Abstract
Ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (anammox) bacteria have emerged as significant factors in the marine nitrogen cycle and are responsible for the oxidation of ammonium to nitrite and dinitrogen gas, respectively. Potential for an interaction between these groups exists; however, their distributions are rarely determined in tandem. Here we have examined the vertical distribution of AOA and anammox bacteria through the Arabian Sea oxygen minimum zone (OMZ), one of the most intense and vertically exaggerated OMZs in the global ocean, using a unique combination of intact polar lipid (IPL) and gene-based analyses, at both DNA and RNA levels. To screen for AOA-specific IPLs, we developed a high-performance liquid chromatography/mass spectrometry/mass spectrometry method targeting hexose-phosphohexose (HPH) crenarchaeol, a common IPL of cultivated AOA. HPH-crenarchaeol showed highest abundances in the upper OMZ transition zone at oxygen concentrations of ca. 5 μ, coincident with peaks in both thaumarchaeotal 16S rDNA and amoA gene abundances and gene expression. In contrast, concentrations of anammox-specific IPLs peaked within the core of the OMZ at 600 m, where oxygen reached the lowest concentrations, and coincided with peak anammox 16S rDNA and the hydrazine oxidoreductase (hzo) gene abundances and their expression. Taken together, the data reveal a unique depth distribution of abundant AOA and anammox bacteria and the segregation of their respective niches by >400 m, suggesting no direct coupling of their metabolisms at the time and site of sampling in the Arabian Sea OMZ.
Keywords: anammox, Arabian Sea, intact polar lipid (IPL), nitrification, oxygen minimum zone (OMZ), thaumarcheota
Introduction
The oxygen minimum zone (OMZ) of the Arabian Sea represents a globally important site for oceanic fixed nitrogen (N) loss (Bange et al., 2000, 2005 and references therein). Its vertical distribution extends from approximately 150 to 1000 m below the sea surface with oxygen concentrations at times as low as 0.1 μ (Morrison et al., 1999; Paulmier and Ruiz-Pino, 2009), rendering it one of the most expansive and intense OMZs globally. N-cycling in the Arabian Sea may even impact the earth's climate due to OMZ intensity-related fluctuations in N-loss via heterotrophic denitrification Suthhof et al., 2001). This process simultaneously liberates the potent greenhouse gas, nitrous oxide (N2O), as a metabolic intermediate, while circumventing the biological pump via respiratory emancipation of carbon dioxide (CO2).
Anaerobic ammonium oxidation (anammox) occurs to a great extent in oxygen-limited waters, where canonical denitrification was conventionally assumed to dominate N-loss (Arrigo, 2005; Brandes et al., 2007 and references therein). This autotrophic metabolism combines ammonium (NH4+) with nitrite (NO2−) to form dinitrogen gas (N2) (Strous et al., 1999), most of which is then lost from the system. Their role in oceanic N-cycling is gradually becoming more clear: anammox may be responsible for up to 40% of N-loss in some anoxic marine environments (Dalsgaard et al., 2003; Kuypers et al., 2003, 2005), although recent evidence suggests that its contribution to N-loss in the Arabian Sea OMZ may be considerably less (Ward et al., 2009). The questions of when, where and under what conditions anammox bacteria thrive are receiving ever-increasing attention as this autotrophic process liberates neither CO2 nor N2O (Lam et al., 2009; Voss and Montoya, 2009).
Ammonia-oxidizing archaea (AOA) also use NH4+ as an electron donor, which is oxidized aerobically to NO2−. The ubiquity and abundance of marine Thaumarchaeota (formerly known as Marine Group I Crenarchaeota: Brochier-Armanet et al., 2008; Spang et al., 2010) and their associated genes coding for the alpha subunit of ammonia monooxygenase enzyme (amoA) in marine waters have suggested their potential importance in oceanic nitrification (Arrigo, 2005), subsequently stimulating a need to define the ecological niches in which they are most likely to persist (Prosser and Nicol, 2008; Erguder et al., 2009).
In contrast to anammox bacteria, which are anaerobic organisms, AOA are aerobic ammonia oxidizers (Francis et al., 2005; Coolen et al., 2007), potentially suggesting that these two groups of microbes will have different niches. However, AOA appear to be also remarkably successful under low-oxygen conditions (Karner et al., 2001; Coolen et al., 2007; Lam et al., 2007; Beman et al., 2008; Molina et al., 2010) and can actually be enriched under such circumstances (Park et al., 2010). Furthermore, anammox bacteria have also been detected in OMZs, where low amounts of oxygen may still be present (Lam et al., 2009). Thus, in low-oxygen environments, they potentially can compete for ammonia, or interact with AOA, providing nitrite to anammox bacteria. Indeed, anammox bacteria and AOA have been reported to co-exist in the same water masses in the Black Sea (Kuypers et al., 2003; Francis et al., 2005; Coolen et al., 2007), in marine sponges (Hoffmann et al., 2009) and in suspended particles in the Namibian upwelling system (Woebken et al., 2007). In addition, Lam et al. (2007) presented evidence that AOA together with nitrifying bacteria provide nitrite for anammox bacteria in the Black Sea. However, it is unknown how widespread this potential interaction is.
