Abstract
The anaerobic oxidation of methane (AOM) with sulfate controls the emission of the greenhouse gas methane from the ocean floor. AOM is performed by microbial consortia of archaea (ANME) associated with partners related to sulfate-reducing bacteria. In vitro enrichments of AOM were so far only successful at temperatures ⩽25 °C; however, energy gain for growth by AOM with sulfate is in principle also possible at higher temperatures. Sequences of 16S rRNA genes and core lipids characteristic for ANME as well as hints of in situ AOM activity were indeed reported for geothermally heated marine environments, yet no direct evidence for thermophilic growth of marine ANME consortia was obtained to date. To study possible thermophilic AOM, we investigated hydrothermally influenced sediment from the Guaymas Basin. In vitro incubations showed activity of sulfate-dependent methane oxidation between 5 and 70 °C with an apparent optimum between 45 and 60 °C. AOM was absent at temperatures ⩾75 °C. Long-term enrichment of AOM was fastest at 50 °C, yielding a 13-fold increase of methane-dependent sulfate reduction within 250 days, equivalent to an apparent doubling time of 68 days. The enrichments were dominated by novel ANME-1 consortia, mostly associated with bacterial partners of the deltaproteobacterial HotSeep-1 cluster, a deeply branching phylogenetic group previously found in a butane-amended 60 °C-enrichment culture of Guaymas sediments. The closest relatives (Desulfurella spp.; Hippea maritima) are moderately thermophilic sulfur reducers. Results indicate that AOM and ANME archaea could be of biogeochemical relevance not only in cold to moderate but also in hot marine habitats.
Keywords: anaerobic methanotrophy, ANME, syntrophy, thermophilic microorganisms
Introduction
Methane, the simplest hydrocarbon, is an atmospheric trace gas of climatic relevance. Vast amounts of methane are formed by anaerobic microbial degradation of organic matter as well as by geothermal processes (Schoell, 1980) and may eventually reach the atmosphere. In the ocean, however, most methane migrating upwards from deep sediment strata is already consumed within the anoxic seafloor at distinct horizons characterized by simultaneous methane and sulfate depletion (Reeburgh, 2007). These horizons are usually populated by archaea (ANME groups) and bacteria of distinct phylogenetic lineages clustering with methanogens and Deltaproteobacteria, respectively (Boetius et al., 2000; Knittel and Boetius, 2009). The ANME archaea are thought to oxidize the methane in principle via a reversal of the reactions of methanogenesis (Zehnder and Brock, 1979; Krüger et al., 2003; Hallam et al., 2004; Scheller et al., 2010). Biochemical studies so far focused on the ANME-associated protein that is closely related to methyl-coenzyme M reductase (Mcr), the nickel enzyme catalyzing the terminal step in methanogenesis (Thauer, 1998; Hallam et al., 2003; Krüger et al., 2003; Scheller et al., 2010). The associated Deltaproteobacteria most likely perform the sulfate reduction, using methane-derived reducing equivalents (Hoehler et al., 1994). Yet, the mechanism of the intercellular transfer of reducing equivalents has not been elucidated, and axenic binary cultures have not been reported to date.
ANME-2 is a prominent phylogenetic archaeal group at most marine cold gas seeps (Orphan et al., 2001; Mills et al., 2003; Wegener et al., 2008b; Knittel and Boetius, 2009) and some sulfate–methane transition zones where the temperature is 4–14 °C (Knittel and Boetius, 2009). Another group, ANME-3, occurs for instance at Haakon Mosby Mud Volcano (Niemann et al., 2006; in situ temperature −1.5 °C) and the Eastern Mediterranean seepages (Omoregie et al., 2008; 14 °C). Cells of both groups form dense consortia with specific bacterial phylotypes clustering with sulfate-reducing Deltaproteobacteria (most often within Desulfosarcinales, Seep-SRB-1a; Schreiber et al., 2010) or relatives of Desulfobulbus (Knittel et al., 2003; Lösekann et al., 2007; Pernthaler et al., 2008; Schreiber et al., 2010), respectively. A third phylogenetic group, ANME-1, is dominant in the microbial mats covering chimney structures at methane seeps in the Black Sea (Michaelis et al., 2002; in situ temperature of ∼10 °C), and in several diffusive methane interfaces (Thomsen et al., 2001; Lanoil et al., 2005; Harrison et al., 2009; Aquilina et al., 2010). The anaerobic oxidation of methane (AOM) habitats with these ANME types exhibit temperatures between −1.5 and 20 °C (Boetius et al., 2009; Supplementary Table 1). First hints of anaerobic methanotrophs thriving at higher temperatures were obtained by identification of 16S rRNA genes of ANME-1 (Teske et al., 2002; Schrenk et al., 2004; Roussel et al., 2008) and ANME-specific core lipids (Schouten et al., 2003) in hydrothermally influenced marine sediments. Also, radiotracer incubation of such sediment indicated thermophilic AOM (Kallmeyer and Boetius, 2004).
Indeed, AOM according to CH4+SO42− → HCO3−+HS−+H2O should in principle be possible also under thermophilic conditions (ΔG with 100 kPa CH4, 0.028 SO42−, 0.03 HCO3− and 0.001 HS− between 25 and 80 °C ranges from −30 to −34 kJ mol−1; for calculations see Materials and methods). Also from an enzymatic point of view, a restriction of AOM only to low temperatures is not expected. Methanogenesis as a metabolically related process (see above) and sulfate reduction with electron donors other than methane have been reported to occur at temperatures up to 100 °C (Jørgensen et al., 1992; Stetter, 1996).
Here, we investigated possible thermophilic AOM and the responsible microorganisms in hydrothermally influenced sediments from the Guaymas Basin, Gulf of California. We combined rate measurements, enrichment cultivation with methane as sole organic substrate, and analyses of molecular markers. Hydrothermal Guaymas Basin sediments are characterized by rapid thermochemical transformation of labile fractions of settled detritus to volatile fatty acids, methane and higher hydrocarbons (Simoneit et al., 1988; Martens, 1990). These compounds act as substrates for diverse microorganisms such as sulfate reducers. The high microbial sulfide production in these sediments fosters patchy mats of sulfide-oxidizing bacteria such as Beggiatoa (Jannasch et al., 1989). From such areas, a variety of sulfate-reducing bacteria (Rueter et al., 1994; Jeanthon et al., 2002), sulfate-reducing archaea (Burggraf et al., 1990; Khelifi et al., 2010) and methanogenic archaea (Kurr et al., 1991) have been isolated.
