Abstract
Major acetate-utilizing bacterial and archaeal populations in methanogenic anaerobic digester sludge were identified and quantified by radioisotope- and stable-isotope-based functional analyses, microautoradiography-fluorescence in situ hybridization (MAR-FISH) and stable-isotope probing of 16S rRNA (RNA-SIP) that can directly link 16S rRNA phylogeny with in situ metabolic function. First, MAR-FISH with 14C-acetate indicated the significant utilization of acetate by only two major groups, unidentified bacterial cells and Methanosaeta-like filamentous archaeal cells, in the digester sludge. To identify the acetate-utilizing unidentified bacteria, RNA-SIP was conducted with 13C6-glucose and 13C3-propionate as sole carbon source, which were followed by phylogenetic analysis of 16S rRNA. We found that bacteria belonging to Synergistes group 4 were commonly detected in both 16S rRNA clone libraries derived from the sludge incubated with 13C-glucose and 13C-propionate. To confirm that this bacterial group can utilize acetate, specific FISH probe targeting for Synergistes group 4 was newly designed and applied to the sludge incubated with 14C-acetate for MAR-FISH. The MAR-FISH result showed that bacteria belonging to Synergistes group 4 significantly took up acetate and their active population size was comparable to that of Methanosaeta in this sludge. In addition, as bacteria belonging to Synergistes group 4 had high Km for acetate and maximum utilization rate, they are more competitive for acetate over Methanosaeta at high acetate concentrations (2.5–10 m). To our knowledge, it is the first time to report the acetate-utilizing activity of uncultured bacteria belonging to Synergistes group 4 and its competitive significance to acetoclastic methanogen, Methanosaeta.
Keywords: acetate-utilizing bacterium, anaerobic digestion, MAR-FISH, RNA-SIP, Synergistes
Introduction
Different groups of anaerobic microorganisms decompose organic matter in a series of steps, which ultimately produce methane and carbon dioxide as terminal products. Methane is formed from two primary substrates, acetate and H2/CO2 (or formate). Acetate is an important intermediate of anaerobic decomposition of organic matter, as about two-third of methane is produced from acetate in anaerobic digestion reactors (McCarty and Smith, 1986). Thus, acetoclastic methanogens including mainly Methanosaeta and Methanosarcina have been well studied (Zinder, 1998). Methanogenic acetate degradation is carried out by either the methanogenic archaea or some anaerobic acetate-oxidizing bacteria. When inorganic electron acceptors other than CO2 are absent, acetate is degraded by syntrophic acetate oxidation coupled to hydrogen-consuming process, for example, hydrogenotrophic methanogenesis. Otherwise, anaerobic acetate oxidation reaction is energetically extremely unfavorable. Only a few syntrophic acetate-oxidizing bacteria have been successfully cultivated to date owing to their slow growth rates and difficulty of reproducing their growth conditions (Schink and Stams, 2006; Hattori, 2008). Therefore, information on the population and diversity of anaerobic acetate-oxidizing bacteria and the competition for acetate with acetate-utilizing methanogen in anaerobic digester sludge is limited.
Microautoradiography combined with fluorescence in situ hybridization (MAR-FISH) is a powerful technique to simultaneously determine the phylogenetic identity and in situ specific metabolic function of microorganisms in complex microbial community at a single-cell resolution (Andreasen and Nielsen, 1997; Okabe et al., 2004). Thus, applying this technique, it is possible to identify and quantify slow-growing acetate-utilizing microorganisms in anaerobic digester sludge without the need to isolate them in culture. However, selection and design of specific FISH probes requires phylogenic information (16S rRNA gene sequences) of cultivated and yet-uncultivated microorganisms, which are actively involved in acetate utilization in advance. For this reason, ribosomal RNA-based stable-isotope probing (RNA-SIP) combined with full-cycle 16S rRNA analysis is useful to identify active acetate-utilizing microorganisms under methanogenic conditions and to design specific FISH probes for the following MAR-FISH analysis. RNA-SIP is a powerful approach to directly identify microbial populations active in a defined metabolic process and has recently been applied to identify acetate-utilizing microbial populations in methanogenic lake sediments (Schwarz et al., 2007). It was found that acetate was predominantly consumed by acetoclastic methanogens in the lake sediment. However, this study did not use FISH or MAR-FISH to quantitatively determine the population sizes of acetate-utilizing methanogens and bacteria.
In this study, first, RNA-SIP combined with full-cycle 16S rRNA analysis was performed to identify active acetate-utilizing bacteria in anaerobic digester sludge, and then MAR-FISH with newly designed FISH probes was conducted to quantitatively investigate their acetate-utilizing activity and competitive relationship with acetoclastic methanogens at different concentrations of acetate. The results of RNA-SIP and MAR-FISH showed that bacteria belonging to the Synergistes group 4 were only dominant acetate-utilizing bacteria in the anaerobic digestersludge and they had lower affinity to acetate and higher utilization rate than Methanosaeta like acetoclastic methanogen at high acetate concentrations.
Materials and methods
Anaerobic sludge samples
Anaerobic sludge samples were taken from the mesophilic anaerobic fed-batch reactors that were operated stably for more than 2 years in our laboratory. The seed sludge for the anaerobic digesters was obtained from an anaerobic mesophilic digester at the Ebetsu municipal wastewater treatment plant located at Ebetsu city, Hokkaido, Japan. Powdered whole-milk (Meiji Dairies Corporation, Tokyo, Japan) composed of carbohydrate (57%), lipid (25%) and protein (13%) was fed every 2 days at a loading rate of 1.5 g COD l−1 day−1. Mineral solution was supplemented with the powdered whole-milk. Other details of the reactor operation and the composition of the mineral solution were described elsewhere (Ariesyady et al., 2007b).
Analytical measurements
CH4, CO2 and H2 were analyzed by gas chromatography (Shimazu, Kyoto, Japan) equipped with a thermal conductivity detector and a 6-m, 2-mm i.d. SHINCARBON T column (Shinwa Chemical, Kyoto, Japan). Volatile fatty acids were determined with an ion chromatograph equipped with an ICE-AS1 column (DX-100, Dionex, Sunnyvale, CA, USA).
MAR-FISH
Incubation with radiolabeled substrates
In all, 10 ml slurry samples were taken from the anaerobic digester and centrifuged at 2500 g for 5 min. Of these, 8 ml of the supernatant was replaced with the mineral solution (without powdered milk). The sludge and mineral solution were gently mixed, and then 3 ml of the mixtures were transferred to 5-ml serum bottles. The serum bottles were sealed with gas-tight rubber stoppers and anaerobically incubated with shaking at 30 r.p.m. at 37 °C with [U-14C]glucose for 1 h, [1-14C]propionate for 2 h or [2-14C]acetate for 5 h. [U-14C]Glucose was added with unlabeled glucose (14C/(12C+14C); 1%) to give a final concentration of 2.5 m. [1-14C]Propionate was added with unlabeled propionate (14C/(14C+12C); 8%) to give a final concentration of 1.5 m. [2-14C]Acetate was added with unlabeled acetate (14C/(12C+14C); 25%) to give final concentrations of 0.5, 1.0, 2.5, 5 and 10 m. Controls were prepared by pasteurizing the sludge at 70 °C for 30 min and run in parallel for all analyses.