A few studies have examined the distribution of AOA and anammox bacteria in the Arabian Sea. A low-resolution study based on the presence of crenarchaeol (a glycerol dialkyl glycerol tetraether membrane lipid thought to be specific to AOA; de la Torre et al., 2008; Pitcher et al., 2010) suggested highest concentrations of AOA just below the photic zone (Sinninghe Damst et al., 2002). In contrast, anammox-specific ladderane core lipids were found to be most abundant within the core of the OMZ at depths of ca. 500 m (Jaeschke et al., 2007). However, Ward et al. (2009) found low anammox bacterial gene abundance and anammox rates up to 250 m depth, around the nitrite maximum, and anammox activity was found to be patchy and unpredictable regarding depth distribution in comparison with denitrification (Bulow et al., 2010).
To gain a better insight into the niches of these two groups within the Arabian Sea OMZ, we used a unique approach based on quantification of both intact polar lipids (IPLs) and DNA/RNA (functional) genes. IPLs consist of the core membrane lipids still covalently bound to polar head groups that are relatively labile (that is, unstable outside of the intact cell; White et al., 1979), and thus more accurately represent lipids synthesized by living or recently living cells. We developed a novel high-performance liquid chromatography/mass spectrometry/mass spectrometry (HPLC/MS/MS) method to target AOA-specific IPLs, as well as using a previously published method targeting a C20-[3]-monoether ladderane lipid containing a phosphatidylcholine (PC) head group (henceforth referred to as ‘PC-monoether ladderane') specific for anammox bacteria (Jaeschke et al., 2009). This study was complemented with the quantification of 16S rDNA and functional genes (AOA amoA and hzo genes of anammox bacteria, which encode for the enzyme hydrazine oxidoreductase converting hydrazine to N2) and their expression (that is, RNA abundance) as an indication of the activity of AOA and anammox bacteria.
Materials and methods
The complete Materials and methods section is provided as Supplementary Information.
Physical properties of the water column
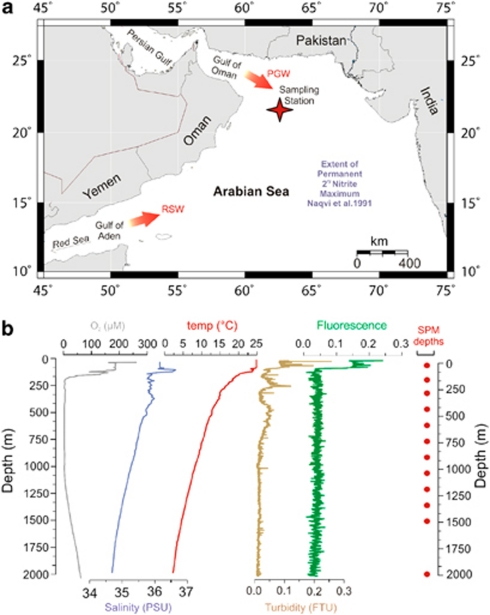
A conductivity–temperature–depth (CTD) system equipped with attached oxygen, turbidity and fluorescence sensors was deployed to record the physical properties of the water column at a depth profile station in the Northern Arabian Sea (lat 21°55.6′, long 63°10.6′) (Figure 1).
Figure 1.
(a) Location of Arabian Sea sampling station (indicated by the star; lat 21°55', long 63°10′), just outside the region containing a quasi-permanent secondary nitrite (NO2−) maximum (Revsbech et al., 2009). Persian Gulf Water (PGW) and to a much lesser extent, Red Sea Water (RSW), influence the OMZ at our study site. (b) Hydrographic characteristics of the water column at our sampling station as recorded from the CTD sensors, and depths where SPM sampling was performed using an in situ pump.
Suspended particulate matter sampling
A depth profile of 12 suspended particulate matter (SPM) samples was collected at our sampling station by large-volume (ca. 200–1700 l) in situ pump filtration onto pre-washed 0.7 μm GF/F filters (Pall Corporation, Port Washington, NY, USA). A total of six deployments of two McLane WTS-LV in situ pumps (McLane Laboratories Inc., Falmouth, MA, USA) were carried out between 14 and 20 January 2009. Upon retrieval of the pumps, GF/F filters containing SPM were removed and immediately frozen at −80 °C. A rosette sampler containing 24 × 12 l Niskin bottles (OceanTest equipment Inc., Fort Lauderdale, FL, USA) was attached to the CTD deployed during each pump cast to collect water from the sampler for inorganic nutrient analysis and onboard filtration of DNA/RNA. For nutrients, ca. 5 ml samples were filtered over 0.45 μm × 25 mm Acrodisc HT Tuffryn Membrane syringe filters (Pall Corporation) into pre-rinsed pony vials. Water samples from the Niskin bottles were filtered onboard for DNA/RNA via clean Teflon tubing into pre-rinsed 20 l Nalgene bottles in a dark climate chamber maintained at 7 °C over 142 mm × 0.2 μm PC filters (Millipore, Billerica, MA, USA) and immediately at −80 °C.