Materials and methods
Sediment characterization and sampling
Samples from a hydrothermal vent site in the Guaymas Basin were obtained during the RV Atlantis cruise AT15-56 in November/December 2009 with the submersible Alvin (Dive 4570; 27°00.437 N, 111°24.548 W) from bacterial mats (Supplementary Figure 1). Temperature profiles were measured in situ and the retrieved sediment was sectioned into the following horizons: (A) 2–13 cm, 4–30 °C; (B) 14–25 cm, 30–60 °C and (C) 26–45 cm, 60–85 °C (Supplementary Figure 1c). The cored material consisted of unconsolidated, methane-rich clays. The sections were transferred to glass bottles, diluted 1:1 with artificial anoxic seawater medium (Widdel and Bak, 1992) and stored anoxically with a methane headspace until further processing.
Determination of sulfate reduction rates
Sulfate reduction rates were determined by injection of 25 μl 35SO42− tracer (Hartmann Analytics, Braunschweig, Germany) so as to achieve ∼75 kBq per horizon (∼30 cm3) into push cores at intervals of 1 cm and incubation at in situ temperature for 12 to 48 h. Cores were sliced into 1 cm sections and transferred to zinc acetate solution (0.9 ) to stop the reaction and fix sulfide. The total reduced inorganic sulfur was separated from the radiolabeled reactant (35SO42−) by reduction with Cr2+ and cold distillation (Kallmeyer et al., 2004). The radioactivity of both pools was quantified by scintillation counting (scintillation cocktail, Lumasafe Plus (Perkin Elmer, Waltham, MA, USA); scintillation counter, 2900TR LSA (Hewlett Packard, Palo Alto, CA, USA)), and rates were calculated as described (Jørgensen and Fenchel, 1974).
Determination of methane oxidation rates
Methane oxidation to inorganic carbon was determined via radiolabeling of homogeneous sediment slurries prepared from defined proportions of seawater medium and sediment. Samples (2 ml slurry; dry weight 195 mg) were distributed to culture tubes (5 ml) and sealed with butyl rubber stoppers inside an anoxic chamber. The headspace was completely filled with methane-saturated (250 kPa) synthetic seawater, while gas was allowed to escape via an inserted hypodermic needle. Samples were pre-incubated for 5 days at the designated temperatures. Control samples were inactivated with formaldehyde solution (final concentration, 20 g l−1). Upon pre-incubation, carrier-free 14CH4 (15 kBq) dissolved in 50 μl of anoxic water was added per tube. Tubes were further incubated for 48 h. Activity was stopped by alkalization with NaOH. The total methane concentration was determined by gas chromatography using a 5890A instrument (Hewlett Packard) equipped with a Porapak-Q column (6 ft, 0.125 in, 80/100 mesh; Agilent, Santa Clara, CA, USA) and a flame ionization detector, and operated at 40 °C with helium as carrier gas. Radioactivity in methane and inorganic carbon was determined after their separation and conversion to gaseous CO2 as described (Treude et al., 2003). Label in the trapped CO2 fractions was determined by scintillation counting (cocktail Ultima Gold XR, Perkin-Elmer; instrument 2900TR LSA, Hewlett Packard). Rates were calculated as described previously (Treude et al., 2003).
Enrichment of anaerobic methanotrophic microorganisms
Microorganisms were enriched in culture vials (156 ml) at 37 °C (samples from horizon A), 50 °C (horizon B) and 60 °C (horizon B) with synthetic seawater medium (Widdel and Bak, 1992) under a headspace (56 ml) of 250 kPa CH4 and 40 kPa CO2. Controls for each temperature were incubated with N2 and CO2. Vials were incubated at the indicated temperatures on rotary shakers at low speed (50 rpm). Activity was followed by quantification of sulfide using a rapid colorimetric test (Cord-Ruwisch, 1985). The sulfidic supernatant was replaced by fresh medium every 40–70 days.
Determination of the methane oxidation to sulfide production ratio
To measure methane consumption coupled to sulfate reduction (and resulting sulfide production), sediment slurries (50 ml) were added to culture vials (250 ml) inside an anoxic chamber. Bottles were completely filled with CH4-saturated (250 kPa) anoxic seawater medium (see above), incubated at 50 °C, and intensely shaken by hand once per day. Per time point, 1 ml supernatant was withdrawn and replaced with the same volume of sterile medium. The sample was injected into rubber-sealed vials (6 ml) containing 0.5 ml ZnCl2 (50 g l−1) to bind sulfide. Methane was quantified in the headspace by gas chromatography (see above). Sulfide was liberated from ZnS (by acidification) and quantified colorimetrically using the methylene blue-forming reaction in a miniaturized assay (Aeckersberg et al., 1991).
DNA extraction, PCR and clone library construction
DNA was extracted as described by Zhou et al. (1996) from a 9-ml subsample of enrichment cultures incubated for 3 and 9 months. After 9 months, only the supernatant after 1 h settling time was used for the extraction. The protocol encompassed three cycles of freezing and thawing, chemical lysis in a high-salt extraction buffer (1.5 NaCl) by heating of the suspension in the presence of sodium dodecyl sulfate and hexadecyltrimethylammonium bromide, and treatment with proteinase K. The following domain-specific primers were used to amplify almost full-length 16S rRNA genes from the extracted chromosomal DNAs: Bacteria, primers GM3F (Muyzer et al., 1995) and EUB1492R (Kane et al., 1993); Archaea, primers 20F (Massana et al., 1997) and Arc1492R (Teske et al., 2002). In addition, fragments of the gene coding for the α subunit of methyl-coenzyme M reductase (mcrA) were amplified using the primers ME1 and ME2 (Hales et al., 1996).
