Abstract
Cyclin-dependent protein kinases (CDKs) play key roles in regulating the eukaryotic cell cycle. We have analyzed the expression of four rice (Oryza sativa) CDK genes, cdc2Os1, cdc2Os2, cdc2Os3, and R2, by in situ hybridization of sections of root apices. Transcripts of cdc2Os1, cdc2Os2, and R2 were detected uniformly in the dividing region of the root apex. cdc2Os1 and cdc2Os2 were also expressed in differentiated cells such as those in the sclerenchyma, pericycle, and parenchyma of the central cylinder. By contrast, signals corresponding to transcripts of cdc2Os3 were distributed only in patches in the dividing region. Counterstaining of sections with 4′,6-diamidino-2-phenylindole and double-target in situ hybridization with a probe for histone H4 transcripts revealed that cdc2Os3 transcripts were abundant from the G2 to the M phase, but were less abundant or absent during the S phase. The levels of the Cdc2Os3 protein and its associated histone H1-kinase activity were reduced by treatment of cultured cells with hydroxyurea, which blocks cycling cells at the onset of the S phase. Our results suggest that domains other than the conserved amino acid sequence (the PSTAIRE motif) have important roles in the function of non-PSTAIRE CDKs in distinct cell-cycle phases.
CDKs are Ser/Thr protein kinases involved in the regulation of the eukaryotic cell cycle (for review, see Solomon, 1993; King et al., 1994; Lees, 1995; Morgan, 1995; Pines, 1995). cDNAs encoding CDKs from many organisms have been isolated. A single major CDK has been identified in the fission yeast Schizosaccharomyces pombe (CDC2) and in the budding yeast Saccharomyces cerevisiae (CDC28) (Hindley and Phear, 1984; Lörincz and Reed, 1984). However, the growing list of CDKs in human cells suggests that during development each CDK in metazoans plays a specific role at a specific time in the cell cycle. CDKs are activated by binding of cyclins and phosphorylation (for review, see Morgan, 1995; Fisher, 1997). Each CDK interacts with a specific subset of cyclins, and the size of this subset varies. For example, CDC28 can associate with many different cyclins, whereas human CDC2 interacts with relatively few (for review, see Nigg, 1995). A short, conserved amino acid sequence in CDKs, PSTAIRE, is responsible for the binding of cyclins that activate CDKs by changing the conformation at the catalytic site (Jeffrey et al., 1995; Morgan, 1996); this sequence also functions in the targeting of CDKs to specific substrates or subcellular locations (Hoffmann et al., 1993; Peeper et al., 1993; Dynlacht et al., 1994).
Plants express different kinds of CDKs; multiple genes for CDKs have been found in Arabidopsis (Ferreira et al., 1991; Hirayama et al., 1991), alfalfa (Hirt et al., 1991, 1993), rice (Oryza sativa; Hata, 1991; Hashimoto et al., 1992; Kidou et al., 1994), soybean (Miao et al., 1993), maize (Colasanti et al., 1991), and snapdragon (Fobert et al., 1994). Recently, Fobert et al. (1996) isolated four cdc2-related genes from snapdragon, and Magyar et al. (1997) described four homologs of cdc2 in alfalfa, in addition to cdc2MsA and cdc2MsB, which had been isolated previously (Hirt et al., 1991, 1993). These findings suggest that different sets of CDK/cyclin pairs might regulate the division of plant cells at each stage of the cell cycle, and that division is not controlled by a single major CDK, as it is in the case of yeast (Doerner, 1994; Ferreira et al., 1994; Murray, 1994; Doonan and Fobert, 1997).
A correlation between the abundance of CDK transcripts and the proliferative state of cells was demonstrated in Arabidopsis, maize, and alfalfa (Colasanti et al., 1991; Hirt et al., 1991; Bergounioux et al., 1992; Martinez et al., 1992; Hemerly et al., 1993). It has also been shown, however, that in Arabidopsis, transcripts of cdc2aAt are localized not only in dividing cells but also in differentiated tissues, such as the parenchyma of the vascular cylinder and the pericycle, which contains cells responsible for the formation of lateral roots (Martinez et al., 1992; Hemerly et al., 1993). Moreover, expression of cdc2aAt could be induced without cell division in suspension cultures (Hemerly et al., 1993). These results suggest that at least some cdc2 transcripts might be correlated with the acquisition of the ability to divide rather than with the actual division of cells.
