Abstract
Microbial ecologists can now start digging into the accumulating mountains of metagenomic data to uncover the occurrence of functional genes and their correlations to microbial community members. Limitations and biases in DNA extraction and sequencing technologies impact sequence distributions, and therefore, have to be considered. However, when comparing metagenomes from widely differing environments, these fluctuations have a relatively minor role in microbial community discrimination. As a consequence, any functional gene or species distribution pattern can be compared among metagenomes originating from various environments and projects. In particular, global comparisons would help to define ecosystem specificities, such as involvement and response to climate change (for example, carbon and nitrogen cycle), human health risks (eg, presence of pathogen species, toxin genes and viruses) and biodegradation capacities. Although not all scientists have easy access to high-throughput sequencing technologies, they do have access to the sequences that have been deposited in databases, and therefore, can begin to intensively mine these metagenomic data to generate hypotheses that can be validated experimentally. Information about metabolic functions and microbial species compositions can already be compared among metagenomes from different ecosystems. These comparisons add to our understanding about microbial adaptation and the role of specific microbes in different ecosystems. Concurrent with the rapid growth of sequencing technologies, we have entered a new age of microbial ecology, which will enable researchers to experimentally confirm putative relationships between microbial functions and community structures.
Keywords: metagenomics, community function, global ecosystems, community structure, environmental microbiology
Introduction
The explosion of metagenomic projects in an increasing variety of terrestrial and marine ecosystems (Tyson et al., 2004; García Martín et al., 2006; Kurokawa et al., 2007; Nealson and Venter, 2007; Vogel et al., 2009) and the availability of new high-throughput sequencing technologies are facilitating our understanding of the ‘black box' of environmental microbial communities. This black box contains a wealth of novel genes that can aid in drug discovery and in a better understanding of processes for climate change, agronomy and pollution degradation. Importantly, this goldmine of biological information is becoming increasingly publically accessible through various databases and annotation platforms (http://metagenomics.anl.gov/; http://camera.calit2.net/; http://img.jgi.doe.gov/cgi-bin/m/main.cgi) and mining these data can aid in both providing answers to and helping to test and create new hypotheses for microbial ecologists. However, the demand for competent bioinformaticians and statistically valid data treatment methods often exceeds supply, thus leaving many microbial ecologists removed from this rush of metagenomic data. Many of the potential insights will come from comparing metagenomic data between ecosystems (Tringe et al., 2005; Dinsdale et al., 2008; Willner et al., 2009). However, currently accessible data are underexploited despite their ecological relevance. Although this lack of data use and the perceived requirement for trained bioinformaticians could raise the question of the value of investing heavily in metagenomics projects (Baveye, 2009), we believe that benefit will come in the form of inter-ecosystem comparisons of microbial functions of interest, intra-ecosystem variations in microbial function, identification of novel genes and correlations between functions (and species) that will shed light on microbial interactions and adaptation.
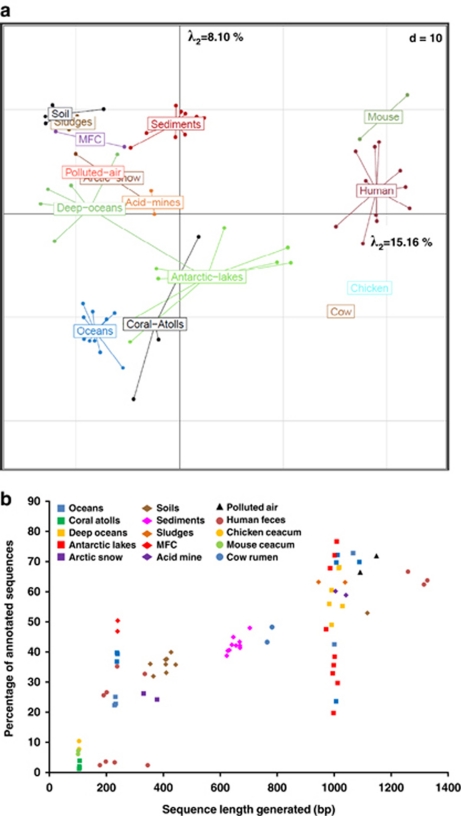
Global metagenomic comparisons can be used to probe for answers to (or tickle the curiosity about) different aspects of microbial ecology by the application of new user-friendly bioinformatics and statistical tools for understanding the strength of observed differences. As an example, we compared the diversity and distribution of 77 metagenomes (most being publically available) corresponding to various projects and environments by using the MG-RAST public platform (Meyer et al., 2008). The relative numbers of sequence reads that were annotated in the metabolic subsystems provided were analyzed by principal component analysis (PCA). In addition, STAMP was used to evaluate the statistical significance of observed differences (Parks and Beiko, 2010). The metagenomes from these different ecosystems (oceans, coral atolls, deep oceans, Antarctic aquatic environments, Arctic snows, soils, hypersaline sediments, sludges, microbial fuel cell biofilms, acid mine biofilms, polluted air and animal microbial populations) are clearly separated (Figure 1a). Significant variations between research labs, sample types, DNA extraction and sequencing techniques for a given ecosystem do not seem to inhibit cross ecosystem comparisons. As an example, for the metagenomes from three ecosystems, ocean, soil and human microbiome, DNA was extracted by different researchers using different methods and different sequencing technologies (three sequential pyrosequencing technologies and the Sanger technology) were used, yet, these metagenomes are still grouped as a function of their original environment (Figure 1a). Thus, although methodological fluctuations exist between laboratories (Leek et al., 2010), these results show a limited (‘batch') effect of methodology (for example, sequencing technology) in high-throughput data when comparing globally different environments. In addition, studies focused on 16S rRNA gene sequencing also clustered animal microbial populations separately from the marine and terrestrial ecosystems (Ley et al., 2008). The exploration of other gene classes responsible for these differences would provide insight into the overall functioning of these ecosystems.