PCR reactions were performed in a Mastercycler Gradient (Eppendorf, Germany) in a 20-μl reaction volume. Each PCR reaction contained: 0.5 μ of each primer, 200 μ of each deoxyribonucleoside triphosphate, 6 μg bovine serum albumin, 1 × PCR buffer (5Prime, Hamburg, Germany), 1 × PCR Enhancer (5Prime), 0.25 U Taq DNA Polymerase (5Prime) and 5–10 ng of template DNA. The following cycling conditions were applied: one initial step at 95 °C for 5 min; 26 cycles, each at 95 °C for 1 min, 42 °C (58 °C for Arch20F/Arc1492R; 48 °C for ME1/ME2) for 1.5 min, and 72 °C for 3 min; and final step at 60 °C for 60 min. After PCR, the DNA of 10 reactions was pooled, gel extracted and purified by using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. The DNA was ligated to the pGEM-T-Easy vector (Promega, Madison, WI, USA) and transformed into Escherichia coli One Shot Top10 cells (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations. Taq cycle sequencing was performed using ABI BigDye Terminator chemistry and an ABI377 sequencer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysis
The phylogenetic affiliation was inferred with the ARB software package (Ludwig et al., 2004) based on Release 102 of the ARB SILVA database (Pruesse et al., 2007). Comparative sequence analyses were performed with representative sequences from the Guaymas Basin enrichments together with sequences of related archaea (Figure 4a), Deltaproteobacteria (Figure 4b) and methyl-coenzyme M reductases (Figure 4c) found in public databases. In total, 877 (deltaproteobacterial) and 1090 (archaeal) nearly full-length 16S rRNA gene sequences (>1350 bp) were used for tree construction. For mcrA tree construction, 483 sequences (>230 amino acids) were considered. Phylogenetic trees were calculated by maximum likelihood analysis (PhyML, RAxML) and the neighbor-joining algorithm. A 50% base frequency filter was used for 16S rRNA gene tree calculation to exclude highly variable positions. The phylogenetic McrA tree was generated from mcrA-deduced amino-acid sequences using PhyML and RAxML with a 30% amino-acid frequency filter. The resulting trees were compared manually and, if necessary, a consensus tree was constructed. Relevant partial sequences were subsequently added to the tree according to maximum parsimony criteria, without allowing changes in the overall tree topology.
Nucleotide sequences have been deposited at EMBL, GenBank and DDBJ under accession numbers FR682488 to FR682496 (archaeal 16S rRNA genes), FR682813 to FR682818 (mcrA) and FR682644 to FR682663 (bacterial 16S rRNA genes).
Design and application of oligonucleotide probes
Oligonucleotide probes HotSeep-1-590, ANME-1-GI812 and ANME-1-GII186 (Supplementary Table 2) were designed using the ARB probe design tool. Specificity of the probes was evaluated by Clone-FISH (fluorescence in situ hybridization) (Schramm et al., 2002) for probe HotSeep-1-590, or directly on enrichment samples for probes ANME-1-GI812 and ANME-1-GII186. To generate melting curves, the probes were hybridized to clones at formamide concentrations of 0%, 10%, 20%, 30%, 40%, 50%, 60% and 70%. To increase the accessibility of the HotSeep-1-590 probe target site on the 16S rRNA, helper oligonucleotides were designed. In addition, the probes were tested for sensitivity (target group hits) and specificity (outgroup hits) in silico with the ARB probe match tool. Probe HotSeep-1-590 has at least three mismatches to non-target sequences. Probes ANME-1-GI812 and ANME-1-GII186 were designed to discriminate between the two ANME-1 Guaymas subclusters: probe ANME-1-GI812 has two mismatches to cluster II and ANME-1-GII186 has five mismatches to cluster I. Oligonucleotide probes were synthesized by Biomers (Ulm, Germany).
Samples for cell hybridization were fixed with formaldehyde (30 g l−1, final concentration) for 2 h at room temperature, washed with 1 × PBS (phosphate-buffered saline), pH 7.2 and stored in PBS:ethanol (1:1) at −20 °C until further processing. Fixed samples were treated by mild sonication for 20 s with a MS73 probe (Sonoplus HD70, Bandelin, Berlin, Germany) at ⩽10 W.
CARD-FISH (catalyzed reporter deposition fluorescence in situ hybridization) of cells was performed as described previously (Pernthaler et al., 2004) with the following modifications: aliquots of the enrichment samples were filtered onto 0.2 μm pore-size polycarbonate filters (GTTP, Millipore, Billerica, MA, USA). For cell wall permeabilization, filters were sequentially incubated in sodium dodecyl sulfate solution (0.5%) for 10 min and proteinase K solution (15 μg ml−1, 0.1 Tris–HCl, 0.05 EDTA, pH 8, 0.5 NaCl) for 2 min at room temperature. Endogenous peroxidases were inactivated by incubating the filters in 0.01 HCl for 5 min at room temperature. Permeabilization with higher HCl concentrations or lysozyme caused damage of cells and disturbance of their arrangement within the ANME-1/HotSeep-1 sheaths. The oligonucleotide probes were applied with formamide concentrations according to literature data or Clone-FISH results, respectively (Supplementary Table 2). For dual-CARD-FISH, peroxidases of initial hybridizations were inactivated by 30 min incubation in 0.3% H2O2 in methanol at room temperature. Catalyzed reporter deposition was performed using the fluorochromes Alexa Fluor 488 and Alexa Fluor 594. Finally, samples were stained with DAPI (4′,6′-diamidino-2-phenylindole). Micrographs were obtained by confocal laser scanning microscopy (LSM510; Zeiss, Oberkochen, Germany).
Lipid extraction and analyses
Total lipid extracts were retrieved by modified Bligh and Dyer extraction (Sturt et al., 2004). Fractions of core glycerol dialkyl glycerol tetraether (GDGT) were purified by preparative liquid chromatography (ThermoFinnigan Surveyor equipped with preparative LiChrosphere Si60 column, 250 mm × 10 mm × 5 μm, Alltech, Unterhaching, Germany) connected to a Gilson FC204 fraction collector (Gilson, Inc., Middleton, WI, USA). Analysis of core GDGTs followed the described procedure (Huguet et al., 2006) with C46-GDGT as injection standard. Analysis was performed by combined liquid chromatography–quadrupole mass spectrometry (1200 series, Agilent), using a Prevail Cyano column (3 × 150 mm2; Grace, Waukegan, IL, USA). GDGTs were eluted isocratically. Compounds were detected by atmospheric pressure positive ion chemical ionization mass spectrometry.