Genes for four different CDKs have been isolated from rice: cdc2Os1, cdc2Os2 (Hashimoto et al., 1992), rcdc2 (designated cdc2Os3 in this report) (Kidou et al., 1994), and R2 (Hata, 1991). cdc2Os1 and cdc2Os2 are homologs of cdc2, and R2 is similar to the gene for the CDK-activating kinase, which is required for activation of CDK by phosphorylation of a conserved Thr residue in the so-called “T” loop (Morgan, 1995; Umeda et al., 1998; Yamaguchi et al., 1999). The deduced amino acid sequence of Cdc2Os3 showed that it is distinct from proteins in the CDC2/CDC28 family (Kidou et al., 1994), whereas cdc2Os1 and cdc2Os2 are closely related to the homologs of cdc2 that have been isolated from various organisms (Hashimoto et al., 1992). The cdc2Os1 gene was able to partially complement a temperature-sensitive mutation in the cdc28 gene in yeast, but cdc2Os2 and R2 were unable to complement the same mutation (Hashimoto et al., 1992).
We analyzed transcript levels of genes for CDKs in rice plants by in situ hybridization and found that cdc2Os3, which has an altered PSTAIRE sequence, was expressed in a cell-cycle-dependent manner, whereas the other two homologs of cdc2 with the conserved PSTAIRE motif were expressed throughout the cell cycle. The transcripts and protein products of cdc2Os3 were abundant from the G2 to the M phase, an indication that the Cdc2Os3 protein might function in mitosis.
MATERIALS AND METHODS
Plant Material
Rice (Oryza sativa L. var. Yamahoushi) seeds were germinated in water, and seedlings were grown in an incubator at 27°C. In general, seedlings were used for in situ hybridization 1 d after germination. Sections of lateral root primordia were prepared from 7-d-old seedlings. For treatment with compounds that interrupt the cell cycle, roots of 1-d-old seedlings after germination were submerged in water that contained 100 mm hydroxyurea or 0.5% (w/v) colchicine for 26 h. Rice cells in suspension culture were maintained in liquid Murashige-Skoog medium (Glab et al., 1994) on a gyratory shaker (80 rpm) at 27°C, and subcultured at weekly intervals. For treatment with compounds that interrupt the cell cycle, cells in a 5-d-old suspension culture were grown in medium that contained 0.05% (w/v) colchicine or 10 mm hydroxyurea for 36 h.
Preparation of Probes
For preparation of specific RNA probes, individual fragments of cDNAs were subcloned into the pBluescript II SK− vector (Stratagene) as described below. A pBluescript plasmid carrying cdc2Os1 cDNA (accession no. X60374) was digested with DraII, and the DraII cDNA fragment was removed to produce a plasmid that contained the 3′-noncoding region (nucleotides 997–1115). A pBluescript plasmid carrying cdc2Os2 cDNA (accession no. X60375) was digested with XhoI, and the XhoI fragment (nucleotides 875–1126) containing the 3′-noncoding region was subcloned into the XhoI site of pBluescript. A pBluescript plasmid carrying cdc2Os3 cDNA (accession no. D64036) was digested with SacII and EcoRI, and the SacII-EcoRI fragment (nucleotides 454–1121) was subcloned into the SacII/EcoRI site of pBluescript. A pBluescript plasmid carrying R2 cDNA (accession no. X58194) was digested with BamHI, and the BamHI cDNA fragment was removed to produce a plasmid that contained the 3′-noncoding region (nucleotides 1421–1764). A cDNA clone encoding histone H4 (accession no. D10397), which had been isolated previously (Uchimiya et al., 1992), was digested with EcoRI and BamHI, and the insert was transferred to the EcoRI/BamHI site of pBluescript.
We used the plasmids to generate sense and antisense RNA probes by transcription from the T7 and T3 promoters of pBluescript II SK−. Digoxigenin- and biotin-labeled probes were generated with a digoxigenin RNA-labeling kit in combination with a digoxigenin RNA-labeling mix and a biotin RNA-labeling mix, respectively (Boehringer Mannheim).
