantokins from Conus parius venom are specific antagonists of N-methyl-D-aspartate receptors. J Biol Chem. 2007;282:36905–36913. doi: 10.1074/jbc.M706611200. [DOI] [PubMed] [Google Scholar]
- Teichert RW, Olivera BM. Natural products and ion channel pharmacology. Future Med Chem. 2010;2(5):731–744. doi: 10.4155/fmc.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiological Reviews. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Tucker JK, Tenorio MJ. Systematic classification of Recent and fossil conoidean gastropods. Conchbooks; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.