The radiolabeled [U-14C]glucose and [2-14C]acetate were purchased from the Amersham Pharmacia Biotech (Buckinghamshire, UK). The radiolabeled [1-14C]propionate was purchased from the American Radiolabeled Chemicals Inc. (St Louis, MO, USA). The specific activity of [U-14C]glucose, [1-14C]propionate and [2-14C]acetate was 11.7 GBq mmol−1, 2.07 GBq mmol−1 and 2.26 GBq mmol−1, respectively.
Liquid scintillation counting
The uptakes of radiolabeled substrates were measured for all cultures by liquid scintillation counting before FISH and microautoradiographic procedures as described by Ariesyady et al. (2007b) and Ito et al. (2002).
Sample fixation, washing and FISH
The incubation was terminated by 4% paraformaldehyde. The fixation of the samples, washing and FISH were conducted according to Okabe et al. (1999) and Ito et al. (2002). After FISH, sample slides were stained with 4′, 6-diamidino-2-phenylindole to determine total cell numbers (Okabe et al., 2007).
FISH probes
Bacteria and Archaea target oligo nucleotide probes used in this study were listed in Table 1. The probes were labeled with fluoresceinisothiocyanate or tetramethylrhodamine 5-isothiocyanate at the 5′ end.
Table 1. FISH oligonucleotide probes used in this study.
| Probe | Sequence (5′–3′) | rRNA target site (Escherichia coli numbering) | Specificity | % FAa | Reference |
|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S (338–355) | Most but not all Bacteria | —b | Amann et al. (1990) |
| EUB338-II | GCAGCCACCCGTAGGTGT | 16S (338–355) | Bacterial groups not covered by EUB338 and EUB338-III | —b | Daims et al. (1999) |
| EUB338-III | GCTGCCACCCGTAGGTGT | 16S (338–355) | Bacterial groups not covered by EUB338 and EUB338-II | —b | Daims et al. (1999) |
| ARC915 | GTGCTCCCCCGCCAATTCCT | 16S (915–934) | Archaea | 35 | Stahl and Amann (1991) |
| MX825 | TCGCACCGTGGCCGACACCTAGC | 16S (825–847) | Some Methanosaetaceae | 50 | Raskin et al. (1994) |
| MS821 | CGCCATGCCTGACACCTAGCGAGC | 16S (821–844) | Methanosarcina | 40 | Raskin et al. (1994) |
| MS1414 | CTCACCCATACCTCACTCGGG | 16S (1414–1434) | Genus I, II, IV and V of Methanosarcinaceae | 50 | Raskin et al. (1994) |
| MB1174 | TACCGTCGTCCACTCCTTCCTC | 16S (1175–1196) | Methanobacteriaceae | 45 | Raskin et al. (1994) |
| MG1200 | CGGATAATTCGGGGCATGCTG | 16S (1200–1220) | Family I, II and III of Methanomicrobiales | 20 | Raskin et al. (1994) |
| ALF1b | CGTTCG(C/T)TCTGAGCCAG | 16S (19–35) | Alphaproteobacteria some other bacteria | 20 | Manz et al. (1992) |
| BET42a | GCCTTCCCACTTCGTTT | 23S (1027–1043) | Betaproteobacteria | 35 | Manz et al. (1992) |
| GAM42a | GCCTTCCCACATCGTTT | 23S (1027–1043) | Gammaproteobacteria | 35 | Manz et al. (1992) |
| SRB385 | CGGCGTCGCTGCGTCAGG | 16S (385–402) | Most Desulfovibrionales and other bacteria | 30 | Amann et al. (1990) |
| SRB385Db | CGGCGTTGCTGCGTCAGG | 16S (385–402) | Desulfovibrionaceae and other bacteria | 30 | Rabus et al. (1996) |
| LGC354a | TGGAAGATTCCCTACTGC | 16S (354–371) | Firmicutes | 35 | Meier et al. (1999) |
| LGC354b | CGGAAGATTCCCTACTGC | 16S (354–371) | Firmicutes | 35 | Meier et al. (1999) |
| LGC354c | CCGAAGATTCCCTACTGC | 16S (354–371) | Firmicutes | 35 | Meier et al. (1999) |
| HGC69a | TATAGTTACCACCGCCGT | 23S (1901–1918) | Actinobacteria | 20 | Roller et al. (1994) |
| CF319a/b | TGGTCCGTRTCTCAGTAC | 16S (319–336) | Most Flavobacteria, some Bacteroidetes, some Sphingobacteria | 35 | Manz et al. (1992) |
| CFB563 | GGACCCTTTAAACCCAAT | 16S (563–580) | Bacteroidetes | 20 | Weller et al. (2000) |
| PLA46 | GACTTGCATGCCTAATCC | 16S (46–63) | Planctomycetales | 30 | Neef et al. (1998) |
| GNSB-941 | AAACCACACGCTCCGCT | 16S (941–957) | Chloroflexi | 35 | Gich et al. (2001) |
| CFX1223 | CCATTGTAGCGTGTGTGTMG | 16S (1223–1242) | Chloroflexi | 35 | Björnsson et al. (2002) |
| Syner195 | GCAGTACTCGCGTACCTT | 16S (195–212) | Synergistes group 4 (Synergistetes PD-UASB-13) | 10–20 | This study |
Abbreviation: FISH, fluorescence in situ hybridization.
FA, formamide concentration in the hybridization buffer.
The probe can be used at any formamide concentrations.
The probe Syner195 was designed using the probe design tool of the ARB software package (Ludwig et al., 2004). Probe sequences were confirmed for specificity using the probe check tool of the Ribosomal Database Project (Cole et al., 2005). Specific formamide concentrations for the 9probes were experimentally determined by performing FISH at different formamide concentrations of 0%, 10%, 15%, 20% and 30%. Although no pure cultures of Syner195 target Synergistes group 4 bacteria have been obtained, the specific signals were identifiable in the digester sludge by the small-rod morphotype and MAR-positive signals with 14C-acetate. Applying this probe to the digester sludge after incubation with 14C-acetate, FISH signals of the MAR-positive cells of the characteristic morphotype became weaker above 10% formamide concentration. Thus, we selected 10% formamide as the optimal formamide concentration. The determined formamide concentration was listed in Table 1.
Autoradiographic developing procedure
Following the FISH, the autoradiographic procedure was performed directly on the cover glasses by using liquid film emulsion (LM-1, Amersham Pharmacia Biotech, Piscataway, NJ, USA) (Lee et al., 1999; Kindaichi et al., 2004). The optimum exposure time was determined to be 5 days for the samples incubated with [2-14C]acetate and [1-14C]propionate, and to be 2 days for the samples incubated with [U-14C]glucose in preliminary experiments.
Microscopy and enumeration by MAR-FISH
A model LSM510 confocal laser scanning microscope (Carl Zeiss, Oberkochen, Germany) equipped with an ultraviolet laser (351 nm and 364 nm), an Ar ion laser (450–514 nm) and two HeNe lasers (543 nm and 633 nm) were used. The formation of silver grains in the autoradiographic film was observed by using the transmission mode of the confocal laser scanning microscope system. A MAR-positive cell was defined as a cell covered with more than four silver grains in this study (Okabe et al., 2005). The numbers of MAR-positive cells and total probe-hybridized cells (or total MAR-positive cells) were determined in triplicate by directly counting at least 1000 silver grain-covered cells in randomly chosen microscopic fields of a few slides prepared for each sample.