In Situ Hybridization
In situ hybridization was performed as described by Hihara et al. (1996) with minor modifications. Tissues were fixed in a solution of 50% ethanol, 5% acetic acid, and 3.7% formaldehyde. Paraffin blocks were cut at 10 μm for cross-sections and at 5 μm for longitudinal sections. The method for double-target in situ hybridization was essentially the same as that described by Kouchi et al. (1995), but we used biotin-labeled RNA probes instead of fluorescein-labeled probes. The biotin-labeled probe for transcripts of histone H4 was detected with a streptavidin-alkaline phosphatase conjugate (Boehringer Mannheim) in combination with Fast Red TR/Naphthol AS-MX (Sigma), and then the digoxigenin-labeled probe for the cdc2Os3 transcript was detected with digoxigenin-specific antibodies conjugated with alkaline phosphatase (Boehringer Mannheim) in combination with a nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate stock solution (Boehringer Mannheim). Sections were counterstained with DAPI (1 μg mL−1 in 0.05 m Tris-HCl, pH 7.0, and 0.5% Triton X-100) after detection of digoxigenin-labeled probes.
Isolation and Purification of GST-Fusion Proteins
The open reading frames of cdc2Os1, cdc2Os2, and cdc2Os3 were amplified by PCR with primers that included recognition sequences for specific restriction enzymes, BamHI, EcoRI, and EcoRI, respectively, at the amino-terminal and carboxy-terminal ends. After digestion with appropriate enzymes, the amplified fragments were ligated to pGEX vectors (Pharmacia). pGEX-2T was digested with BamHI, pGEX-1λT with EcoRI, and pGEX-1λT with EcoRI, and the fragments were introduced into Escherichia coli BL21 cells. The nucleotide sequences of the amplified fragments were confirmed for each construct. The E. coli cells were grown in a Luria-Bertani medium to an A600 of 0.6 at 27°C, and expression of GST-fusion proteins was induced by the addition of 0.4 mm isopropyl-β-d-galactoside and allowed to continue for 4 h at 27°C. The GST-fusion proteins were purified with glutathione Sepharose 4B (Pharmacia) according to the protocol from the manufacturer.
Immunoblotting with Antibodies against Rice CDKs
Total protein was extracted from rice suspension-cultured cells as described by Magyar et al. (1997). Proteins were fractionated by SDS-PAGE on a 12% polyacrylamide gel and subjected to immunoblotting with an ECL western-blotting detection system (Amersham). Polyclonal antibodies were raised in rabbits against the internal peptides CPEFAKNPTLI and SPDFKNHRIV of Cdc2Os1 and Cdc2Os2, respectively, and against the carboxy-terminal peptide PYFNDVNKELY of Cdc2Os3.
Assay for Histone H1 Kinase
An aliquot of 100 μg of total protein extracted from suspension-cultured cells was incubated with 10 μL of antiserum for 2 h at 4°C, and immune complexes were precipitated with 30 μL of a 50% suspension of protein A-agarose (GIBCO-BRL) for 1 h at 4°C. The immunoprecipitates were washed three times with bead buffer (50 mm Tris-HCl, 5 mm NaF, 250 mm NaCl, 0.1% [w/w] Nonidet P-40, 0.1 mm Na3VO4, 5 mm EDTA, and 5 mm EGTA, pH 7.5) containing 10 μg mL−1 leupeptin and 0.1 mm benzamidine, and once with kinase buffer (50 mm Tris-HCl, 15 mm MgCl2, 5 mm EGTA, and 1 mm DTT, pH 7.8). A phosphorylation reaction was conducted with each immunoprecipitate in kinase buffer that contained 0.5 mg mL−1 histone H1 as a substrate, 0.01 mm ATP, 0.185 MBq of [γ-32P]ATP (167 TBq/mmol, ICN), and 60 μg mL−1 cAMP-dependent protein kinase inhibitor (Sigma). After incubation for 15 min at room temperature, the reaction was stopped by the addition of sample buffer for SDS-PAGE, boiled for 5 min, and loaded onto a 12% polyacrylamide gel. Phosphorylated proteins were detected with an imaging plate scanner (BAS1000, Fuji, Tokyo, Japan).
RESULTS
Expression of cdc2Os1, cdc2Os2, and R2 throughout the Dividing Region of Rice Roots
cDNAs encoding four different CDKs have been isolated from rice, and the corresponding genes have been designated cdc2Os1, cdc2Os2, rcdc2, and R2, respectively (Hata, 1991; Hashimoto et al., 1992; Kidou et al., 1994). In this paper rcdc2 is referred to as cdc2Os3, since it was the third homolog of cdc2 to be identified. To investigate the level of transcripts of these genes, we prepared root sections from rice seedlings and subjected them to in situ hybridization. RNA probes were prepared from cDNAs such that the respective probes were specific for each CDK, as determined by northern analysis (M. Umeda, unpublished data), and labeled with digoxigenin.