Extraction and analysis of IPLs
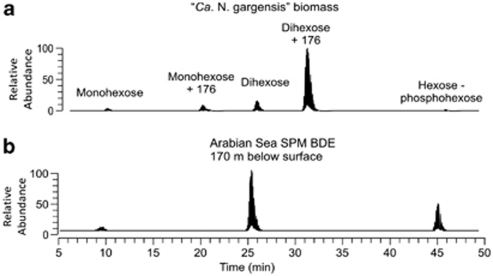
IPLs were extracted from freeze-dried filters using a modified Bligh and Dyer technique (Bligh and Dyer, 1959) as described by Schouten et al. (2008). Analysis of crenarchaeol-based IPLs (crenarchaeol-hexose, crenarchaeol-dihexose, crenarchaeol-hexose–phosphohexose, crenarchaeol-hexose-‘176', crenarchaeol-dihexose-‘176' ‘176' represents an unknown headgroup with a mass of 176 Da) and crenarchaeol-hexose-‘180' (representing a major IPL in Nitrosopumilus maritimus SCM1; Prosser and Nicol, 2008) was achieved by HPLC/electrospray ionization/MS/MS in selected reaction monitoring (SRM) mode using chromatographic conditions and source settings as described by Schouten et al. (2008). SRM transitions were optimized by direct infusion of an IPL extract from ‘Ca. Nitrososphaera gargensis' biomass (Pitcher et al., 2010), resulting in the conditions listed in Supplementary Table S2. The SRM transition for crenarchaeol-hexose-‘180' was based on those for crenarchaeol-hexose-‘176' as the IPL extract of N. maritimus available for SRM optimization was insufficient. IPLs were quantified as peak area response L−1, due to the lack of quantitative standards. For more details on this HPLC/MS-MS method, see Supplementary methods. The C20-[3]-monoether ladderane lipid containing a PC headgroup (PC-monoether ladderane) (Supplementary Figure S1) was analyzed by HPLC/electrospray ionization/MS/MS using an SRM method described previously (Jaeschke et al., 2009).
DNA extraction
DNA was extracted from SPM filtered onto 142-mm × 0.2-μm polycarbonate filters. Filters were cut into ∼0.5 × 0.5 cm squares. Cells were lysed by bead beating with 1.5 g of sterile 0.1-mm zirconium beads (Biospec, Bartlesville, OK, USA) in an extraction buffer containing 10 m Tris-HCl pH 8, 25 m Na2EDTA pH 8, 1% (v/v) sodium dodecyl sulfate (SDS), 100 m NaCl and molecular biology grade water at 70 °C for 30 min and then extracted with phenol-chloroform and precipitated using ice-cold ethanol.
RNA extraction and reverse transcription
RNA was extracted from a set of 142 mm × 0.2 μm polycarbonate filters. Cells were lysed by bead beating in RLT buffer (Qiagen Inc., Valencia, CA, USA) supplemented by 1/100 vol β-mercaptoethanol. Lysate was purified and concentrated twice with RNeasy Mini kit (Qiagen Inc.). The extracted RNA was treated with Rnase-free DNase (DNA-free, Ambion Inc., Austin, TX, USA). RNA quality and concentration were estimated using the Experion RNA StdSens Analysis Kit (Bio-Rad Laboratories, Hercules, CA, USA). Reverse transcription was performed with an Enhanced Avian First Strand synthesis kit (Sigma-Aldrich, St Louis, MO, USA) using random nonamers.
Q-PCR analysis
All Q-PCR analyses were performed on a Biorad CFX96 Real-Time System/C1000 Thermal cycler equipped with CFX Manager Software. The copy numbers of archaeal 16S rDNA were estimated by using the 16S rDNA specific primers Parch519F and ARC915R (Coolen et al., 2004); Thaumarchaeota (MCG1) 16S rDNA with MCGI-391F and MCGI-554R primers as described by Coolen et al. (2007); archaeal amoA with primers CrenAmoAQ-F and CrenAmoAModR (Mincer et al., 2007); and 16S rDNA anammox with Brod541F and Amx820R as described by Li et al. (2010). For the quantification of the hzo gene, hzo primers hzo_LV2F and hzo_LV1R (this study) were designed by aligning n=50 sequences obtained from the cloning of the PCR product hzo1F-hzo1R (Lam et al., 2007). For more details on the DNA, RNA extraction, Q-PCR standard preparation and Q-PCR procedure (including efficiencies and correlation coefficients), see Supplementary methods.