Figure 1.
(a) PCA based on the relative distribution of annotated sequences (E-value<10−5) categorized in 838 different functional subsystems detected in the 77 metagenomes. Distributions were normalized as a function of the number of annotated sequences for each metagenome. The percentages of the illustrated two major axes correspond to the fraction of the total variance that they represent (see insert showing all of the axes and their percentage of the overall variance). (b) Relationship between average sequence length and the percentage of annotated functions (E-value<10−5) for the metagenomes used here. The different average sequence sizes are due in part to variations in sequencing technology. In addition, ocean and Antarctic metagenomes have annotations varying considerably for the same average sequence length. This fluctuation is due in part to the presence of sequences related to eukaryotic and virus sequences for oceans and Antarctic aquatic environments.
A considerable limit of MG-RAST and other annotation platforms is the use of ‘annotated' sequences, that is, those that have been classified as belonging (with a fixed probability of similarity, in our case here we used an E-value limit of 10−5) to some established functional subsystems (for example, carbohydrate metabolism) or other databases, and the exclusion of non-annotated sequences, which might provide both novel functions and important differences between ecosystems. These non-annotated sequences provide a tremendous resource for future functional experiments and protein modeling. Some novel and potentially ecologically important functional groups are not being identified because of the dependence of current platforms on the already sequenced (and hopefully well annotated) genomes of mostly cultivated microorganisms. An added caveat to the discovery and exploitation of non-annotated sequences (including possible gene assembly) is the dependence of sequence length on the percentage of annotated sequences (Figure 1b). In panel 1b, a clear correlation between the percentage of annotated sequences and sequence length is shown; however, there seems to be a confounding effect, which is the proportion of Eukarya- or virus-related sequences in the metagenomic data set. For example, the percentage of annotated sequences in some ocean metagenomes (sequence length of about 1000 nt) is negatively correlated to the proportion of eukaryotic sequences (R2>0.86, when using 59 metagenomes from the global ocean survey). Due to important annotation fluctuations independent of microbial community structure (Figure 1b), both functional and taxonomical distributions were normalized as a function of the number of annotated sequences as was carried out for Figure 1a and not the entire data set.