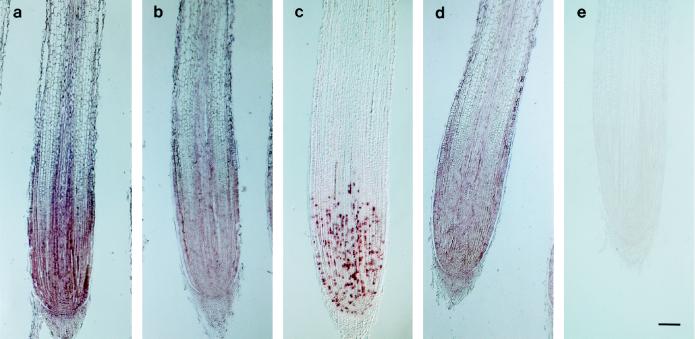
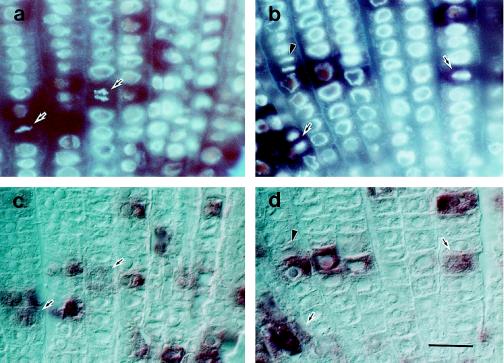
As shown in Figure 1, signals corresponding to transcripts of cdc2Os1, cdc2Os2, and R2 were detected in the root apex. A relatively uniform distribution of signals was observed for the transcripts of these three genes. In the upper parts of the root, no signals were detected in the cortex. For a more detailed investigation, root cross-sections were prepared and allowed to hybridize with each probe. Figure 2a shows the results for cdc2Os1. On sections that included the root cap and the quiescent center, signals were detected in the dividing cells of the root cap but not in the quiescent center or in the differentiated cells of the root cap (section 5). Uniform signals were observed in the dividing region of the root apex (sections 3 and 4). However, in the upper region, cdc2Os1 transcripts were restricted to the sclerenchyma and to inner files of cells that included the pericycle and central cylinder (section 2). No signals were detected in the xylem poles. The same pattern of distribution of transcripts was observed with the probe for transcripts of cdc2Os2, but signals were weaker than those for the cdc2Os1 probe (data not shown). These results suggest that cdc2Os1 and cdc2Os2 were expressed in the sclerenchyma, pericycle, and parenchyma of the central cylinder in the differentiated zone of roots, as well as in the dividing region. The uniform signals due to each transcript indicated that cdc2Os1, cdc2Os2, and R2 were probably expressed throughout the cell cycle. The control sense probe gave no signals on either longitudinal sections or cross-sections (Figs. 1e and 2c).
Figure 1.
In situ hybridization of rice root apices with probes specific for transcripts of cdc2Os1, cdc2Os2, cdc2Os3, and R2. Longitudinal sections of roots were allowed to hybridize with digoxigenin-labeled RNA probes. Hybridization signals are visible as brownish-purple staining. a, cdc2Os1 antisense probe; b, cdc2Os2 antisense probe; c, cdc2Os3 antisense probe; d, R2 antisense probe; and e, cdc2Os1 sense probe. Bar = 100 μm.
Figure 2.
In situ hybridization of cross-sections of rice roots with probes specific for transcripts of cdc2Os1 and cdc2Os3. Cross-sections were prepared from each part of the root, as shown schematically by red shading in the region of dividing cells in the drawing on the left. Numbers indicate positions from which sections were prepared. Pc, Pericycle; Qc, quiescent center; Rc, root cap; Sc, sclerenchyma; Xy, xylem. a, cdc2Os1 antisense probe; b, cdc2Os3 antisense probe; and c, cdc2Os1 sense probe. Bar = 100 μm.
Dependence of Expression of cdc2Os3 Transcripts on the Phase of the Cell Cycle
When the probe was specific for transcripts of cdc2Os3, we observed a patchy pattern of signals in the root apex where the other transcripts had given uniform signals (Fig. 1c). The level of cdc2Os3 transcripts was higher than those of cdc2Os1 and cdc2Os2. A patchy pattern was also apparent in the region near the shoot meristem at the base of the stem (Fig. 3a). Moreover, the primordium for lateral root formation gave a similar patchy pattern in a small number of dividing cells (Fig. 3b). In the actively dividing region, the division of cells was poorly synchronized, so neighboring cells were unlikely to be at the same stage of the cell cycle. Accordingly, these results suggest that cdc2Os3 is expressed at particular phases of the cell cycle.