Incubation with 13C-labeled substrate ([13C6]glucose and [13C3]propionate)
In total, 25 ml slurry samples were taken from the anaerobic reactor, transferred to a 50 ml polycarbonate tube and centrifuged at 2500 g for 5 min. The centrifugation resulted in approximately 5 ml of sludge and 20 ml of supernatant. After replacing the gas phase of the sample tube with N2 and CO2 (80:20) gas, the sample tube was transferred to an anaerobic chamber containing N2 and CO2 (80:20). The following preparation processes and incubation were conducted in the anaerobic chamber. The supernatant was first replaced with 20 ml of the mineral solution, and then the mixture of the mineral solution and sludge was transferred to a 30-ml glass vial. For incubation with 13C-labeled glucose (13C6, >99 at% 13C; Isotec, Miamisburg, OH, USA), the vial was incubated with 2.5 m of glucose for 48 h at 37 °C. The 2.5m glucose was degraded to CH4 and CO2 via acetate and propionate as detectable intermediates during the 48-h incubation. For the incubation with 13C-labeled propionate (13C3, >99 at% 13C; Isotec), 13C3-propionate dissolved in the mineral solution was continuously fed into the vial at a loading rate of 0.18 mmol l−1 h−1 by a microsyringe pump (IC3100; KD Scientific Inc., Holliston, MA, USA) to keep propionate concentration around 0.5 m. The loading rate was determined at a higher propionate degradation rate of the digester sludge of the anaerobic reactor. The vial was incubated at 37 °C for 58 h under anoxic condition (in an anoxic grove box). During the 58-h incubation period, the concentration of propionate was monitored every 3–5 h and was confirmed to be in the range of 0.1–0.5 m.
RNA extraction and fractionation
The sludge sample (25 ml) was centrifuged at 15 000 g for 10 min immediately after the incubation with either 13C6-glucose or 13C3-propionate. Total RNA was extracted from the entire harvested pellet with the FastRNA Pro Soil-Direct Kit (Qbiogene Inc., Irvine, CA, USA). The extracted RNA was purified before DNase I digestion (Lueders et al., 2004a, 2004b). The purified RNA (12 μg) was subsequently loaded with cesium trifluoroacetate equilibrium density gradient in 2.0-ml Beckman Quick-Seal polyallomer Bell Top tubes, and subjected to density gradient centrifugation with Optima TLX (Beckmann Coulter, Tokyo, Japan) at 64 000 r.p.m. and 20 °C for 36 h (Manefield et al., 2002a, 2002b). Centrifuged gradients were fractionated into 20 gradient fractions with the fraction recovery system (Bechmann Coulter) at the flow rate of 3.3 μl s−1 by displacement with ddH2O using a syringe pump (Manefield et al., 2002b). In fractions 1 to 20, fraction 1 was the first faction collected from the bottom of the gradient. The amount of RNA of each gradient fraction was quantified fluorometrically by RiboGreen assay (Invitrogen, Carlsbad, CA, USA) (Lueders et al., 2004a). In control experiment with unlabeled RNA from the digester sludge, RiboGreen measurements showed that unlabeled RNA enriched between fractions 10 and 15 (with peak fraction 12). After the incubation with either 13C6-glucose or 13C3-propionate, RNA also appeared between fractions 5 and 7 (with peak fraction 6).
Reverse transcription-polymerase chain reaction, cloning, sequencing and phylogenetic analysis
For cloning and sequencing analysis, the fraction 6 that contained heavy 13C-labeled RNA was amplified with the Superscript III one-step reverse transcription-polymerase chain reaction kit (Invitrogen) using bacterial primer pair 8f and 1492r (Lane, 1991). The reverse transcription-polymerase chain reaction was carried out with the following amplification program: one cycle consisting of 55 °C for 30 min (reverse transcription) and 94 °C for 2 min, and then 40 cycles consisting of 94 °C for 15 s; 54 °C for 30 s and 68 °C for 2 min, followed by final extension at 68 °C for 7 min. The reverse transcription-polymerase chain reaction product was gel-purified and cloned by using a TOPO XL PCR cloning kit (Invitrogen). Randomly selected clones were sequenced on an ABI model 3100-Avant genetic analyzer with a BigDye terminator Ready Reaction kit (Applied Biosystems, Foster City, CA, USA). The sequences obtained were compared with reference 16S rRNA gene sequences available in the GenBank/EMBL/DDBJ databases using the BLAST search (Altschul et al., 1997). Sequences with 97% or greater similarity were grouped into operational taxonomic units. Phylogenetic analysis was performed using the MEGA3 software package (Kumar et al., 2004) after multiple alignments of data by CLUSTAL W (Thompson et al., 1994). The phylogenetic trees were constructed using neighbor-joining and maximum-parsimony methods. The confidence level for nodes was ascertained by performing a bootstrap analysis (1000 replications).
Determination of acetate degradation rates
Acetate degradation rates of the anaerobic sludge at different acetate concentrations were determined with [2-14C]acetate by liquid scintillation counting. The experimental setup (the incubation condition) was the same as that for MAR-FISH (see MAR-FISH—Incubation with radiolabeled substrates). [2-14C]Acetate was added with unlabeled acetate to give final concentrations of 0.5 m, 1.0 m, 2.5 m, 5 m, 10 m and 20 m. Acetate is not only degraded, but also produced in the anaerobic sludge. For example, acetate could be produced by anaerobic self-degradation of the sludge and also produced by anaerobic degradation of residual substrates. Measuring the amount of [2-14C]acetate makes it possible to exclude acetate production by these other processes.
Culture samples were taken at the incubation times of 0 h, 2 h, 4 h, 6 h, 8 h and 10 h. After centrifugation of the samples at 15 000 g for 10 min, a trace amount of sulfuric acid was added to the supernatant to de-gas 14C-carbon dioxide that could be produced from the degradation of 14C-acetate. Then, 14C in the supernatant, that is, residual 14C-acetate in the supernatant, was determined by liquid scintillation counting. The addition of sulfuric acid did not influence the measurements of acetate concentrations. Acetate degradation rates (μmol gVSS−1 h−1) at different acetate concentrations were calculated from the slopes of liner fitting for time-dependent changes in acetate concentrations during the incubation. Km and Vmax were determined by the Lineweaver–Burke plot of the rates of acetate degradation versus acetate concentrations.
Acetate degradation rate of two probe-defined Synergistes and Methanosaeta groups were determined by multiplying the acetate degradation rates of the anaerobic digester sludge by the fraction of [14C]acetate-utilizing MAR-positive Synergistes group 4 and Methanosaeta populations at different acetate concentrations. In this study, the acetate uptake activity (that is, the number of silver grains accumulated on the cells) of both Synergistes and Methanosaeta is more closely related to the cell number than the cell size, even though the cell size of filamentous Methanosaeta was bigger than one of Synergistes. Therefore, we used the fraction of cell numbers instead of the cell area.
Nucleotide sequence accession numbers
Sequences were deposited in the GenBank/EMBL/DDBJ database under accession numbers AB603808–AB603841.