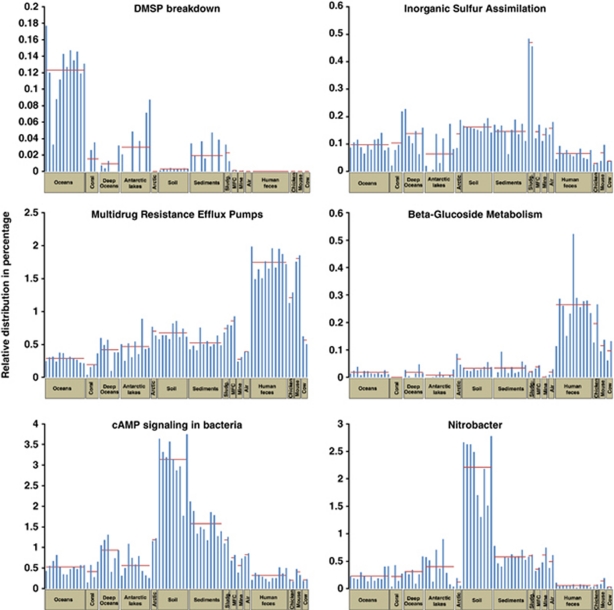
Although comparing ecosystems based on their relative sequence (‘reads') frequencies in different metabolic subsystems can provide insight into functional differences, specific functions (or species) can be and should be individually examined in order to answer specific questions or to test hypotheses. Comparing large numbers of metagenomes can highlight unusual functional and phylogenetic distributions either between or within ecosystems. We provide a few examples of this approach to emphasize its significance (Figure 2). Oceans possess the highest relative number of metagenomic sequences related to dimethylsulfoniopropionate (DMSP) breakdown (Figure 2). DMSP occurs in considerable amounts in marine algae, for which this molecule and its breakdown products probably serve as an antioxidant system (Sunda et al., 2002). But more importantly, its degradation can release dimethyl sulfide molecules (DMS) into the atmosphere, where they might improve cloud formation and limit solar radiation at the planet surface (Charlson et al., 1987). This functional subsystem is distinctly more abundant in the ocean ecosystem than in any of the 14 other environments. In ocean-related metagenomes and within this subsystem (‘DMSP'), sequences corresponding to DmdA (DMSP demethylase) and to DmdB2 (DMSP breakdown hydrolase) were found. On the other hand, inorganic sulfur assimilation-associated sequences are not particularly higher in the oceans than in other ecosystems and are on the same order as that for DMSP degradation in the ocean (about 0.1% of annotated sequences). Inorganic sulfur assimilation is more highly represented in the two activated sludge metagenomes, corresponding to ∼0.5% of the annotated sequences (Figure 2). The sulfur in sewage sludge can form gaseous SO2 and cause associated acid-rain problems, if incorporated in sludge co-combustion processes. Therefore, biological mechanisms involved in sulfur cycling have immediate impacts on environmental processes.
Figure 2.
Comparison of the relative distribution in percentage (based on the annotated sequences (E-value<10−5)) of five functional classes and one genus (SEED annotation) among the 77 metagenomes deposited in MG-RAST. The horizontal line corresponds to the average of the relative distribution for each of the 15 environments.
Another example focuses on multidrug resistance efflux pumps involved in antibiotic resistance that have been extensively studied in pathogens (Li and Nikaido, 2004). However, these pumps are present in all living organisms and are not restricted to antibiotic compounds (Martinez et al., 2009). They are also capable of extruding heavy metals, solvents and antiseptics (Pumbwe et al., 2007). They are thought to be largely present in soil and in association with plants (Konstantinidis and Tiedje, 2004). However, we found that they appear to be more represented in human feces and chicken and mouse cecum (Figure 2), where they could have an important role in pathogen antibiotic resistance. These pumps are present in all the environments studied, confirming their multifunction role, but are relatively limited in oceans, deep oceans, polluted airs and acid mine drainage biofilms, where they are apparently less dominant. Although correlations can be calculated between functions that are relatively dominant in the same ecosystem (for example, beta-glucosides for animal-associated microbial communities), these correlations do not necessarily have any mechanistic value, but could be simply co-correlated to other phenomena. To provide more clear relationships, the presence of these two targeted subsystems on the same sequence (whether on a read or a contig) is required. The cAMP signaling is another example where we find more sequences related to bacterial cAMP signaling in soil than elsewhere (Figure 2).
The cAMP is an important secondary messenger in all three domains of life. Interestingly, as a cAMP subversion mechanism, some bacterial pathogens inject adenylate cyclase protein toxins into plants (Agarwal and Bishai, 2009). Thus, soil microbial communities appear to possess a considerable potential for deceiving plant signaling mechanisms, if cAMP is involved in pathogenicity; however, its role in other metabolic functions cannot be disregarded.
Metagenomic sequence data can also be used to evaluate the microbial community structure. In metagenomes without targeted gene amplification, the number of housekeeping genes present that can be used to assess which species are present is rather limited. On the other hand, all annotated sequences could be assigned to a closest related species and used to define the community structure with the caveat that only known species will be defined. An example is the apparent distribution of the genus Nitrobacter (known to oxidize nitrite to nitrate; Schmidt, 1978) in different ecosystems and its relative dominance in soil (Figure 2).