Figure 3.
In situ hybridization of a region near the shoot meristem and the primordium for formation of lateral roots with a probe specific for transcripts of cdc2Os3. Longitudinal sections were allowed to hybridize with the digoxigenin-labeled cdc2Os3 antisense probe. a, Region near the shoot meristem. b, Primordium arising from the pericycle in the initial stages of formation of a lateral root. Bars = 100 μm.
Cross-sections of roots also gave patchy patterns of cdc2Os3 signals throughout the dividing region of the root apex (Fig. 2b, sections 3–5); however, when we investigated the differentiated cells in the upper region, we found almost no signals on cross-sections (Fig. 2b, sections 1 and 2). Therefore, it is likely that transcripts of cdc2Os3 were restricted to dividing cells of roots and were not expressed in the differentiated cells. By contrast, cdc2Os1 and cdc2Os2 were expressed in several differentiated tissues as well as in dividing regions (Fig. 2a).
Detection of Abundant Transcripts of cdc2Os3 from the G2 to the M Phase of the Cell Cycle
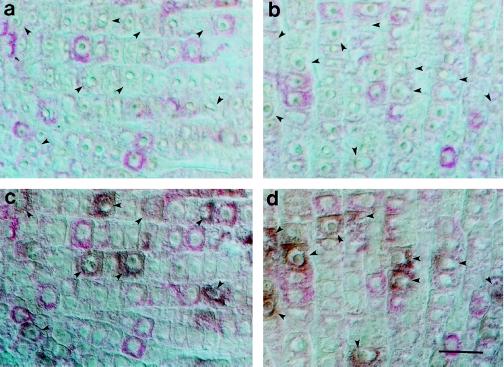
To identify the stage of the cell cycle at which cdc2Os3 was expressed, we counterstained sections with the DNA-specific dye, DAPI (Nacalai, Kyoto, Japan), which reveals the extent of condensation of nuclei. As shown in Figure 4, mitotic cells with condensed nuclei were usually positive for cdc2Os3 transcripts, although the levels of the transcript seemed to depend on the stage of mitosis. When 200 metaphase cells with condensed chromosomes at the equatorial plane were counted for signals corresponding to cdc2Os3 transcripts, almost all of the cells (99%) contained significant amounts. Similarly, almost all of the anaphase cells (99%) with two daughter chromosomes also contained the transcripts, but signals were weaker than those in metaphase cells. Moreover, signals from cells that were forming cell plates were much less intense than signals from cells at the early stage of mitosis (Fig. 4). These results suggest that expression of cdc2Os3 might extend to the M phase and end with the completion of mitosis. By contrast, about 90% of the cells that expressed cdc2Os3 transcripts did not contain condensed nuclei, an indication that cdc2Os3 was also expressed during the preceding G2 phase and the prophase of mitosis.
Figure 4.
Correlation between expressions of cdc2Os3 and mitosis. Sections of root apex were probed with the digoxigenin-labeled cdc2Os3 antisense probe and then counterstained with DAPI. Images were viewed by epifluorescence (a and b) and bright-field (c and d) microscopy. Arrows indicate mitotic cells with condensed nuclei, and arrowheads indicate cells forming cell plates. Bar = 20 μm.
To determine whether cdc2Os3 was expressed during the S phase, we performed double-labeling experiments in which each section was hybridized with probes specific for transcripts of a gene for histone H4 and cdc2Os3. The probe for histone H4 transcripts was labeled with biotin and positive signals were recognized as having a red coloration; the cdc2Os3 probe was labeled with digoxigenin, and positive signals were recognized as having a brownish-purple coloration. Figure 5 shows examples of sections after color development. We allowed many sections to hybridize with both probes. Almost all cells (99%) with signals corresponding to cdc2Os3 transcripts were negative for histone H4 transcripts, suggesting that the timing of the expression of the two genes did not overlap or overlapped for only a very short period of the cell cycle. Our results indicated that transcripts of cdc2Os3 were abundant from the G2 to the M phase but not during the S phase.
Figure 5.