Results
In situ detection of acetate-utilizing archaea and bacteria
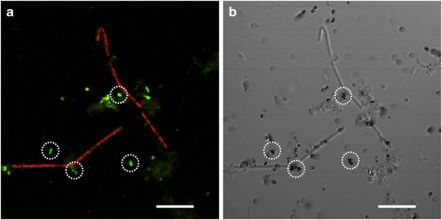
The populations of anaerobic acetate-utilizing bacteria and archaea in anaerobic digester sludge were investigated by microautoradiography-FISH technique with 14C-acetate and domain bacteria and archaea-specific oligonucleotide probes. Small-rod bacterial cells and filamentous archaeal cells were mainly detected as MAR-positive with 14C-acetate (Figure 1). The MAR-positive archaeal cells were morphologically Methanosaeta-like cells. A significant uptake of 14C-acetate was found on the small rod bacterial cells that showed bright fluorescent signals with EUB338-mixed probe, which holds the majority of MAR-positive bacterial cells. However, these EUB338-mixed probe-hybridized small rod cells could not be hybridized with general phylum and subphylum level FISH probes (Table 1). The MAR-positive cells with 14C-acetate accounted for 15% of total cells.
Figure 1.
MAR-FISH image of acetate-utilizing bacterial and archaeal cells in the anaerobic digester sludge. (a) FISH image and (b) MAR image. In situ hybridization was performed with fluorescein isothiocyanate (FITC)-labeled EUB338-mixed probe and tetramethylrhodamine 5-isothiocyanate (TRITC)-labeled ARC915 probe. After incubating with 2.5 m glucose for 1 h, 740 kBq [2-14C]acetate was injected into the sample (the radiolabeled acetate concentration was 18% of total acetate concentration), and the samples were incubated for 2 h. Dotted circles indicate MAR-positive bacterial cells. Filamentous archaeal cells are also MAR-positive. Bar represents 10 μm.
Phylogenetic analysis of heavy 13C-labeled bacterial 16S rRNA
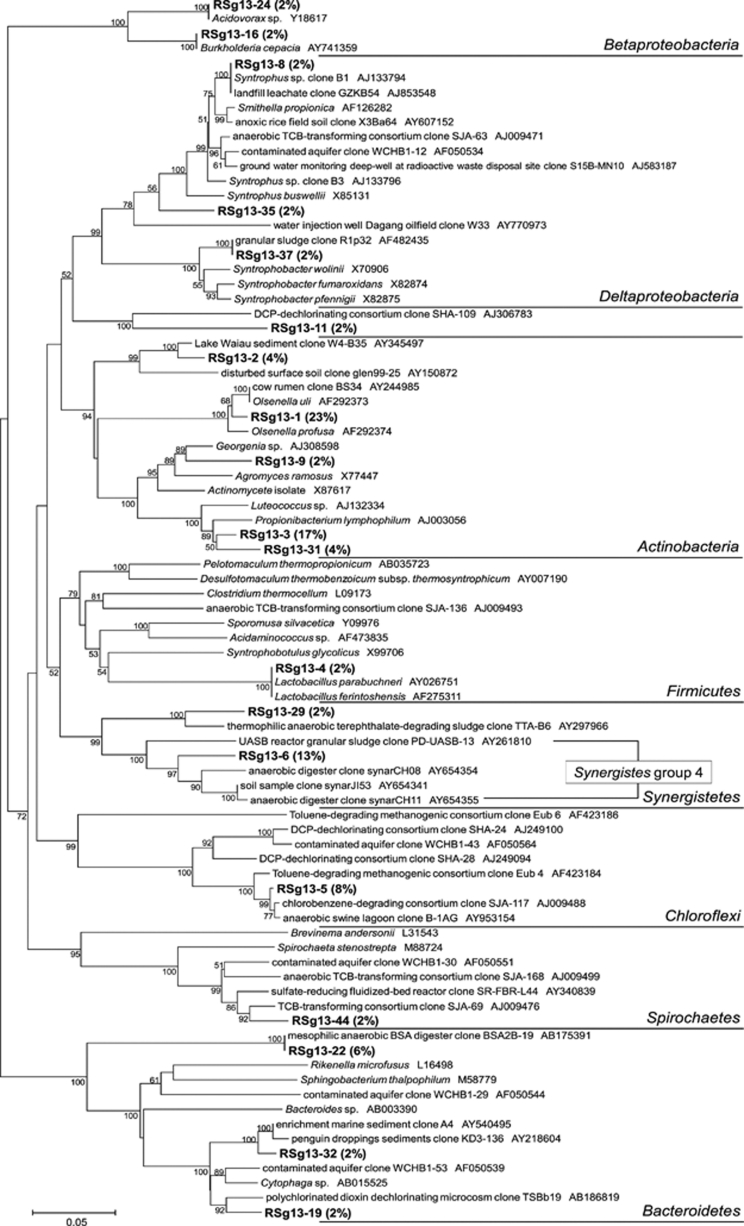
The incubation conditions with [13C6]glucose and [13C3]propionate were carefully adjusted in order that the initial 13C substrates were degraded down to methane via acetate and that RNA molecules of acetate utilizers were sufficiently labeled with produced 13C-acetate. The phylogenetic affiliation of 16S rRNA sequences was analyzed for the total 48 clones of heavy 13C-labeled bacterial rRNA derived from the anaerobic digester sludge incubated with 2.5 m [13C6]glucose for 48 h (Figure 2). Twenty-four heavy 16S rRNA clones (50%) belonged to Actinobacteria, in which Olsenella (11 clones, 23%) and Propionibacterium (8 clones, 17%) were predominant. Synergistes (7 clones, 15%), Bacteroides (5 clones, 10%) and Chloroflexi (4 clones, 8%) followed. Six clones among the seven clones of the Synergistes constituted a monophyletic cluster in the Synergistes group 4. Two clones (4%) were affiliated with propionate-degrading Syntrophobacter and Smithella, respectively.
Figure 2.
A phylogenetic tree showing the affiliation of 16S rRNA clones retrieved from ‘heavy' fraction of the RNA, which was extracted from the anaerobic digester sludge incubated with 2.5 m [13C6]glucose for 48 h (batch incubation). The numbers near branching points indicates bootstrap values. The scale bar represents 5% sequence divergence.