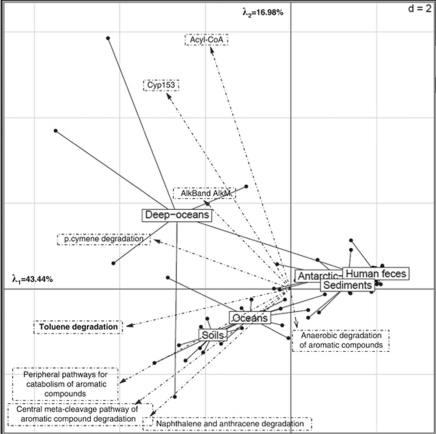
Another approach for comparing ecosystem metagenomes could be the combination of results from annotation platforms and the number of sequences determined by using BLAST for specific genes (Altschul et al., 1990). This BLAST approach can be applied by tagging metagenomes and developing ‘in house' annotation systems that researchers can create for specific questions. For example, a sample from the Gulf of Mexico is among the ocean metagenomes, therefore, these metagenomes could be screened for their relative petroleum hydrocarbon degrading abilities. Using BLAST, pooled and tagged metagenomes were screened for sequences similar to those associated with the degradation of oil compounds. For example, we looked at genes that code for the AlkB and AlkM enzymes, which are capable of degrading aliphatic oil compounds. Other genes including those associated with cytochrome P450 (CYP153 family), which has also been implicated in aliphatic hydrocarbon degradation, were included in our metagenomic screening. The relative distribution of these genes was used to predict variations in hydrocarbon degradation potential among ecosystems. These distributions were normalized as a function of the number of annotated sequences on annotation platforms as carried out for the MG-RAST subsystems. We simultaneously compared metagenomes from some of these ecosystems using both functional subsystems associated with hydrocarbon degradation and the specific hydrocarbon degradation genes cited above using a PCA approach. Antarctic aquatic environments, human feces and hypersaline sediments were similar in their general lack of sequences (other than the presence of those associated with anaerobic aromatic compound metabolism) associated with hydrocarbon degradation (Figure 3). The presence of anaerobic aromatic compound metabolism sequences was, however, observed in all ecosystems, but this type of metabolism is not limited to petroleum hydrocarbons. The location of the different functional genes can be projected on the same PCA plot in order to provide a visual clue as to which functional genes are associated with which ecosystems (Figure 3). For example, the majority of deeper ocean samples (from at least 500 m depth) have relatively more aliphatic oil-degrading genes than the surface ocean and soil, which contain more aromatic oil-degrading genes. The important fluctuations observed for deep oceans are due to the limited number of functions compared in this PCA. In addition, based on these selected hydrocarbon degradation functions, oceans and soil are relatively similar.
Figure 3.
PCA of six selected ecosystems based on their number of sequences associated with petroleum hydrocarbon degradation functions (E-value<10−5). The functional classes as provided by MG-RAST and the local blasts are plotted on the same PCA as the samples in order to observe relationships between function and environment.
This rapidly growing metagenomic sequence data from different environments can also help researchers target microbial communities that might have roles in a range of important functions. Although gene presence per se is not indicative of enzymatic activity, and the ecosystems compared here are not equal in amount of sequence data, understanding the relative proportions of these genes in specific ecosystems might provide better insight into their relative importance. Perhaps metatranscriptomic and metaproteomic approaches will help to understand the temporal nature of the specific activities and functions that are expressed. After such primary analysis, clone libraries could be constructed (and are being constructed in some cases) and probed for the sequences of interest, after which the genetic environment of these functional genes could be elucidated.
We have shown only a very limited analysis of existing metagenomic data here in order to illustrate existing resources available to microbial ecologists today. These resources are being constantly replenished by increasing data sets and sequenced ecosystems. The distribution of every defined function and species can already be evaluated at different taxonomical levels in hundreds of metagenomes using both annotation platforms and local BLAST for specific questions. Differences in functional gene families and specific functions (or target sequences) between metagenomes from different environments can aid our understanding of how microbial communities function. The beauty of this influx of metagenomic data is that so much remains to be discovered. As an example, estimations established that between 104 (Torsvik et al., 2002) and 107 (Gans et al., 2005) different species can be present in 1 g of soil. In addition, we have emphasized the considerable difficulties in accessing soil genetic richness that limit the number of detected species when using only one DNA extraction approach (Delmont et al., 2011). As a consequence, currently available metagenomes related to soil and other ecosystems represent only a fraction of their existing genetic potential.