Double-labeling of the root apex for transcripts of cdc2Os3 and of a gene for histone H4. a and b, Detection of hybridization signals with the biotin-labeled antisense probe for histone H4 transcripts (red). c and d, Same section after detection of the digoxigenin-labeled cdc2Os3 probe (brownish-purple). Arrowheads indicate cells labeled with the cdc2Os3 antisense probes. Bar = 20 μm.
Differential Expression of cdc2Os3 in Response to Compounds That Interrupt the Cell Cycle
We analyzed the transcription regulation of cdc2Os3 in further detail with compounds that interrupt the cell cycle. One-day-old seedlings were transferred to water containing hydroxyurea, which blocks cycling cells at the onset of the S phase, or containing colchicine, which inhibits the formation of spindle fibers and blocks cells in mitosis. After treatment for 26 h, root sections were prepared and subjected to in situ hybridization. Roots treated with hydroxyurea showed no signals in the region of dividing cells (Fig. 6b) where a patchy pattern had been observed on sections of untreated seedlings (Fig. 6a). By contrast, after treatment with colchicine, cdc2Os3 transcripts were still detected at the root tips (Fig. 6c). Since colchicine caused radial expansion close to the root tip and promoted the vacuolation of cells, the signals corresponding to cdc2Os3 transcripts were restricted to a small region in the root apex. These results support the hypothesis that cdc2Os3 is expressed from the G2 to the M phase but is not expressed during the transition from the G1 to the S phase.
Figure 6.
In situ hybridization of root apices after treatment with cell-cycle blockers. Longitudinal sections of root apices were prepared from seedlings that had been treated with hydroxyurea or colchicine, and probed with the digoxigenin-labeled cdc2Os3 antisense probe. a, Control root (not treated with blockers of the cell cycle); b, root treated with hydroxyurea; and c, root treated with colchicine. Details of treatments are given in the text. Bar = 100 μm.
To analyze expression at the protein level, we raised polyclonal antibodies in rabbits against peptides specific to each cdc2 homolog. Since the Cdc2Os1-specific antibodies produced a high background on immunoblots, we chose antibodies against Cdc2Os2 and Cdc2Os3 for further analysis. When we used recombinant Cdc2 proteins fused to GST for immunoblotting, the preparations of antibodies specifically recognized GST-Cdc2Os2 and GST-Cdc2Os3, respectively (Fig. 7a). Neither preparation of antibodies cross-reacted with GST (data not shown). Therefore, we used these antibodies to investigate the differential expression of Cdc2Os2 and Cdc2Os3 proteins.
Figure 7.
Changes in the level of the Cdc2Os3 protein in response to cell-cycle blockers. a, Specific cross-reactions of the GST-Cdc2Os2 and GST-Cdc2Os3 fusion proteins with antibodies. One microgram of each GST-fusion protein was subjected to immunoblotting with antibodies against Cdc2Os2 or Cdc2Os3. Lanes 1, GST-Cdc2Os1; lanes 2, GST-Cdc2Os2; and lanes 3, GST-Cdc2Os3. b, Immunological detection of Cdc2Os2 (p40) and Cdc2Os3 (p36) in suspension-cultured rice cells that had been treated with cell-cycle blockers. Total protein was extracted from cultured cells after no treatment (NT) and after treatment with colchicine (CO) or hydroxyurea (HU). Twenty micrograms of each sample was subjected to immunoblotting with Cdc2Os2- or Cdc2Os3-specific antibodies. c, Histone H1-kinase activities in the immunoprecipitates obtained with the antibodies. Total protein was extracted from cultured cells as described above and used for immunoprecipitation with Cdc2Os2- or Cdc2Os3-specific antibodies. Immunoprecipitates were subjected to the histone H1-kinase assay, and phosphorylated histone H1 was detected.
Rice cells in suspension culture were treated with hydroxyurea or colchicine, and total protein was extracted from cells and subjected to immunoblotting. As shown in Figure 7b, antibodies against Cdc2Os2 and Cdc2Os3 immunoreacted with proteins of 40 and 36 kD, respectively. The level of Cdc2Os2 (p40) protein was unchanged after treatment with the two compounds (Fig. 7b). By contrast, hydroxyurea, which arrests cells at the onset of the S phase, reduced the level of Cdc2Os3 (p36). Colchicine had no effect on the level of Cdc2Os3 protein (Fig. 7b). Next we immunoprecipitated endogenous Cdc2 proteins with the antibodies, and subjected the immunoprecipitates to kinase assays with histone H1 as the substrate. Although the kinase activity associated with Cdc2Os2 was detected in both samples, Cdc2Os3-specific antibodies allowed recovery of histone H1-kinase activity from colchicine-treated cells but not from hydroxyurea-treated cells (Fig. 7c). These results suggest that the levels of Cdc2Os3 protein and its associated histone H1-kinase activity were high in G2/ M-arrested cells but low at the entry to the S phase.