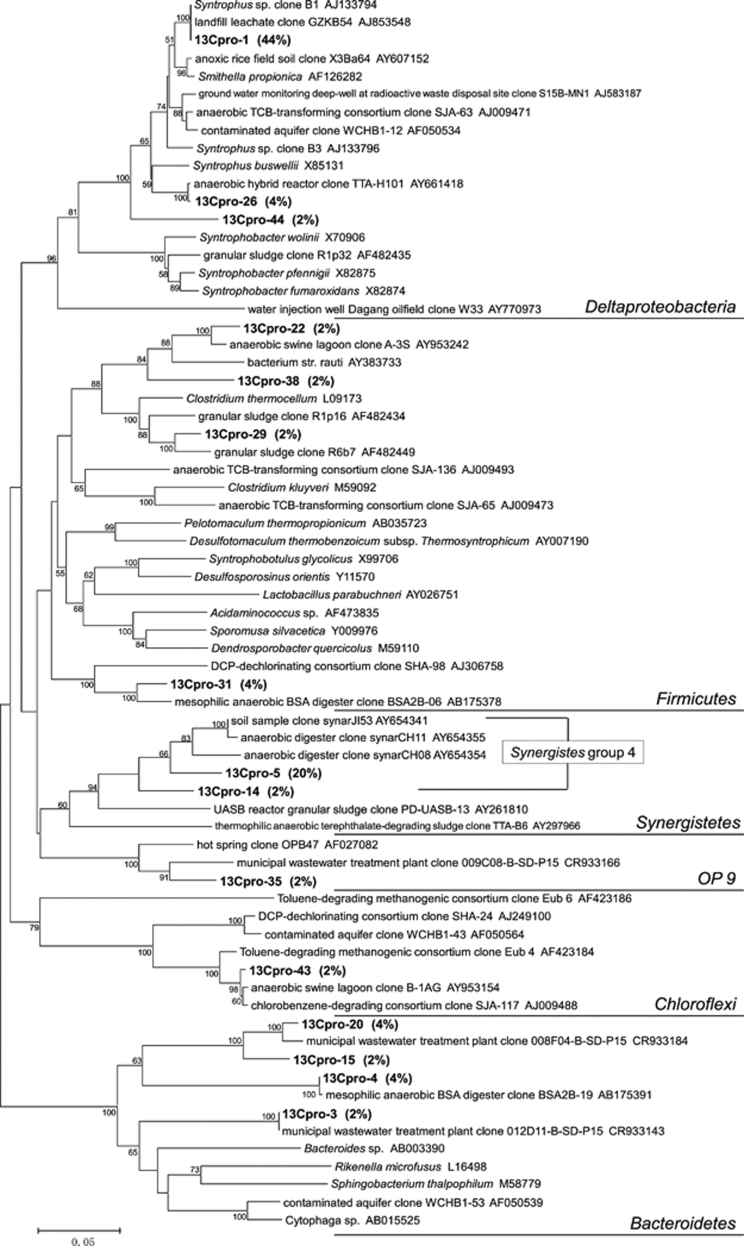
The phylogenetic affiliation of 16S rRNA sequences was also analyzed for the total 45 clones of heavy 13C-labeled bacterial rRNA derived from the anaerobic digester sludge incubated with 0.5 m [13C3]propionate for 58 h (Figure 3). First, 20 heavy 16S rRNA clones (44%) belonged to the Smithella lineage of Deltaproteobacteria. Second, predominant clones belonged to Synergistes group 4 (9 clones, 20%). Six clones (13%) and five clones (11%) were widely distributed in Bacteroidetes and Firmicutes, respectively.
Figure 3.
A phylogenetic tree showing the affiliation of 16S rRNA clones retrieved from ‘heavy' fraction of the RNA, which was extracted from the anaerobic digester sludge incubated at 0.5 m [13C3]propionate for 58 h (continuous-flow incubation). The numbers near branching points indicates bootstrap values. The scale bar represents 5% sequence divergence.
Identification of acetate-utilizing bacteria
As the results of phylogenetic analyses of heavy 16S rRNA derived from the sludge incubated with 13C-glucose and 13C-propionate, Synergistes group 4 was detected in both clone libraries, suggesting that this group could be a dominant acetate degrader. Therefore, FISH probe specific for the Synergistes group 4 was designed (Syner195 in Table 1) and used for MAR-FISH with [2-14C]acetate to confirm that the Synergistes group 4 was an acetate-utilizing bacterium in the anaerobic digester sludge. FISH and MAR-FISH analyses revealed the Syner195 probe-hybridized bacteria belonging to the Synergistes group 4 were abundantly present in the digester sludge. The number of Syner195 probe-hybridized cells was 1.2 × 109 cells mg-VSS−1, whereas the number of MX825 probe-hybridized cells was 1.4 × 109 cells mg-VSS−1. The morphotype of the probe Syner195 probe-hybridized cells was a short rod, 2 μm length and 1 μm width. Dense silver grains covered on the Syner195 probe-hybridized short-rod cells (Figure 4), indicating a significant utilization of 14C-acetate. This result clearly indicated that bacteria belonging to the Synergistes group 4 were acetate utilizers. The Syner195 probe-hybridized Synergistes cells were all MAR-negative when incubated with either 14C-propionate or 14C-glucose. Therefore, it is concluded that Synergistes can use neither glucose nor propionate. The Methanosaeta-like filamentous archaeal cells, which were hybridized with Methanosaetaceae-specific MX825 probe, were also MAR-positive (Figure 4). The number of silver grains accumulated on the Methanosaeta-like filamentous archaea was similar to that on bacteria belonging to the Synergistes group 4.
Figure 4.
MAR-FISH images of acetate-utilizing archaeal and bacterial cells present in the anaerobic digester sludge incubated for 5 h at 0.5 m acetate containing 19% [2-14C]acetate. In these images, MAR-positive cells are identified with genus-specific probes, which are yellowish owing to the cross-hybridization with domain probes. (a) FITC-labeled MX825 probe-stained filamentous MAR-positive cells. The sample was simultaneously hybridized with TRITC-labeled ARC915 probe. (b) FITC-labeled Syner195 probe-stained MAR-positive cells. The sample was simultaneously hybridized with TRITC-labeled EUB338-mixed probe. Bars represent 10 μm.
The involvement of Synergistes group 4 and Methanosaeta-like filamentous archaea in glucose degradation to methane and carbon dioxide was tested by MAR-FISH after incubation with [U-14C]glucose for 1 h, 3 h, 12 h and 36 h. The time-course analysis by MAR-FISH with the specific probes revealed that the number of MAR-positive cells belonging to the Synergistes group 4 and Methanosaeta increased with time, indicating that glucose was degraded to methane via acetate and acetate was mainly utilized by both groups in the anaerobic digester sludge (data not shown).
Acetate utilization by Synergistes group 4 and Methanosaeta
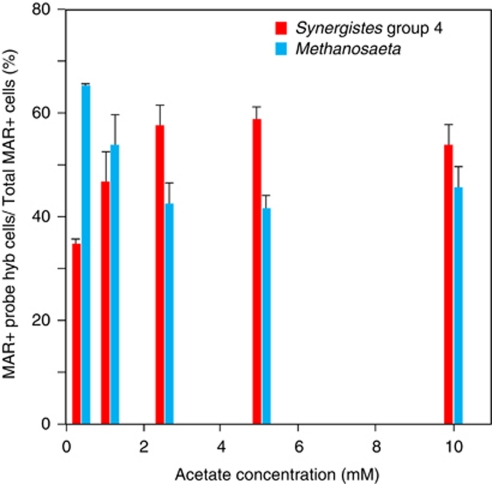
Effect of acetate concentrations on the activities of the Synergistes group 4 and Methanosaeta-like filamentous archaea was investigated by MAR-FISH with the initial acetate concentrations of 0.5 m, 1 m, 2.5 m, 5 m and 10 m, respectively (Figure 5). Incubation time was 5 h at each acetate concentration. At 0.5 m acetate, the Syner195 probe-hybridized Synergistes cells accounted for approximately 35% of the total MAR-positive cells, whereas the probe MSX825-hybridized Methanosaeta cells accounted for 65% of the total MAR-positive cells. At 1.0 m acetate, the percentages of the MAR-positive Synergistes cells and Methanosaeta cells were approximately 45% and 55%, respectively. However, the MAR-positive Synergistes cells were more abundant than the probe MSX825-hybridized Methanosaeta cells at 2.5 m, 5 m and 10 m acetate. Fractions of total MAR-positive cells of total cells were almost constant values of 3–4% at all acetate concentrations. The result indicated that the activities of the two groups were dependent on the acetate concentrations. Other MAR-positive microorganisms were negligible and only these two groups were active acetate utilizers at all acetate concentrations.