Results
Hydrographic setting and water column chemistry
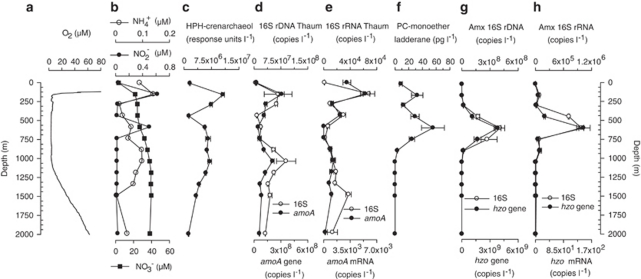
In January 2009, the water column of the Arabian Sea at the southeast slope of the Murray Ridge, where the water depth reaches 3010 m, was investigated by a range of oceanographic and microbial ecology methods. Dissolved oxygen concentration, as measured by the CTD oxygen sensor, decreased from fully saturated at the surface to <2.5 μ (that is, at the detection limit of the CTD oxygen sensor real oxygen concentrations are likely to be substantially lower; see Revsbech et al., 2009; Stolper et al., 2010) within the core of the OMZ and increased again with depth starting at 1050 m (Figure 1b). Sulfide was not detected, indicating the absence of euxinic conditions. Salinity increased at 95 and 325–400 m, corresponding to the influx of Arabian Sea high salinity water and the Persian Gulf outflow water masses, respectively (Shetye et al., 1994). Particulate matter, as indicated by turbidity, was concentrated towards the sea surface where fluorescence was also high, but also showed increased amounts between 170–300 and 450–750 m (Supplementary Table S1). Ammonium (NH4+) concentrations values ranged between 0–0.14 μ with a maximum value at 170 m depth (0.14 μ). Elevated concentrations were also found from 900 to 1200 m depth (0.08–0.1 μ). Nitrite (NO2−) concentrations showed slight peaks at 170 m (0.62 μ) and 600 m (0.50 μ) (Supplementary Table S1). SPM was sampled for IPL and DNA/RNA analyses at 12 depths in the water column (Figure 1b).
Distribution and abundance of lipids and genes
We developed a new Selective Monitoring Reaction method for the main crenarchaeol-based IPLs synthesized by ‘Ca. N. gargensis' (Pitcher et al., 2011) with six different head groups (Figure 2a, Supplementary Table S1). Of these IPLs, three were detected at high levels in the Arabian Sea SPM: the monohexose, dihexose and hexose-phosphohexose (HPH)-crenarchaeol (Figure 2b), each of which was present at all depths (for example, Figure 3b). Trace amounts of the hexose+'180' crenarchaeol IPL found in N. maritimus SCM1 (Schouten et al., 2008) were also sometimes observed. For comparison with archaeal genes, we focused on HPH-crenarchaeol as opposed to the hexose-based IPLs because it is an abundant IPL in all screened AOA thus far (Schouten et al., 2008; Pitcher et al., 2010, 2011) and is likely to be the best biomarker for putative AOA due to the labile nature of the phosphate-ester bond compared with the glycosidic ether bond (Harvey et al., 1986; Schouten et al., 2010).
Figure 2.
SRM of crenarchaeol-based IPLs with headgroups (labeled peaks; see Supplementary Table S1 for specific monitored transitions). Total ion current of the SRM traces of (a) ‘Ca. N. gargensis' biomass and (b) total ion current of the SRM trace of suspended particulate matter collected at 170 m from the Arabian Sea.
Figure 3.
Depth profile at sampling station: (a) oxygen (O2); (b) ammonium (NH4+) and nitrite (NO2−); (c) HPH-crenarchaeol; (d) thaumarchaeotal 16S rDNA and amoA gene abundances; (e) thaumarchaeotal 16S rRNA and amoA mRNA abundances; (f) PC-monoether ladderane abundance; (g) anammox bacteria 16S rDNA and hydrazine oxidoreductase (hzo) gene abundances; and (h) anammox bacteria 16S rRNA and hydrazine hzo mRNA abundances.
HPH-crenarchaeol showed a substantial increase from the surface waters to 170 m, where its relative abundance reached the highest detected levels and then decreased rapidly to 450 m (Figure 3c). HPH-crenarchaeol abundance was somewhat higher again between 600 and 1200 m. Copy numbers and expression (that is, RNA abundance) of thaumarchaeotal 16S rDNA and amoA genes followed HPH-crenarchaeol concentration, also peaking at 170 m water depth, and a subtle increase was observed at greater depth (Figure 3d). However, 16S rDNA and amoA DNA copy numbers showed a more pronounced peak at 1050 m water depth than both HPH-crenarchaeol and RNA concentrations.
Anammox bacterial markers, that is, the PC-monoether ladderane, anammox 16S rRNA and hzo genes (both DNA and RNA quantification), co-varied well throughout the water column (Figures 3f–g). Full-scan MS analysis showed that additional lipids in the water column (for example, PC-lysolipids) contributed significantly to the SRM signal of the PC monoether assay at 170 m depth, resulting in an overestimation of its abundance. In contrast to AOA, maximum abundances of anammox markers were observed in the core of the OMZ between 450 and 750 m. Both PC-monoether ladderanes and anammox genes were low at concentrations outside this depth range.