For isotopic analysis, GDGTs were subjected to ether cleavage using BBr3 (Sigma-Aldrich, St Louis, MO, USA) in dichloromethane (Bradley et al., 2009). Carbon isotopic composition of produced biphytanes was determined using gas chromatography combustion isotope ratio mass spectrometry (gas chromatography: Hewlett Packard 5890 series II equipped with a 30-m TRX-5MS fused silica column; combustion: ThermoFinnigan Combustion Interface-II: mass spectrometry: Finnigan MAT252). Carbon isotopic values are expressed in per mil (‰) deviations from the Pee Dee Belemnite standard. The analytical error was <1‰.
Thermodynamic calculations
The standard free energy change of AOM was calculated from free energies of formation (Stumm and Morgan, 1996). The free energy change for non-standard conditions was calculated using the van t'Hoff equation, concentrations in the medium at the beginning of incubation and activity coefficients from the IUPAC project on ionic strength corrections for stability constants (http://www.iupac.org/web/ins/2000-003-1-500).
Results and discussions
Sediment cores were taken from underneath a bacterial mat (identified as Beggiatoa by microscopy, and in prior analyses by Jannasch et al. (1989); Supplementary Figures 1a and b). The abundance of Beggiatoa was indicative of high rates of sulfate reduction. Sulfate reduction rates determined in replicate cores peaked between 3 and 6 cm with 1500 nmol per day per ml (average throughout the upper 15 cm: 530 nmol per day per ml, corresponding to roughly 0.25 μmol per day per gdw; Supplementary Figure 1d).
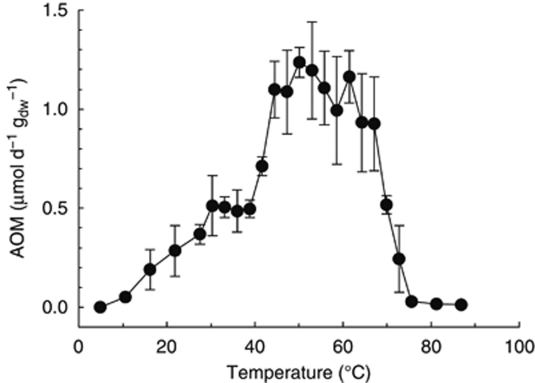
Homogenized material from the three horizons was further diluted, equilibrated with methane (250 kPa) and incubated without headspace in a temperature gradient block (0 to 90 °C; Kallmeyer et al., 2003). After 5 days of pre-incubation, methane oxidation rates were determined using 14C-methane. We measured substantial AOM rates between 4 and 70 °C (Figure 1). Highest rates occurred between 42 and 65 °C, with ∼1.2 μmol per day per gdw. AOM was not observed at ⩾75 °C.
Figure 1.
Rates of AOM in Guaymas Basin sediment at different temperatures measured as 14CH4 conversion to 14CO2. Homogenous samples (2–45 cm sediment depth, in situ temperature, 4–85 °C) were pre-incubated for 5 days at designated temperatures followed by incubation with labeled methane for 48 h. Error bars indicate s.d. from triplicates.
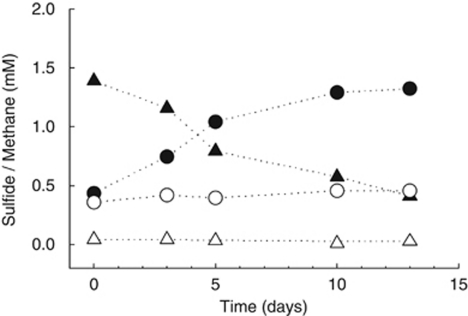
To confirm the stoichiometry of methane oxidation and sulfate reduction in our enrichments, methane consumption and sulfide production were chemically quantified in headspace-free subsamples (2 months after methane had been added to start AOM activity) at 50 °C. Within 2 weeks methane consumption occurred simultaneously with sulfide production (Figure 2). As in previous low-temperature enrichments (Nauhaus et al., 2002; Holler et al., 2009), the ratio between methane consumed and sulfide formed (corrected against the background) was in accordance with the expected stoichiometry of 1 mol CH4 oxidized per mol SO42− reduced. The proportion of methane-derived reducing equivalents channeled into biosynthesis and therefore not available for sulfate reduction was previously shown to be extremely low (around 1% Nauhaus et al., 2007; Wegener et al., 2008a) and can be neglected in evaluating the AOM stoichiometry. In controls without methane, no production of methane (methanogenesis) was detected and sulfide production was only ∼10% of rates in the presence of methane. Hence, these measurements gave clear evidence for thermophilic AOM in the natural enrichment.
Figure 2.
Time course experiment of AOM enrichment incubated without headspace at 50 °C. Sulfide formation (black circles) and methane consumption (black triangles) in the enrichment. A control without methane addition (open triangles, background methane) showed only minor sulfide formation (open circles).
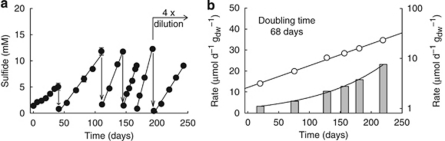
To allow further increase of the biomass and activity of the organisms responsible for thermophilic AOM, horizons A (37 °C) and B (50 and 60 °C) were incubated for 10 months in anoxic synthetic seawater medium with methane as sole organic substrate. To avoid the inhibition of growth by accumulated sulfide (Boetius et al., 2009), the supernatant was replaced by fresh medium at sulfide concentrations of 10 m, which was every 40–70 days (Figure 3a). After each addition of new medium (dry weight-related) sulfide production with methane was faster than in the previous period (Figures 3a and b). Over the total incubation time of 250 days, methane-dependent sulfate reduction rates of the 50 °C culture increased exponentially from 3 to 34 μmol per day per gdw (Figure 3b). This indicated significant increase of active methanotrophic biomass. The apparent doubling time was 68 days. However, within each individual incubation periods, the increase in sulfide appeared constant (‘linear' growth). This indicated that the inhibitory effect of accumulating sulfide and increase in the catalytic microbial biomass counteracted such that the actual activity was constant. The doubling time at 50 °C was shorter than in incubations at 37 °C (77 days) and at 60 °C (112 days) (see Supplementary Figure 2). Thus, we assume an apparent optimum for the growth of thermophilic methanotrophs around 50 °C. In all control experiments without methane, sulfate reduction declined and was below detection limit after 5 months of incubation (data not shown).
Figure 3.