Figure 5.
Relative abundance of [14C]acetate-utilizing Methanosaeta and Synergistes group 4 populations at 0.5 m, 1.0 m, 2.5 m, 5 m and 10 m acetate determined by MAR-FISH using probes of MSX825 and Syner195, respectively. The samples were incubated for 5 h at each acetate concentration containing 19% [2-14C]acetate. The genus-specific probes were always combined with domain-specific probes, and the sample was counter-stained with 4′, 6-diamidino-2-phenylindole (DAPI). Error bars represent the standard errors of duplicated measurements.
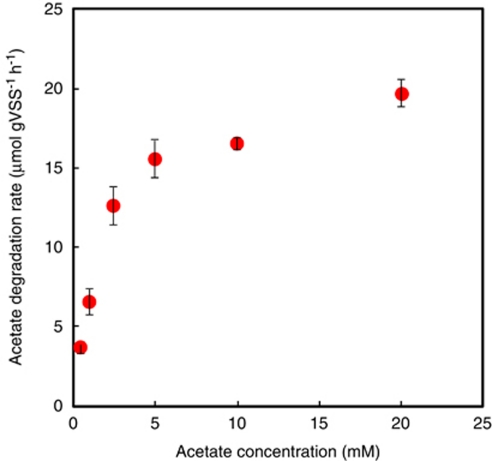
14C-acetate degradation rates of the sludge
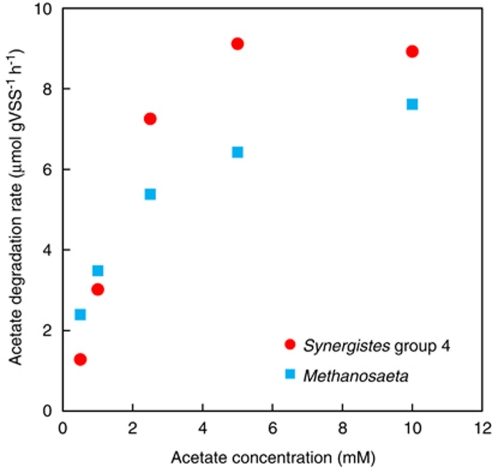
14C-acetate degradation rates of the digester sludge were determined from batch experiments for 10-h incubation at the acetate concentrations ranging from 0.5 m to 20 m containing 185 kBq 14C-acetate (Figure 6). The acetate degradation rates enzyme kinetically increased with increasing acetate concentration. The Km and Vmax values of this anaerobic digester sludge were determined by the Lineweaver–Burke plot of the acetate degradation rates to be 2.8 m and 24 μmol g-VSS−1 h−1, respectively (Table 2). As Synergistes group 4 and Methanosaeta were dominant acetate utilizers in this anaerobic digester sludge, the overall acetate degradation rate is considered to be the sum of acetate degradation rates of Synergistes group 4 and Methanosaeta. The number of silver grain is theoretically proportional to the amount of incorporated (assimilated) 14C atom into cells under the same incubation and microautoradiographic conditions. The number of silver grains accumulated on the Methanosaeta-like filamentous cells was similar to that on bacteria belonging to the Synergistes group 4 (Figure 4), indicating that the acetate degradation by MAR-positive cells of Synergistes group 4 is the same as that by Methanosaeta. It should be also assumed that the incorporation rate is proportional to the degradation rate for the Synergistes group 4 and Methanosaeta, and this relationship is held at all acetate concentrations. Based on these assumptions, the individual acetate degradation rates by the Synergistes group 4 and Methanosaeta (Figure 7) could be roughly estimated from the relative abundance of [14C]acetate-utilizing MAR-positive Synergistes group 4 and Methanosaeta populations (Figure 5) and specific acetate degradation rates (μmol gVSS−1 h−1) of the anaerobic digester sludge (Figure 6). The Km and Vmax values for Synergistes group 4 were 13 m and 36 μmol gVSS−1 h−1, whereas those for Methanosaeta were 1 m and 8 μmol gVSS−1 h−1, respectively (Figure 7 and Table 2).
Figure 6.
Acetate degradation rates of the anaerobic digester sludge incubated at 0.5 m, 1.0 m, 2.5 m, 5 m, 10 m and 20 m acetate containing 185 kBq [2-14C]acetate for 10 h, which was determined by liquid scintillation counting. Error bars represent the standard errors of duplicated measurements.
Table 2. Km and Vmax values for acetate catabolism by the anaerobic digester sludge, Methanosaeta and Synergistes group 4.
| Km(m) | Vmax(μmol gVSS−1 h−1) | |
|---|---|---|
| Anaerobic digester sludge (lab reactor) | 2.8 | 24 |
| Methanosaeta | 1 | 8 |
| Synergistes group 4 | 13 | 36 |
Figure 7.
Acetate degradation rates of Methanosaeta and Synergistes group 4 in the anaerobic digester sludge at 0.5 m, 1.0 m, 2.5 m, 5 m and 10 m acetate concentrations. The rates were calculated from the values of Figures 5 and 6.
Discussion
Identification of major acetate-utilizing microorganisms in anaerobic digester sludge
Cultivation and isolation of anaerobic acetate-utilizing bacteria have been very difficult owing to their slow and syntrophic growth with hydrogenotrophic methanogens. Therefore, abundance, diversity and phylogenetic affiliations of anaerobic acetate-utilizing bacteria in anaerobic environments including digester sludge remain largely unknown. Recent 16S rRNA gene-based phylogenetic analyses revealed the presence of a vast diversity of microorganisms in anaerobic digester sludge, the majority of which have not yet been cultivated and characterized (Chouari et al., 2005; Sekiguchi, 2006; Ariesyady et al., 2007a; Narihiro et al., 2009).
This study was originally designed to identify bacterial groups that are responsible for degradation of glucose, propionate and acetate in anaerobic digester sludge. Therefore, we have performed RNA-SIP combined with full-cycle 16S rRNA analysis using 13C-glucose, 13C-propionate and 13C-acetate as sole carbon source, respectively. However, the RNA-SIP with 13C-acetate was failed because the incorporation rate of 13C in RNA was very slow, and enough heavy RNA could not be obtained with incubation of 1–3 days. Major acetate-utilizing bacterial and archaeal populations in methanogenic anaerobic digester sludge were, therefore, identified by finding microbial groups commonly detected in heavy 16S rRNA gene clone libraries derived from 13C-propionate and 13C-glucose. A specific FISH probe was newly designed for the commonly detected microbial group and used for MAR-FISH to confirm acetate utilization of this bacterial group.