Discussion
Depth distributions of AOA
The results of our IPL and gene-based analyses show that AOA are abundant in the OMZ of the Arabian Sea and confirm previous evidence for the occurrence of planktonic Thaumarchaeota in the Arabian Sea based on the recovery of fossil crenarchaeol (Schouten et al., 2010). The specificity of crenarchaeol to AOA (de la Torre et al., 2008; Pitcher et al., 2010), coupled with the occurrence of HPH-crenarchaeol in all screened AOA to date (Schouten et al., 2008; Pitcher et al., 2010, 2011), renders HPH-crenarchaeol as the most fitting biomarker lipid for tracking AOA. Similarities between AOA genes and IPL profiles (Figure 3) indicate that our AOA-specific crenarchaeol-based IPL SRM is a suitable tool for tracking active AOA occurrence in the marine water column. Previous studies have observed that the degradation rate of IPLs is different according to the environmental conditions and the polar head group, for example, glycosidic ether lipids degrading much slower than phosphoester lipids (Harvey et al., 1986; Schouten et al., 2010). Therefore, HPH-crenarchaeol is likely the best marker for living AOA. Phosphoester lipids have been shown to degrade substantially within 2–4 days, though small percentages remain even after several weeks (White et al., 1979; Harvey et al., 1986). Thus, IPL turnover occurs likely on a scale of days to weeks (that is, similar to DNA), whereas RNA (especially mRNA) will have a much shorter half-life on the order of minutes to hours (Takayama and Kjelleberg, 2000).
Thaumarchaeotal amoA and 16S rDNA gene abundances were positively correlated, suggesting that the majority of the Thaumarchaeota are ammonia oxidizers. The archaeal amoA:16S rDNA ratio (average 2–3) is comparable to the values observed in other environmental studies (for example, Wuchter et al., 2006; Beman et al., 2008; Galand et al., 2009; Molina et al., 2010). The detection of a putative AOA community deep in the Arabian Sea is in agreement with recent studies implicating AOA in deep-sea nitrification (Mincer et al., 2007; Konstantinidis et al., 2009; Church et al., 2010; Santoro et al., 2010). The coincidence of peak HPH-crenarchaeol and AOA 16S rRNA and amoA DNA gene abundances and expression, predominantly at 170 m and, to a lesser extent, around 1050 m, points to the OMZ transition zones as possible preferred niches for Arabian Sea Thaumarchaeota and indicates that they are adapted to cope with low oxygen concentrations, as at both depths oxygen concentration was ∼5 μ (Figure 3a; Supplementary Table S1). Other water column studies have revealed similar subsurface peaks in putative AOA abundance at or near the onset of low oxygen concentrations (Coolen et al., 2007; Beman et al., 2008; Lam et al., 2009, 2011; Molina et al., 2010). Although AOA have also been recovered in high abundance from oxygenated marine waters (for example, Karner et al., 2001; Wuchter et al., 2006; Herfort et al., 2007) including the 20-m SPM sample at our station, and have been grown successfully under fully oxic culture conditions (Könneke et al., 2005; de la Torre et al., 2008; Hatzenpichler et al., 2008), their abundance and gene expression was much higher at 170 m relative to the fully oxygenated surface water and ventilated bottom waters (Figure 3). This is in agreement with recent enrichment studies that showed preferential growth of Thaumarchaeota at low oxygen concentrations (Park et al., 2010). Furthermore, a recent study by Stolper et al. (2010) showed that aerobic metabolisms, such as ammonia oxidation, can potentially proceed at very low oxygen concentrations. It should be noticed that AOA 16S rDNA and amoA DNA copy abundances showed a much stronger peak at 1050 m depth, in contrast to AOA IPLs and RNA copy numbers. This might indicate that AOA are less metabolically active at this depth in comparison with the population occupying the niche at 170 m.
Within the core OMZ, between 600–750 m, AOA reach their lowest values where oxygen concentrations are minimal. This, together with low levels of amoA mRNA, could be an indication that oxygen levels are perhaps too low within the core of the OMZ to support a strong aerobic AOA activity. If so, then the AOA in the Arabian Sea exhibit an especially narrow range of preferred oxygen conditions (that is, 5 μ>O2>2.5 μ). Indeed, low oxygen concentration has been shown to inhibit growth of N. maritimus SCM1 in culture (Km=0.13 μ), suggesting a limited activity under very low-oxygen/anoxic conditions in nature (Martens-Habbena et al., 2009). Although high copy numbers of AOA 16S rDNA have been previously detected in suboxic areas of the Black Sea (O2 concentrations below detection limit; Coolen et al., 2007; Lam et al., 2007), low amoA expression was also observed. These observations indicate that under low-oxygen conditions (<2.5 μ) AOA might survive, but metabolic activity is reduced. Furthermore, the presence of AOA in the mid-OMZ may also be restricted by competition for NH4+ (and/or O2) with other microbes, such as anammox bacteria. NH4+ concentrations throughout the OMZ were lower but near the Km values reported for N. maritimus SCM1 (Martens-Habbena et al., 2009), indicating that AOA should not be limited by NH4+ concentrations at mid-OMZ depths (assuming environmental AOA are also adapted to low-substrate conditions). Thus, the pointed decrease in AOA abundance towards the core OMZ suggests that oxygen may be a more important factor in determining their depth distribution at this time and location.