Long-term enrichment of anaerobic methanotrophs at 50 °C. (a) Sulfide formation (black circles) in the initial sediment suspension and upon five exchanges of the sulfide-rich supernatant. Medium exchanges are indicated with arrows. (b) Linear (vertical bars) and semi-logarithmic (open circles) illustration of methane-dependent sulfide production of the different incubation periods over time; normalized by dry weights.
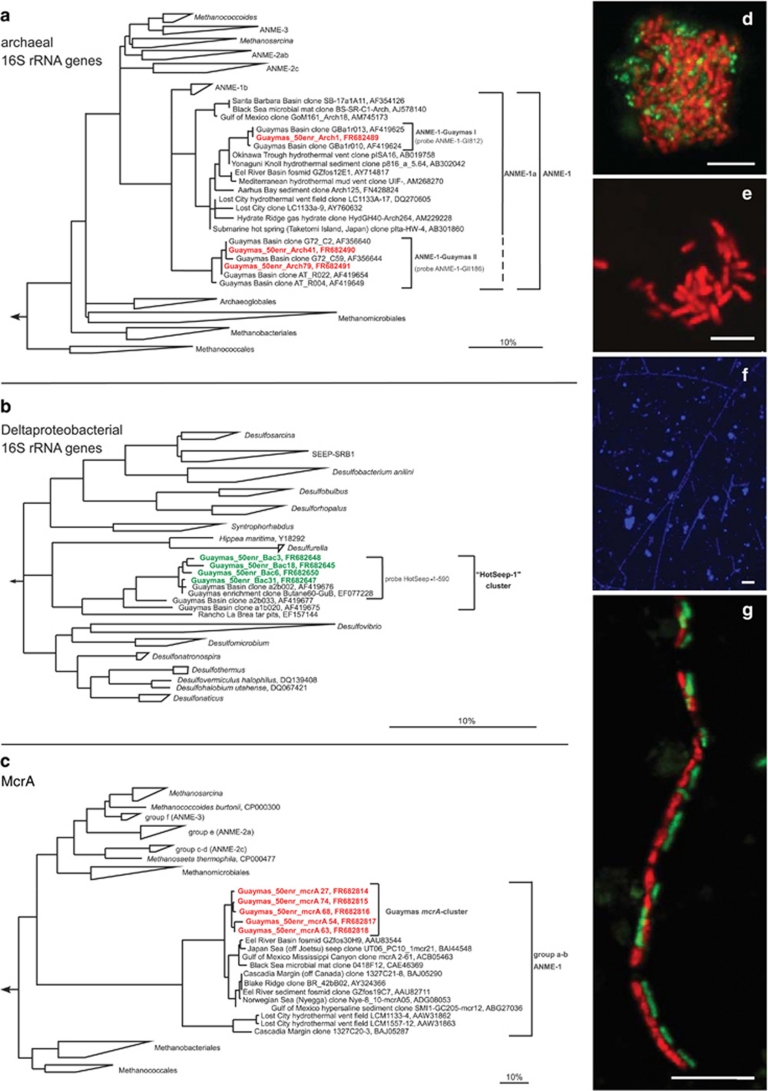
To identify phylotypes responsible for AOM with sulfate at 50 °C, the enrichment was examined by cloning 16S rRNA genes and genes encoding methyl-coenzyme M reductase (Mcr) subunit A (mcrA). The most frequently obtained archaeal 16S rRNA genes and transcribed mcrA genes were affiliated with mcrA of ANME-1 (Figures 4a and c; Table 1), matching sequences previously isolated from Guaymas Basin sediment (Teske et al., 2002). The retrieved ANME-1 16S rRNA gene sequences cluster together with other sequences previously retrieved from Guaymas Basin (Teske et al., 2002; clone G72_C2 and G72_C59, Longnecker and Reysenbach, database release) and form two Guaymas-specific subclusters named ANME-1-Guaymas I and II (Figure 4a; Table 1). Sequence similarity within subcluster ANME-1-Guaymas I ranged between 97.9% and 99.9% and within subcluster Guaymas II between 97.9% and 99.8%. The retrieved mcrA-derived sequences form a separate cluster with at least 6% amino-acid divergence to other sequences within the mcrA groups a–b (Figure 4c). In comparison to mcrA from methanogenic archaea, the present sequence contains a modification (motif VX2CCX4CX5C), which has been previously observed in the abundant nickel protein of ANME-1 consortia in a cold habitat (Krüger et al., 2003). The most frequently retrieved bacterial 16S rRNA clone sequences represent a lineage distantly related to Desulfurella spp., sulfur-reducing thermophilic Deltaproteobacteria (Figure 4b; Table 1). 16S rRNA gene sequences representing this lineage were previously obtained from surficial Guaymas hydrothermal sediments rich in ANME-1, but without detectable ANME-2 (Teske et al., 2002). The same group was found in a heterogeneous sulfate-reducing enrichment culture from Guaymas Basin sediment amended with butane and grown at 60 °C (Kniemeyer et al., 2007). However, the presently obtained AOM enrichment characterized by a high proportion of related 16S rRNA genes did not show detectable sulfate reduction activity with butane instead of methane within an incubation time of 7 weeks. Corresponding to the naming of bacterial seep clusters identified at cold seeps (‘Seep-SRB-1 to -4' and subgroups; Knittel et al., 2003; Schreiber et al., 2010), we named this new group of thermophilic, ANME-associated bacteria ‘HotSeep-1'.
Figure 4.
Phylotypes and cell aggregates in the methane-oxidizing anaerobic-enrichment culture grown at 50 °C. Phylogenetic trees showing the affiliations of 16S rRNA gene sequences retrieved from Guaymas methane-oxidizing enrichments with selected reference sequences of (a) Euryarchaeota and (b) Deltaproteobacteria. Sequences from this study are printed in bold red (archaea) and bold green (bacteria). Probe specificity is indicated with brackets. Bar=10% estimated sequence divergence. (c) Phylogenetic tree of amino acid sequences of the α subunit of the methyl-coenzyme M reductase (mcrA), (d–g) cell aggregates of ANME-1 visualized by CARD-FISH. Scale bars=10 μm. (d, e, g) Confocal laser scanning micrographs. (f) Regular epifluorescence micrograph. (d) Spherical ANME-1/HotSeep-1 aggregates stained with probe ANME-1-350 (red) and probe HotSeep-1-590 (green). (e) Monophyletic ANME-1 aggregate stained with probe ANME-1-350. (f) DAPI staining showing long chain-forming ANME-1 aggregates. (g) Chain-forming ANME-1/HotSeep-1 aggregates. ANME-1 cells were identified as members of subcluster ANME-1-Guaymas I (probe ANME-1-GI812, red) and bacterial partners as members of the HotSeep-1 cluster (probe HotSeep-1-590, green).