DISCUSSION
Although synchronization of rice cells in suspension culture has been reported (Ohtsubo et al., 1993; Sauter, 1997), it is difficult to investigate phase-specific expression of genes in such cells because of low efficiency of synchronization. Sauter (1997) partially synchronized rice cells in suspension culture and performed northern hybridizations with some rice cdc2 genes as probes. Levels of cdc2Os2 and R2 transcripts were slightly elevated after the release from a hydroxyurea block, but the changes did not prove unequivocally that cdc2Os2 and R2 were expressed in a G1/S-phase-specific manner (Sauter, 1997). Moreover, the mitotic index of the cell culture was below 5% (Sauter, 1997). We analyzed the expression of rice genes for CDKs by in situ hybridization of root sections, and found that transcripts of cdc2Os1, cdc2Os2, and R2 were uniformly detectable in dividing cells of roots. Thus, accumulation of cdc2Os1, cdc2Os2, and R2 transcripts was not strictly related to particular stages of the cell cycle, although the levels might change slightly during the cell cycle. Our results indicate that in situ hybridization is a powerful tool for studies of the cell cycle in rice plants, as it is in snapdragon (Fobert et al., 1994, 1996) and soybean (Kouchi et al., 1995).
We found that cdc2Os1 and cdc2Os2 were expressed also in the sclerenchyma, pericycle, and parenchyma of the central cylinder in the differentiated zone of roots. In parts of Arabidopsis roots beyond the apical meristem, expression of cdc2aAt is restricted to the parenchyma of the vascular cylinder and to the pericycle cells (Martinez et al., 1992; Hemerly et al., 1993). Rice cdc2Os1 and cdc2Os2 are closely related to cdc2aAt of Arabidopsis at the amino acid level (Fig. 8), and may also be correlated with the ability of these cells to divide (Hemerly et al., 1993). The pericycle is a differentiated tissue, but retains the potential to divide and is responsible for lateral root formation. The expression of cdc2Os1 and cdc2Os2 in the parenchyma of the central cylinder suggests that this cell layer might engage in some mitotic activity that contributes to the thickening of primary roots in rice plants. The rice-specific expression of these two cdc2 genes in the sclerenchyma remains to be investigated.
Figure 8.
Phylogenetic tree for members of the CDK protein family. The tree was constructed using the CLUSTAL software program (Higgins et al., 1992), with sequences selected from the databases. Rice CDKs are boxed. Amcdc2a-d, CDKs of snapdragon; Cdc2aAt and Cdc2bAt, CDKs of Arabidopsis; Cdc2MsA-F, CDKs of alfalfa; NtCdc2, CDK of tobacco; ZmCdc2, CDK of maize; ScCdc28, Cdc28 of S. cerevisiae; SpCdc2, Cdc2 of S. pombe; Cdk2–7, human CDKs.
In contrast to transcripts of cdc2Os1, cdc2Os2, and R2, transcripts of cdc2Os3 were distributed with a patchy pattern in the dividing region of the root apex. Such a pattern was also observed in the region near the shoot meristem and in the primordia for lateral root formation. Counterstaining of sections with DAPI indicated that almost all of the cells with mitotic nuclei contained cdc2Os3 transcripts, while cells forming cell plates had trace levels of transcripts. In double-labeling experiments with probes specific for transcripts of a gene for histone H4 and cdc2Os3, signals did not overlap, an indication that expression of cdc2Os3 does not extend to the S phase. Furthermore, treatment of seedlings with hydroxyurea, which blocks cells in the early S phase, inhibited the patchy expression of cdc2Os3 at the root apex, whereas transcripts were still detectable in roots treated with colchicine, which blocks cells in mitosis. The patchy pattern on colchicine-treated sections may have reflected the partial synchrony of cell division in this case.
Transcripts of cdc2Os3 appeared to be abundant from the G2 to the M phase but almost disappeared when cells had completed mitosis at telophase. However, we cannot exclude the possibility that cdc2Os3 might be expressed more than once during the cell cycle but discontinuously. Transcripts of cdc2Os3 were detected in the dividing region of the root, whereas the expression was observed in a wide region near the shoot meristem. More detailed experiments are required to investigate whether cdc2Os3 is specifically expressed in dividing cells.