Depth distributions of anammox bacteria
Anammox 16S rRNA and hzo gene abundances, the expression of these genes and PC-monoether ladderane profiles co-vary well (Figures 3f–h) and the maximum abundance of anammox genes and IPLs occurred between 450 and 750 m, evidencing a prominent community of active anammox bacteria over this depth range. The substantially lower abundance of IPLs and genes, and their expression, at other depths suggests that mid-OMZ depths are optimal for anammox bacteria. Anammox bacteria have been closely associated with NO2− maxima previously in the Black Sea (Kuypers et al., 2005; Lam et al., 2007) and in the Arabian Sea (Ward et al., 2009; Bulow et al., 2010). Therefore, we did not expect to find such high abundances of anammox bacteria in the absence of a strong secondary nitrite maximum (SNM) at our sampling station; within the OMZ NO2− was only slightly elevated (0.5 μ) at a single depth (600 m) (Figure 3b). This observation suggests that significant anammox communities may also exist elsewhere where NO2− is present in lower concentrations, particularly because in situ concentration may not be representative of the actual flux or turnover. Interestingly, previous core ladderane analyses from Arabian Sea SPM also showed maximum lipid abundances at ca. 600 m, at multiple stations off the Omani coast (Jaeschke et al., 2007). Although core lipids were used in this case (as opposed to IPLs) and the sampling resolution was much lower, this nevertheless substantiates our findings and the likelihood of anammox communities existing outside/below the SNM of the Arabian Sea (and potentially other low-NO2− marine environments). If so, the presence of a nitrite maximum should not necessarily be taken as a likely spot for the presence and activity of anammox bacteria (cf. Ward et al., 2009). Indeed, Lam et al. (2011) have recently observed that the strong SNM can be a signature of an aged water mass with nitrate-reducing conditions that has experienced past nitrogen loss but can no longer support in situ N-loss activity.
The anammox 16S rRNA gene abundances observed between 450 and 750 m (1.7–4.1 × 108 16S rDNA anammox copies l−1) are comparable to those observed by Ward et al. (2009) at shallower depths (80–250 m) where NO2− was ca. 5–10 μ. As Ward et al. (2009) did not report results of samples deeper than the SNM for genes and anammox activity, it is not possible to say whether a significant community of anammox bacteria was present from 450 to 700 m at the stations they studied. Nevertheless, our findings indicate that the distribution of anammox bacteria is not patchy as previously suggested (Bulow et al., 2010) and they may be important in removing nitrogen from core OMZ depths in the Arabian Sea in addition to any denitrification, which may also be occurring. Possibly, the low oxygen concentrations at these depths might promote a cryptic sulfur cycle that in turn stimulates the activity of anammox bacteria, as has been suggested for the Peruvian OMZ (Canfield et al., 2010).
The Arabian Sea SNM has been attributed to denitrification activity in the Arabian Sea (that is, accumulated NO2− from NO3− reduction). The absence of a strong SNM at our site suggests the possibility that denitrification may not have been intense, resulting in only scant amounts of denitrification-derived NO2− as a substrate for anammox. However, depending on the activity level of local anammox bacteria, rapid conversion of NO2− could also mask intensely high denitrification. Water between 250 and 400 m at our sampling station did show an increase in salinity characteristic of Persian Gulf Water (Figure 2). From the present data, it is not possible to determine whether the Persian Gulf Water and/or local denitrification were influencing the anammox community at our site. Nevertheless, the identification of abundant biomarker lipids, specific genes and their gene expression pointing to a substantial active community of anammox bacteria in the Arabian Sea at the core of the OMZ in the absence of a prominent SNM is an intriguing observation that could imply a much larger potential contribution of anammox bacteria to N-loss than is currently estimated.