Table 1. Numbers of retrieved clones for genes encoding 16S rRNA and McrA (subunit A of methyl-coenzyme M reductase) from a methane-oxidizing anaerobic enrichment culture analyzed after 3 months (enrichment subsample) and 9 months (supernatant subsample) of incubation at 50 °C.
| Phylogenetic affiliation | 16S rRNA (3 months) | 16S rRNA (9 months) | mcrA |
|---|---|---|---|
| Archaea | |||
| Euryarchaeota | |||
| ANME-1 | 64 | ||
| ANME-1-Guaymas I | 19 | ||
| ANME-1-Guaymas II | 27 | ||
| Methanosarcinales | |||
| ANME-2c | 3 | ||
| Others | 1 | ||
| Thermoplasmatales | 6 | ||
| Halobacteriales | 4 | ||
| Total archaeal clones analyzed | 56 | n.a. | 68 |
| Bacteria | |||
| Proteobacteria | |||
| Betaproteobacteria | 3 | ||
| Gammaproteobacteria | 1 | 10 | |
| Deltaproteobacteria | |||
| HotSeep-1-cluster | 53 | 71 | |
| DSS group (Desulfosarcinales) | 1 | ||
| Desulfuromonadales | 1 | ||
| Syntrophobacterales | 1 | ||
| Others | 2 | 12 | |
| Acidobacteria | 1 | ||
| Actinobacteria | 3 | ||
| Bacteriodetes | 1 | ||
| Candidate division OD1 | 5 | 1 | |
| Candidate division OP3 | 3 | 2 | |
| Candidate division OP8 | 8 | 13 | |
| Chloroflexi | 3 | 5 | |
| Deferribacteres | 1 | ||
| Planctomycetes | 2 | ||
| Unaffiliated | 2 | 2 | |
| Total bacterial clones analyzed | 80 | 127 | |
Abbreviations: n.a., not analzyed.
Fluorescence in situ hybridization with specific 16S rRNA targeting oligonucleotide probes (Supplementary Table 2) revealed spherical ANME-1 cell aggregates associated with Deltaproteobacteria of the HotSeep-1 group after 2 months of incubation (Figure 4d). The consortia varied in size and harbored between a few and several hundred cells. ANME-1 cells in these aggregates were identified by the new probe ANME-1-GII186 as members of subgroup ANME-1-Guaymas II. In addition, hybridization showed ANME-1 aggregates without partner bacteria (Figure 4e). ANME-1 cells in these aggregates typically have a length of 1.2 μm and a width of 0.3–0.4 μm. After 5 months, unique chain-forming aggregates with >100 ANME-1 cells appeared (Figures 4f and g) associated in a 1:1 ratio with thinner (0.4 μm in width) rod-shaped bacteria (Figure 4g). The ANME-1 cells in these chain-forming aggregates are larger than those in spherical aggregates with a length of 2–2.5 μm and a width of 0.7 μm. Hybridization with the new probe, ANME-1-GI812, identified chain-forming members of subgroup ANME-1-Guaymas I (Figure 4g). The partner bacteria were attached to the ANME-1 cells, and both cell types were enclosed in a common sheath. We observed further growth of these ANME-1 chains. The longest chains observed after 11 months of incubation had a length of >250 μm. Aggregations of coiled ANME-1 chains have also been detected. Hybridization with probe HotSeep-1-590 showed that these partner bacteria belong as well to the HotSeep-1 cluster. Based on the observation of significant increase of activity, and ANME-1 HotSeep-1 chains in the 50 °C enrichments, we propose that members of the ANME-I Guaymas clades and their partner bacteria of the HotSeep-1 cluster are thermophiles and oxidize methane with sulfate at an apparent temperature optimum of 45–60 °C.
As an independent complementary method for the chemotaxonomic identification of ANME, we analyzed the archaeal GDGT lipids in the enrichment. The intact lipids contained mostly diglycosides, which are abundant in lipids in ANME-1 dominated habitats (Rossel et al., 2011). The GDGT cores were dominated by chains without or with four or five cyclopentane rings. GDGTs with one to three rings, which were previously assigned to thermophilic ANME-1 (Schouten et al., 2003), were significantly depleted in 13C as compared with typical planktonic signals (Table 2). The isoprenoids prepared from the GDGTs, especially mono- and bicyclic biphytane, were depleted in 13C (δ13C, −49‰). This indicates incorporation of methane-derived carbon as shown for lipids of anaerobic methanotrophs (Wegener et al., 2008a). These results are consistent with earlier analyses of hydrothermally influenced Guaymas Basin sediment (Schouten et al., 2003).
Table 2. Concentrations of archaeal glycerol dialkyl glycerol tetraether (GDGT) in the enrichment culture (top) and stable carbon isotopic compositions of GDGT-derived biphytane derivatives obtained by ether cleavage (bottom).
| Tetraethers | Concentration μg gdw−1 | Rel. abundance (%) |
|---|---|---|
| GDGT-0 | 1.90 | 25 |
| GDGT-1 | 0.49 | 6 |
| GDGT-2 | 0.70 | 9 |
| GDGT-3 | 0.47 | 6 |
| GDGT-4 | 1.78 | 23 |
| GDGT-cr. | 2.24 | 29 |
| GDGT-cr. (iso) | 0.07 | 1 |
|
Isoprenoids |
Isotopic composition (δ13C vs. PDB) |
Rel. abundance (%) |
| Acyclic biphytane | −25.2‰ | 40 |
| Monocyclic biphytane | −49.1‰ | 13 |
| Bicyclic biphytane | −41.6‰ | 29 |
| Tricyclic biphytane | −20.0‰ | 18 |
Structures of the GDGTs are shown in Schouten et al. (2003).