In the future, continuous advances in sequencing technologies (and sequenced genomes) will generate not only more, but also longer sequences, thus increasing significantly metagenomic sensitivity and possibly the percentage of annotated sequences (Figure 1b). The re-annotation of metagenomes when additional reference genomes become available will also stimulate and improve annotations, if those sequenced genomes are accurately annotated themselves. Increased number and length of metagenomic sequences will also lead to genome assembly and possible improved cultivation techniques. In addition to this revolution, the continuous increase in metagenome sequencing projects (for example, TARA, Earth Microbiome Project, Terragenome and Microbial Earth project; Gilbert et al. 2011) and new global metagenomic comparison tools are aiding researchers enter a new age of microbial ecology. However, experiments (including metatranscriptomic and metaproteomic analyses) are becoming essential to confirm the biological roles of annotated functions (and microorganisms) in situ and to increase our knowledge concerning the vast quantity of non-annotated sequences.
Acknowledgments
TOD was supported by the Rhône-Alpes Region and part of this work was supported by the French National Research Agency (Agence National de Recherche) ANR Genomique programme: METASOIL project.
Appendix
Oceans
4441573.3, 4441574.3, 4441576.3, 4441577.3, 4441591.3, 4443688.3, 4443697.3, 4443713.3, 4443714.3, 4443716.3, 4443725.3, 4443729.3.
Coral attols
4440279.3, 4440037.3, 4440039.3, 4440041.3.
Deep oceans
4441619.3, 4441656.4, 4441620.3, 4442503.3, 4441663.3, 4442500.4.
Antarctic aquatic environments
4443683.3, 4443680.3, 4443682.3, 4443684.3, 4443679.3, 4443686.3, 4443685.3, 4443687.3, 4443681.3.
Arctic snows
4443128.3, 4443127.3.
Soils
4441091.3, 4446153.3, http://metasoil.univ-lyon1.fr/ for metagenomes corresponding to Rothamsted Park Grass soil experiment.
Sediments
4440964.3, 4440963.3, 4440965.3, 4440966.3, 4440967.3, 4440969.3, 4440970.3, 4440968.3, 4440971.3, 4440972.3.
Phosphorus removing sludges
4441092.3, 4441093.3.
Microbial fuel cells
4447261.3, 4447259.3.
Acid Mine Drainage Biofilms
4441137.3, 4441138.3.
Singapore indoor polluted airs
4447940.3, 4447941.3.
Human feces
4440825.3, 4440460.5, 4440614.3, 4440611.3, 4440613.3, 4440616.3, 4440595.4, 4440452.7, 4440939.3, 4440942.3, 4440943.3.
Chicken Cecum
4440283.3, 4440284.3.
Mouse cecum
4440463.3, 4440464.3.
Cow rumen
4441679.3, 4441680.3.
These accession numbers correspond to metagenomes available on MG-RASTv2 server (http://metagenomics.anl.gov/v2/).
References
- Agarwal N, Bishai WR. cAMP signaling in Mycobacterium tuberculosis. Indian J Exp Biol. 2009;47:393–400. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baveye PC. To sequence or not to sequence the whole-soil metagenome. Nat Rev Microbiol. 2009;756:757. doi: 10.1038/nrmicro2119-c2. [DOI] [PubMed] [Google Scholar]
- Charlson R, Lovelock J, Andreae M, Warren S. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. [Google Scholar]
- Delmont TO, Robe P, Cecillon S, Clark IM, Constancias F, Simonet P, et al. Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol. 2011;77:1315–1324. doi: 10.1128/AEM.01526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, et al. Functional metagenomic comparison profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- García Martín H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Gilbert J, O'Dor R, Vogel TM. Survey data are still vital to science. Nature. 2011;469:162. doi: 10.1038/469162a. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. P Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The Metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Venter JC. Metagenomics and the global ocean survey: what's in it for us, and why should we care. ISME J. 2007;1:185–187. doi: 10.1038/ismej.2007.43. [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Pumbwe L, Skilbeck CA, Wexler HM. Induction of multiple antibiotic resistance in Bacteroides fragilis by benzene and benzene-derived active compounds of commonly used analgesics, antiseptics and cleaning agents. J Antimicrob Chemoth. 2007;60:1288–1297. doi: 10.1093/jac/dkm363. [DOI] [PubMed] [Google Scholar]
- Schmidt EL.1978Nitrifying microorganisms and their methodologyIn: Schlessinger D (ed.).Microbiology—1978 American Society for Microbiology: Washington, DC; 288–291. [Google Scholar]
- Sunda W, Kieber DJ, Kiene RP, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Tringe SG, Mering CV, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, van Elsas JD, et al. TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol. 2009;7:2. [Google Scholar]
- Willner D, Thurber RV, Rohwer F. Metagenomic signatures of 86 microbial and viral metagenomes. Env Microbiology. 2009;11:1752–1766. doi: 10.1111/j.1462-2920.2009.01901.x. [DOI] [PubMed] [Google Scholar]