This study revealed for the first time that the predominant acetate-utilizing bacterium was belonging to an as-yet-unidentified Synergistes group 4. Synergistetes-affiliated microorganisms composed one core group in anaerobic digester sludge with other five groups affiliated with Chloroflexi, Betaproteobacteria, Bacteroidetes (Rivière et al., 2009). Our Synergistes group 4 clones were affiliated with the phylum Synergistetes (formally ‘Synergistes') subdivision E, in which the genus Aminobacterium was classified (Jumas-Bilak et al., 2009). However, Synergistes group 4 clones were distantly related to the amino acids-degrading Aminobacterium, and closely related to many anaerobic digester clones in the Synergistetes subdivision E. In fact, a part of the Synergistetes subdivision E has been described as ‘Group 4 anaerobic digester' (Godon et al., 2005), and more recent phylogenetic analysis reclassified the ‘Group 4 anaerobic digester' into the groups PD-UASB-13 and HA73 (Hugenholtz et al., 2009). Our clones were closely related to the group PD-UASB-13 (Figures 2 and 3). The bacteria belonging to the Synergistes have been frequently found in other full-scale anaerobic digester treating municipal wastewater sludge (Chouari et al., 2005; Rivière et al., 2009). However, the function of the Synergistes in the anaerobic digesters has not been reported yet until now, as the group PD-UASB-13 has no cultured representatives to date, and their metabolic function is presently unknown. Thus, the ability to degrade acetate by Synergistes group 4 would be valuable information for future cultivation and characterization of this bacterial group.
Acetate utilization by Synergistes group 4 is probably syntrophic acetate oxidation coupled with hydrogenotrophic methanogens. We determined 13CH4 production from degradation of [1-13C]acetate (CH313COOH) by a gas chromatography–mass spectrometry. The result of gas chromatography–mass spectrometry analysis revealed about 10% of the degraded acetate was converted to 13CH4 in anaerobic digester sludge containing 0.5 m acetate (data not shown). 13CH4 could be produced by syntrophic acetate oxidation coupled with hydrogenotrophic methanogens, but not by aceticlastic methanogenesis (Shigematsu et al., 2004). In this study, bacteria belonging to the Synergistes group 4 were only predominant acetate-utilizing bacteria. Based on archaeal 16S rRNA gene clone analysis, several clones closely related to Methanoculleus (2 out of total 35 clones) and Methanosarcina (5 out of total 35 clones) were retrieved from the anaerobic digester sludge (data not shown). The genus Methanoculleus use hydrogen to reduce CO2 to CH4 (Boone et al., 1993). Methanosarcina is known as acetate-utilizing methanogen, whereas many Methanosarcina spp. can also grow by using hydrogen to reduce CO2 to CH4 (Boone et al., 1993). In MAR-FISH analysis with [2-14C]acetate, Methanosarcina was not detected as MAR-positive. Methanosarcina might have a role as hydrogenotrophic methanogen associated with syntrophic acetate-oxidizing bacterium, Synergistes group 4, in the anaerobic digester sludge.
Acetoclastic methanogenesis convert methyl-based carbon (CH3−) of acetate into methane (Zehnder et al., 1980; Zinder, 1998). Therefore, we used [2-14C]acetate, but not [1-14C]acetate, because acetoclastic methanogens produce 14CH4, but not 14CO2, from [2-14C]acetate. Thus, feeding of [2-14C]acetate minimizes the false MAR-positive by 14CO2 cross-feeding to non-acetate-utilizing bacteria. On the other hand, when acetate-utilizing bacteria produced 14CO2 from [1-14C]acetate, homoacetogens can utilize the produced 14CO2 with hydrogen and produce 14C-labeled acetate, which might be another possibility that non-acetate-utilizing bacteria become MAR-positives. In fact, no other bacteria than Synergistes group 4 and Methanosaeta were found as active acetate utilizers in the MAR-FISH experiments with either [2-14C]acetate or [1-14C]acetate. Hence, it was negligible that 14CO2-utilizing bacteria such as homoacetogens appeared as MAR-positives under the experimental condition applied in this study (that is, 5-h incubation with [2-14C]acetate). Syner195 probe-hybridized cells were MAR-negative when cultured for 5 h with 14C-bicarbonate, [1-14C]propionate or [U-14C]glucose (data not shown). When the incubation with 14C-glucose was prolonged till 12 h, 24 h and 48 h, MAR-positive Syner195 probe-hybridized cells increased gradually. This is probably because 14C-glucose was degraded to 14C-acetate, and then the produced 14C-acetate was utilized by Synergistes group 4. These results also confirm the conclusion that the Synergistes group 4 utilized acetate during degradation of glucose.
Acetate utilization by Synergistes group 4 and Methanosaeta
The Km value (1 m) for Methanosaeta (Table 2) is close to the values (0.5–0.9 m) reported in the literature (summarized by Zinder, 1998). This result indicates that Synergistes group 4 is more competitive for acetate over Methanosaeta at high concentrations of acetate. Quantitative MAR-FISH (Nielsen et al., 2003) for each microorganism should be performed to determine the more quantitative contribution of two acetate utilizers. Quantitative MAR-FISH can directly determine in situ acetate degradation rates in the mixed microbial populations, which would provide the more precise contribution of two acetate utilizers. Furthermore, isolation and characterization of Synergistes group 4 is undoubtedly necessary for understanding of their physiology.
Periodical change in acetate concentration in an anaerobic batch reactor, ranging from 0 m to 15 m within 2 days (Ariesyady et al., 2007b), is probably the reason that both acetate utilizers, Synergistes group 4 and Methanosaeta, were present in the same culture, even though their Km values were 10 times different. Methanosaeta and Synergistes group 4 seem to be not competitive, but cooperative for fluctuating concentration of acetate in the anaerobic batch reactor used in this study.
Four distinct bacterial species have been found as major propionate degraders in the anaerobic digester sludge (Ariesyady et al., 2007b), whereas this study revealed that only two microorganisms, Synergistes group 4 and Methanosaeta, were involved in acetate degradation in the same anaerobic digester sludge. Therefore, it should be noted that acetate-degrading microbial community had less diversity than propionate-degrading community.
In conclusion, this study revealed for the first time that bacteria belonging to an as-yet-unidentified Synergistes group 4 were the major acetate-utilizing bacterial populations in methanogenic anaerobic digester sludge. Furthermore, this bacteria group had lower affinity to acetate and higher acetate utilization rate than Methanosaeta-like acetoclastic methanogen. The bacteria belonging to the Synergistes group 4 has been frequently found as one of core microbial groups in anaerobic digester sludge, but their function has been totally unknown. It should be noted that the combination of stable-isotope and radioisotope tracer experiments and molecular analyses (that is, RNA-SIP with full-cycle 16S rRNA analysis and MAR-FISH) is a very powerful tool to identify the in situ function and activity of as-yet-unidentified bacteria in mixed populations.