Apart from the hot Guaymas Basin sediments and some calcified gas vents in cold to moderately thermophilic habitats such as the Black Sea (Michaelis et al., 2002) and the Lost City Hydrothermal Field (Schrenk et al., 2004), most shallow gas-rich habitats investigated so far were dominated by the methanotrophic clades ANME-2 and ANME-3 (Knittel and Boetius, 2009). Also, all prior cultivation attempts using environmental samples from marine cold seeps selected for these groups (Boetius et al., 2009). Here, propagation of methane oxidation (Figures 1, 2 and 3) as well as comparative sequence analysis, hybridization and membrane lipid information demonstrate that thermophilic AOM up to at least 60 °C is mediated by members of the ANME-1 group. Hence, considering previous findings of ANME sequences and lipids in hot environments (Schouten et al., 2003; Schrenk et al., 2004; Roussel et al., 2008) together with our results, we suggest that AOM could be widespread in hot marine habitats including, for example, hydrothermal crusts, brine lakes and subsurface gas reservoirs. Furthermore, it would be interesting to investigate whether thermophilic ANME groups and their associated bacterial partners may be involved in the widely observed transformation of anhydrite to calcite according to
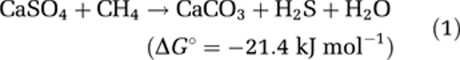 |
in evaporitic cap rocks of moderately heated gas reservoirs (Werner et al., 1988), which would extend the importance of ANME driven methanotrophy in the global carbon cycle and in deep subsurface diagenesis.
Acknowledgments
We are indebted to the crew and pilots of Research Vessel Atlantis and Research Submersible Alvin. We thank Nicole Rödiger, Viola Krukenberg, Philipp Hach, Viola Beier, Gabriele Schüssler, Dennis Enning and Florin Musat for technical support. This study was supported by Grant NSF OCE-0647633 to AT, by the Leibniz program of the Deutsche Forschungsgemeinschaft to AB, the DFG Research Center and Cluster of Excellence MARUM, and the Max Planck Society.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aeckersberg F, Bak F, Widdel F. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch Microbiol. 1991;156:5–14. [Google Scholar]
- Aquilina A, Knab NJ, Knittel K, Kaur G, Geissler A, Kelly SP, et al. Biomarker indicators for anaerobic oxidizers of methane in brackish-marine sediments with diffusive methane fluxes. Org Geochem. 2010;41:414–426. [Google Scholar]
- Boetius A, Holler T, Knittel K, Felden J, Wenzhöfer F.2009The seabed as natural laboratory: lessons from uncultivated methanotrophsIn Epstein SS (ed),Microbiology Monographs: Uncultivated Microorganisms Springer: Berlin; 59–82. [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Bradley AS, Fredricks HF, Hinrichs K-U, Summons RE. Structural diversity of diether lipids in carbonate chimneys at the lost city hydrothermal field. Org Geochem. 2009;40:1169–1178. [Google Scholar]
- Burggraf S, Jannasch HW, Nicolaus B, Stetter KO. Archaeoglobus profundus sp. nov., represents a new species within the sulfate-reducing archaebacteria. Syst Appl Microbiol. 1990;13:24–28. [Google Scholar]
- Cord-Ruwisch R. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods. 1985;4:33–36. [Google Scholar]
- Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl Environ Microbiol. 2003;69:5483–5491. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Harrison BK, Zhang H, Berelson W, Orphan VJ. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California) Appl Environ Microbiol. 2009;75:1487–1499. doi: 10.1128/AEM.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehler TM, Alperin MJ, Albert DB, Martens CS. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem Cycles. 1994;8:45–464. [Google Scholar]
- Holler T, Wegener G, Knittel K, Boetius A, Brunner B, Kuypers MMM, et al. Substantial 13C/12C and D/H fractionation during anaerobic oxidation of methane by marine consortia enriched in vitro. Environ Microbiol Rep. 2009;1:370–376. doi: 10.1111/j.1758-2229.2009.00074.x. [DOI] [PubMed] [Google Scholar]
- Huguet C, Hopmans E, Febo-Ayala W, Thompson D, Damstè JSS, Schouten S. An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org Geochem. 2006;37:1036–1041. [Google Scholar]
- Jannasch HW, Nelson DC, Wirsen CO. Massive natural occurrence of unusually large bacteria (Beggiatoa sp.) at a hydrothermal deep-sea vent site. Nature. 1989;342:834–836. [Google Scholar]
- Jeanthon C, L'Haridon S, Cueff V, Banta A, Reysenbach AL, Prieur D. Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int J Syst Evol Microbiol. 2002;52:765–772. doi: 10.1099/00207713-52-3-765. [DOI] [PubMed] [Google Scholar]
- Jørgensen BB, Fenchel T. The sulfur cycle of a marine sediment model system. Mar Biol. 1974;24:189–201. [Google Scholar]
- Jørgensen BB, Isaksen MF, Jannasch HW. Bacterial sulfate reduction above 100 °C in deep-sea hydrothermal vent sediments. Science. 1992;258:1756–1757. doi: 10.1126/science.258.5089.1756. [DOI] [PubMed] [Google Scholar]
- Kallmeyer J, Boetius A. Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl Environ Microbiol. 2004;70:1231–1233. doi: 10.1128/AEM.70.2.1231-1233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer J, Ferdelman TG, Jansen K-H, Jørgensen BB. A high pressure thermal grandient block for investigating microbial activity in multiple deep-sea samples. J Microbiol Methods. 2003;55:165–172. doi: 10.1016/s0167-7012(03)00138-6. [DOI] [PubMed] [Google Scholar]
- Kallmeyer J, Ferdelman TG, Weber A, Fossing H, Jørgensen BB. A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol Oceanogr Methods. 2004;2:171–180. [Google Scholar]
- Kane MD, Poulsen LK, Stahl DA. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi N, Grossi V, Hamdi M, Dolla A, Tholozan J-L, Ollivier B, et al. Anaerobic oxidation of fatty acids and alkenes by the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus. Appl Environ Microbiol. 2010;76:3057–3060. doi: 10.1128/AEM.02810-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniemeyer O, Musat F, Sievert SM, Knittel K, Wilkes H, Blumenberg M, et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature. 2007;449:898–901. doi: 10.1038/nature06200. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A, Lemke A, Eilers H, Lochte K, Pfannkuche O, et al. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon) Geomicrobiol J. 2003;20:269–294. [Google Scholar]
- Krüger M, Meyerdierks A, Glöckner FO, Amann R, Widdel F, Kube M, et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature. 2003;426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- Kurr M, Huber R, König H, Jannasch HW, Fricke H, Trincone A, et al. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110 °C. Arch Microbiol. 1991;156:239–247. [Google Scholar]