Implications for potential metabolic coupling of archaeal ammonia-oxidation and anammox
AOA could theoretically provide substrates for anammox in the form of NO2−. NH4+, however, is required by both and, therefore, direct coupling of these metabolisms would require competition, and/or alternate sources of this substrate. Despite the fact that NH4+ oxidation requires oxygen, and anammox bacteria are inhibited by as little as 1 μ of oxygen in culture (Strous et al., 1997), anammox bacteria have been recovered from water with an oxygen concentration of 9 μ (Kuypers et al., 2005), and putative AOA are commonly recovered from near-suboxic waters. Recent work suggested that indeed AOA and ammonia oxidizing bacteria may contribute up to 40% of the NO2− required by anammox in the Black Sea (Lam et al., 2007). Although AOA and anammox bacteria could theoretically occupy similar niches in the Arabian Sea, our results indicate that this is not the case, as evidenced mainly by the large vertical segregation (>400 m) of their respective niches. Ward et al. (2009) also hypothesized a non-existent relationship between aerobic ammonium oxidation and anammox activity at shallower depths, although this study was based on another sampling time and stations. However, if AOA at the time of their sampling campaign would have exhibited the same distribution as we observed, it is possible that the anammox bacteria observed at 200 m by Ward et al. (2009) could have indeed been coupled to archaeal ammonia oxidation as we clearly find a maximum in abundance and activity of AOA at the top of the OMZ.
Our results thus indicate that, despite the potentially suitable conditions, AOA and anammox bacteria do not necessarily occupy similar niches, at least not at our site in the Arabian Sea, and the cooperation/competition between these two groups may not be widespread in anoxic basins or OMZs.
Acknowledgments
Sabine Lengger and Lara Pozzatto are thanked for assistance sampling during the PASOM cruise. The PASOM cruise was funded by the Netherlands Organization for Scientific Research (NWO) under number 817.01.015. Additional thanks are due to the captain and crew of the R/V Pelagia for assistance and technical support. Bob Koster and Jan van Ooyen are thanked for assistance with CTD operations and nutrient analyses. Judith van Bleijswijk and Elda Panoto are thanked for assistance with molecular analyses. This is publication number DW-2011-1006 of the Darwin Center for Biogeosciences, which partially funded this project.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:349–355. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- Bange HW, Rixen T, Johansen AM, Siefert RL, Ramesh R, Ittekkot V, et al. A revised nitrogen budget for the Arabian Sea. Global Biogeochem Cy. 2000;14:1283–1297. [Google Scholar]
- Bange HW, Naqvi SWA, Codispoti LA. The nitrogen cycle in the Arabian Sea. Prog Oceanogr. 2005;65:145–158. [Google Scholar]
- Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brandes JA, Devol AH, Deutsch C. New developments in the marine nitrogen cycle. Chem Rev. 2007;107:577–589. doi: 10.1021/cr050377t. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Bulow SE, Rich JJ, Naik HS, Pratihary AK, Ward BB. Denitrification exceeds anammox as a nitrogen loss pathway in the Arabian Sea oxygen minimum zone. Deep-Sea Res I. 2010;57:384–393. [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- Church MJ, Wai B, Karl DM, Delong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MJL, Hopmans EC, Rijpstra WIC, Muyzer G, Schouten S, Volkman JK, et al. Evolution of the methane cycle in Ace Lake (Antarctica) during the Holocene: response of methanogens and methanotrophs to environmental change. Org Geochem. 2004;35:1151–1167. [Google Scholar]
- Coolen MJL, Abbas B, van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG, et al. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Cuna-Gonzalez J. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature. 2003;422:606–608. doi: 10.1038/nature01526. [DOI] [PubMed] [Google Scholar]
- de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing Archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Lovejoy C, Hamilton AK, Ingram RG, Pedneault E, Carmack EC. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ Microbiol. 2009;11:971–980. doi: 10.1111/j.1462-2920.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- Harvey HR, Fallon RD, Patton JS. The effect of organic matter and oxygen on the degradation of bacterial membrane lipids in marine sediments. Geochim Cosmochim Acta. 1986;50:795–804. [Google Scholar]
- Hatzenpichler R, Lebecleva EV, Spieck E, Stoecker K, Richter A, Daims H, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfort L, Schouten S, Abbas B, Veldhuis MJW, Coolen MJL, Wuchter C, et al. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol Ecol. 2007;62:242–257. doi: 10.1111/j.1574-6941.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, Rapp HT, et al. Complex nitrogen cycling in the sponge Geodia barretti. Environ Microbiol. 2009;11:2228–22243. doi: 10.1111/j.1462-2920.2009.01944.x. [DOI] [PubMed] [Google Scholar]
- Jaeschke A, Hopmans EC, Wakeham SG, Schouten S, Sinninghe Damste JS. The presence of ladderane lipids in the oxygen minimum zone of the Arabian Sea indicates nitrogen loss through anammox. Limnol Oceanogr. 2007;52:780–786. [Google Scholar]