Acknowledgments
We gratefully thank the Central Institute of Isotope Science, Hokkaido University, for providing the facilities for the isotope experiments. We acknowledge Hiroko Osakabe for technical support. This study was supported by NEDO and JSPS.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen K, Nielsen PH. Application of microautoradiography to the study of substrate uptake by filamentous microorganisms in activated sludge. Appl Environ Microbiol. 1997;63:3662–3668. doi: 10.1128/aem.63.9.3662-3668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariesyady HD, Ito T, Okabe S. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 2007a;41:1554–1568. doi: 10.1016/j.watres.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Ariesyady HD, Ito T, Yoshiguchi K, Okabe S. Phylogenetic and functional diversity of propionate-oxidizing bacteria in an anaerobic digester sludge. Appl Microbiol Biotechnol. 2007b;75:673–683. doi: 10.1007/s00253-007-0842-y. [DOI] [PubMed] [Google Scholar]
- Björnsson L, Hugenholtz P, Tyson GW, Blackall LL. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology. 2002;148:2309–2318. doi: 10.1099/00221287-148-8-2309. [DOI] [PubMed] [Google Scholar]
- Boone DR, Whitman WB, Rouvuère P.1993Diversity and taxonomy of methanogensIn: Ferry JG (ed).Methanogenesis Chapman & Hall: New York; 35–80. [Google Scholar]
- Chouari R, Paslier DL, Daegelen P, Ginestet P, Weissenbach J, Sghir A. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ Microbiol. 2005;7:1104–1115. doi: 10.1111/j.1462-2920.2005.00795.x. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Gich F, Garcia-Gil J, Overmann J. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch Microbiol. 2001;177:1–10. doi: 10.1007/s00203-001-0354-6. [DOI] [PubMed] [Google Scholar]
- Godon JJ, Moriniere J, Moletta M, Gaillac M, Bru V, Delgenes JP. Rarity associated with specific ecological niches in the bacterial world: the ‘Synergistes' example. Environ Microbiol. 2005;7:213–224. doi: 10.1111/j.1462-2920.2004.00693.x. [DOI] [PubMed] [Google Scholar]
- Hattori S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 2008;23:118–127. doi: 10.1264/jsme2.23.118. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Hooper SD, Kyrpides NC. Focus: Synergistetes. Environ Microbiol. 2009;11:1327–1329. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Nielsen JL, Okabe S, Watanabe Y, Nielsen PH. Phylogenetic identification and substrate uptake patterns of sulfate-reducing bacteria inhabiting an oxic–anoxic sewer biofilm by combining microauto radiography and fluorescent in situ hybridization. Applied Environ Microbiol. 2002;68:356–364. doi: 10.1128/AEM.68.1.356-364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumas-Bilak E, Roudiere L, Marchandin H. Description of ‘Synergistetes' phyl. nov. and emended description of the phylum ‘Deferribacteres' and of the family Syntrophomonadaceae, phylum ‘Firmicutes'. Int J Syst Evol Microbiol. 2009;59:1028–1035. doi: 10.1099/ijs.0.006718-0. [DOI] [PubMed] [Google Scholar]
- Kindaichi T, Ito T, Okabe S. Eco–physiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms. Appl Environ Microbiol. 2004;70:1641–1650. doi: 10.1128/AEM.70.3.1641-1650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds).Nucleic Acid Techniques in Bacterial Systematics John Wiley and Sons: Chichester, UK; 115–175. [Google Scholar]
- Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH, et al. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure–function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T, Manefield M, Friedrich MW. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol. 2004a;6:73–78. doi: 10.1046/j.1462-2920.2003.00536.x. [DOI] [PubMed] [Google Scholar]
- Lueders T, Wagner B, Claus P, Friedrich MW. Stable-isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ Microbiol. 2004b;6:60–72. doi: 10.1046/j.1462-2920.2003.00535.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002a;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M, Whiteley AS, Ostle N, Ineson P, Bailey MJ. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun Mass Spectrom. 2002b;16:2179–2183. doi: 10.1002/rcm.782. [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- McCarty PL, Smith DP. Anaerobic wastewater treatment. Environ Sci Technol. 1986;20:1200–1206. [Google Scholar]
- Meier H, Amann R, Ludwig W, Schleifer KH. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- Narihiro T, Terada T, Kikuchi K, Iguchi A, Ikeda M, Yamauchi T, et al. Comparative analysis of bacterial and archaeal communities in methanogenic sludge granules from upflow anaerobic sludge blanket reactors treating various food-processing, high-strength organic wastewaters. Microbes Environ. 2009;24:88–96. doi: 10.1264/jsme2.me08561. [DOI] [PubMed] [Google Scholar]
- Neef A, Amann R, Schlesner H, Schleifer KH. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology. 1998;144:3257–3266. doi: 10.1099/00221287-144-12-3257. [DOI] [PubMed] [Google Scholar]
- Nielsen JL, Christensen D, Kloppenborg M, Nielsen PH. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ Microbiol. 2003;5:202–211. doi: 10.1046/j.1462-2920.2003.00402.x. [DOI] [PubMed] [Google Scholar]
- Okabe S, Itoh T, Satoh H, Watanabe Y. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl Environ Microbiol. 1999;65:5107–5116. doi: 10.1128/aem.65.11.5107-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Kindaichi T, Ito T. MAR-FISH—an ecophysiological approach to link phylogenetic affiliation and in situ metabolic activity of microorganisms at a single-cell resolution. Microbes Environ. 2004;19:83–98. [Google Scholar]
- Okabe S, Kindaichi T, Ito T. Fate of 14C-labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl Environ Microbiol. 2005;71:3987–3994. doi: 10.1128/AEM.71.7.3987-3994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Odagiri M, Ito T, Satoh H. Succession of sulfur-oxidizing bacteria in the microbial community on corroding concretes in sewer systems. Appl Environ Microbiol. 2007;73:971–980. doi: 10.1128/AEM.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus R, Fukui M, Wilkes H, Widdle F. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Stromley JM, Rittmann BE, Stahl DA. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, et al. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009;3:700–714. doi: 10.1038/ismej.2009.2. [DOI] [PubMed] [Google Scholar]
- Roller C, Wagner M, Amann R, Ludwig W, Schleifer KH. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- Schink B, Stams AJM.2006Syntrophism among prokaryotesIn: Dworkin M, Rosenberg E, Schleifer K-H, Stackebrandt E (eds).The Prokaryotes3rd edn, vol. 2.Springer: New York; 309–335. [Google Scholar]
- Schwarz JIK, Lueders T, Eckert W, Conrad R. Identification of acetate-utilizing Bacteria and Archaea in methanogenic profundal sediments of Lake Kinneret (Israel) by stable isotope probing of rRNA. Environ Microbiol. 2007;9:223–237. doi: 10.1111/j.1462-2920.2006.01133.x. [DOI] [PubMed] [Google Scholar]
- Sekiguchi Y. Yet-to-be cultured microorganisms relevant to methane fermentation processes. Microbes Environ. 2006;21:1–15. [Google Scholar]
- Shigematsu T, Tang Y, Kobayashi T, Kawaguchi H, Morimura S, Kida K. Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl Environ Microbiol. 2004;70:4048–4052. doi: 10.1128/AEM.70.7.4048-4052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Amann R.1991Development and application of nucleic acid probesIn: Stackebrandt E, Goodfellow M (eds).Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons Inc: New York; 205–248. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R, Glöckner FO, Amann R. 16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga–Flavobacterium–Bacteroides. System Appl Microbiol. 2000;23:107–114. doi: 10.1016/S0723-2020(00)80051-X. [DOI] [PubMed] [Google Scholar]
- Zehnder AJ, Huser BA, Brock TD, Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980;124:1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- Zinder SH.1998Physiological ecology of methanogensIn: Ferry JG (ed.).Methanogenesis Chapman & Hall: New York, London; 128–206. [Google Scholar]