- Lanoil BD, Duc MT, Wright M, Kastner M, Nealson KH, Bartlett D. Archaeal diversity in ODP legacy borehole 892b and associated seawater and sediments of the Cascadia Margin. FEMS Microbiol Ecol. 2005;54:167–177. doi: 10.1016/j.femsec.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Lösekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, et al. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl Environ Microbiol. 2007;73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens CS. Generation of short chain acid anions in hydrothermally altered sediments of the Guaymas Basin, Gulf of California. Appl Geochem. 1990;5:71–76. [Google Scholar]
- Massana R, Murray AE, Preston CM, DeLong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis W, Seifert R, Nauhaus K, Treude T, Thiel V, Blumenberg M, et al. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science. 2002;297:1013–1015. doi: 10.1126/science.1072502. [DOI] [PubMed] [Google Scholar]
- Mills HJ, Hodges C, Wilson K, MacDonald IR, Sobecky PA. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol Ecol. 2003;46:39–52. doi: 10.1016/S0168-6496(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Muyzer G, Teske A, Wirsen CO, Jannasch HW. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- Nauhaus K, Albrecht M, Elvert M, Boetius A, Widdel F. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ Microbiol. 2007;9:187–196. doi: 10.1111/j.1462-2920.2006.01127.x. [DOI] [PubMed] [Google Scholar]
- Nauhaus K, Boetius A, Krüger M, Widdel F. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ Microbiol. 2002;4:296–305. doi: 10.1046/j.1462-2920.2002.00299.x. [DOI] [PubMed] [Google Scholar]
- Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K, et al. Novel microbial communities of the Haakon Mosby Mud Volcano and their role as a methane sink. Nature. 2006;443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- Omoregie EO, Mastalerz V, de Lange G, Straub KL, Kappler A, Røy H, et al. Biogeochemistry and community composition of iron- and sulfur-precipitating microbial mats at the Chefren Mud Volcano (Nile Deep Sea Fan, Eastern Mediterranean) Appl Environ Microbiol. 2008;74:3198–3215. doi: 10.1128/AEM.01751-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R.2004Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganismsIn Kowalchuk GA, Bruijn FJd, Head IM, Akkermans AD, van Elsas JD (eds),Molecular Microbial Ecology Manual2nd edn.Kluwer Academic Publishers: Dordrecht,711–726. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeburgh WS. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- Rossel PE, Elvert M, Ramette A, Boetius A, Hinrichs K-U. Factors controlling the distribution of anaerobic methanotrophic communities in marine environments: evidence from intact polar membrane lipids. Geochim Cosmochim Acta. 2011;75:164–184. [Google Scholar]
- Roussel EG, Bonavita M-AC, Querellou J, Cragg BA, Webster G, Prieur D, et al. Extending the sub-sea-floor biosphere. Science. 2008;320:1046–1056. doi: 10.1126/science.1154545. [DOI] [PubMed] [Google Scholar]
- Rueter P, Rabus R, Wilkes H, Aeckersberg F, Rainey FA, Jannasch HW, et al. Anaerobic oxidation of hydrocarbons in crude oil by new types of sulphate-reducing bacteria. Nature. 1994;372:455–458. doi: 10.1038/372455a0. [DOI] [PubMed] [Google Scholar]
- Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature. 2010;465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- Schoell M. The hydrogen and carbon isotopic composition of methane from natural gases of various origins. Geochim Cosmochim Acta. 1980;44:649–661. [Google Scholar]
- Schouten S, Wakeham SG, Hopmans EC, Damsté JSS. Biogeochemical evidence that thermophilic archaea mediate the anaerobic oxidation of methane. Appl Environ Microbiol. 2003;69:1680–1686. doi: 10.1128/AEM.69.3.1680-1686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol. 2002;4:713–720. doi: 10.1046/j.1462-2920.2002.00364.x. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Holler T, Knittel K, Meyerdierks A, Amann R. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ Microbiol. 2010;12:2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x. [DOI] [PubMed] [Google Scholar]
- Schrenk MO, Kelley DS, Bolton SA, Baross JA. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ Microbiol. 2004;6:1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- Simoneit BRT, Kawka OE, Brault M. Origin of gases and condensates in the Guaymas Basin hydrothermal system (Gulf of California) Chem Geol. 1988;71:169–182. [Google Scholar]
- Stetter KO. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- Stumm W, Morgan JJ.1996Aquatic Chemistry3rd edn.John Wiley & Sons: New York [Google Scholar]
- Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs K-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry -- new biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18:617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- Teske A, Hinrichs K-U, Edgcomb V, de Vera Gomez A, Kysela D, Sylva SP, et al. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl Environ Microbiol. 2002;68:1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiol. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- Thomsen TR, Finster K, Ramsing NB. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl Environ Microbiol. 2001;67:1646–1656. doi: 10.1128/AEM.67.4.1646-1656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treude T, Boetius A, Knittel K, Wallmann K, Jørgensen BB. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar Ecol Prog Ser. 2003;264:1–14. [Google Scholar]
- Wegener G, Niemann H, Elvert M, Hinrichs K-U, Boetius A. Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ Microbiol. 2008a;10:2287–2298. doi: 10.1111/j.1462-2920.2008.01653.x. [DOI] [PubMed] [Google Scholar]
- Wegener G, Shovitri M, Knittel K, Niemann H, Hovland M, Boetius A. Biogeochemical processes and microbial diversity of the Gullfaks and Tommeliten methane seeps (Northern North Sea) Biogeosciences. 2008b;5:1127–1144. [Google Scholar]
- Werner ML, Feldman MD, Knauth LP. Petrography and geochemistry of water-rock interactions in Richton Dome cap rock (southeastern Mississippi, USA) Chem Geol. 1988;74:113–135. [Google Scholar]
- Widdel F, Bak F.1992Gram-negative mesophilic sulfate-reducing bacteriaIn Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds),The Prokaryotes2nd edn.Springer: New York,3352–3378. [Google Scholar]
- Zehnder AJB, Brock TD. Methane formation and methane oxidation by methanogenic bacteria. J Bacteriol. 1979;137:420–432. doi: 10.1128/jb.137.1.420-432.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.