- Jaeschke A, Rooks C, Trimmer M, Nicholls JC, Hopmans EC, Schouten S, et al. Comparison of ladderane phospholipid and core lipids as indicators for anaerobic ammonium oxidation (anammox) in marine sediments. Geochim Cosmochim Acta. 2009;73:2077–2088. [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Könneke ME, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific Subtropical Gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jorgensen BB, Kuenen JG, et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature. 2003;422:608–611. doi: 10.1038/nature01472. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci USA. 2005;102:6478–6483. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Kock A, Lettmann KA, Plancherel Y, Lavik G, et al. Origin and fate of the secondary nitrite maximum in the Arabian Sea. Biogeosciences Discuss. 2011;8:2357–2402. [Google Scholar]
- Li M, Hong YG, Klotz MG, Gu JD. A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol. 2010;86:781–790. doi: 10.1007/s00253-009-2361-5. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, Delong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Molina V, Belmar L, Ulloa O. High diversity of ammonia-oxidizing Archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ Microbiol. 2010;12:2450–2465. doi: 10.1111/j.1462-2920.2010.02218.x. [DOI] [PubMed] [Google Scholar]
- Morrison JM, Codispoti LA, Smith SL, Wishner K, Flagg C, Gardner WD, et al. The oxygen minimum zone in the Arabian Sea during 1995. Deep Sea Res II. 1999;46:1903–1931. [Google Scholar]
- Park B-J, Park S-J, Yoon D-N, Schouten S, Sinninghe Damsté JS, Rhee S-K. Autotrophic ammonia oxidizing Group I.1a crenarchaea in marine sediments. Appl Environ Microbiol. 2010;76:7575–7587. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulmier A, Ruiz-Pino D. Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr. 2009;80:113–128. [Google Scholar]
- Pitcher A, Rychlik N, Hopmans EC, Spieck E, Rijpstra WI, Ossebaar J, et al. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic Group I.1b Archaeon. ISME J. 2010;4:542–552. doi: 10.1038/ismej.2009.138. [DOI] [PubMed] [Google Scholar]
- Pitcher A, Hopmans EC, Mosier AC, Park S-J, Rhee S-K, Francis CA, et al. 2011Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing Archaea enriched from marine and estuarine sediments Appl Environ Microbioldoi: 10.1128/AEM.02758-10 [DOI] [PMC free article] [PubMed]
- Prosser JI, Nicol GW. Relative contributions of Archaea and Bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- Revsbech NP, Larsen LH, Gundersen J, Dalsgaard T, Ulloa O, Thamdrup B. Determination of ultra-low oxygen concentrations in oxygen minimum zones by the STOX sensor. Limnol Oceanogr Methods. 2009;7:371–381. [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying Archaea and Bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Schouten S, Hopmans EC, Baas M, Boumann H, Standfest S, Könneke M, et al. Intact membrane lipids of ‘Candidatus Nitrosopumilus maritimus,' a cultivated representative of the cosmopolitan mesophilic group I crenarchaeota. Appl Environ Microbiol. 2008;74:2433–2440. doi: 10.1128/AEM.01709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten S, Middelburg JJ, Hopmans EC, Sinninghe Damste JS. Fossilization and degradation of intact polar lipids in deep subsurface sediments: a theoretical approach. Geochim Cosmochim Acta. 2010;74:3806–3814. [Google Scholar]
- Shetye SR, Gouveia AD, Shenoi SSC. Circulation and water masses of the Arabian Sea. Proc Ind Acad Sci-Earth Plant Sci. 1994;103:107–123. [Google Scholar]
- Sinninghe Damsté JS, Rijpstra WI, Hopmans EC, Prahl FG, Wakeham SG, Schouten S. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl Environ Microbiol. 2002;68:2997–3002. doi: 10.1128/AEM.68.6.2997-3002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Stolper DA, Revsbech NP, Canfield DE. Aerobic growth at nanomolar oxygen concentrations. Proc Natl Acad Sci USA. 2010;107:18755–18760. doi: 10.1073/pnas.1013435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, van Gerven E, Kuenen JG, Jetten M. Effects of aerobic and microaerobic conditions on anaerobic ammonium-oxidizing (Anammox) sludge. Appl Environ Microbiol. 1997;63:2446–2448. doi: 10.1128/aem.63.6.2446-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, et al. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- Suthhof A, Ittekkot V, Gaye-Haake B. Millennial-scale oscillation of denitrification intensity in the Arabian Sea during the late Quaternary and its potential influence on atmospheric N2O and global climate. Global Biogeochem Cy. 2001;15:637–649. [Google Scholar]
- Takayama K, Kjelleberg S. The role of RNA stability during bacterial stress responses and starvation. Environ Microbiol. 2000;2:355–365. doi: 10.1046/j.1462-2920.2000.00119.x. [DOI] [PubMed] [Google Scholar]
- Voss M, Montoya JP. Nitrogen cycle oceans apart. Nature. 2009;461:49–50. doi: 10.1038/461049a. [DOI] [PubMed] [Google Scholar]
- Ward BB, Devol AH, Rich JJ, Chang BX, Bulow SE, Naik H, et al. Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature. 2009;461:78–81. doi: 10.1038/nature08276. [DOI] [PubMed] [Google Scholar]
- White DC, Davis WM, Nickels JS, King JD, Bobbie RJ. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- Woebken D, Fuchs BM, Kuypers MM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol. 2007;73:4648–4657. doi: 10.1128/AEM.02774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.