Both cdc2Os1 and cdc2Os2 include the characteristic PSTAIRE domain (Hashimoto et al., 1992) and are classified as PSTAIRE CDKs on the phylogenetic tree (Fig. 8). PSTAIRE CDKs, such as the products of cdc2aAt in Arabidopsis, Amcdc2a and Amcdc2b in snapdragon, and cdc2MsA and cdc2MsB in alfalfa are expressed throughout the cell cycle (Martinez et al., 1992; Fobert et al., 1996; Magyar et al., 1997). Therefore, the products of cdc2Os1 and cdc2Os2 can also be classified as PSTAIRE CDKs in terms of their pattern of expression. Several plant CDKs with altered PSTAIRE motifs have been reported: the products of Amcdc2c and Amcdc2d in snapdragon (Fobert et al., 1996); cdc2MsC, cdc2MsD, cdc2MsE, and cdc2MsF in alfalfa (Magyar et al., 1997); and cdc2bAt in Arabidopsis (Imajuku et al., 1992). Rice cdc2Os3 encodes a PPTALRE sequence that is the same as those of AmCdc2c, Cdc2MsD, and Cdc2bAt (Table I). However, when Cdc2Os3 was compared with the other non-PSTAIRE CDKs in the whole region, it was close to Amcdc2d and cdc2MsF, rather than to AmCdc2c, Cdc2MsD, or Cdc2bAt (Fig. 8).
Table I.
Amino acid sequences in the PSTAIRE region of planta non-PSTAIRE CDKs
| CDKs expressing during the G2-to-M phase | |
| Cdc2Os3 | EGVPPTALREVSLLRM |
| AmCdc2d | EGVPPTTLREVSLLRM |
| Cdc2MsF | EGVPPTTLREVSILRM |
| CDKs expressing during the S-to-M phase | |
| AmCdc2c | EGVPPTALREVSLLQM |
| Cdc2MsD | EGVPPTALREVSLLQM |
| Cdc2bAt | EGIPPTALREISLLQM |
| CDKs expressing throughout the cell cycle | |
| R2 | EGVNFTALREIKLLKE |
| Cdc2MsC | EGFPITAIREILILKK |
| Cdc2MsE | DGVSPTAIREIMLLRE |
Amino acids that are conserved in the PSTAIRE sequence are shown in bold type. Amcdc2c-d, CDKs of snapdragon; Cdc2bAt, CDK of Arabidopsis; Cdc2MsC-F, CDKs of alfalfa.
Transcripts of cdc2s belonging to the group including cdc2Os3 are abundant from the G2 to the M phase (this study; Fobert et al., 1996; Magyar et al., 1997). On the other hand, Amcdc2c is expressed from the mid-S phase to the early-M phase (Fobert et al., 1996), and cdc2bAt is preferentially expressed during the S and G2 phases (Segers et al., 1996). Transcripts of cdc2MsD are abundant at the G2/M phase and are also detected just after alfalfa cells are released from arrest by aphidicolin (Magyar et al., 1997). Accordingly, we propose that the products of cdc2Os3, Amcdc2d, and cdc2MsF form a distinct subclass of non-PSTAIRE CDKs that are preferentially expressed from the G2 to the M phase. Our results also indicate that domains other than the PSTAIRE region are important for the function of non-PSTAIRE CDKs in distinct cell-cycle phases. The alfalfa genes cdc2MsC and cdc2MsE and the rice gene R2, which encode divergent PSTAIRE sequences, are expressed throughout the cell cycle, and the encoded amino acid sequences are distinct from those of other CDKs (Table I; Fig. 8; Magyar et al., 1997).
The level of the Cdc2Os3 protein (p36) was reduced by treatment of cultured cells with hydroxyurea but not with colchicine. The histone H1-kinase activity associated with Cdc2Os3 was correlated with the level of the protein. However, small amounts of Cdc2Os3 protein (p36) were still detectable in hydroxyurea-treated cells. We do not know whether Cdc2Os3 protein is actually present at the onset of the S phase or if the partial synchronization allowed detection of a small amount of the protein. Nevertheless, it seems that the level of Cdc2Os3 fluctuates during the cell cycle according to the level of the transcript. It is likely that expression of cdc2Os3 is controlled at the transcriptional level and that the Cdc2Os3 protein accumulates from the G2 to the M phase.
Yeast cdc2/cdc28 mutants have been rescued by the overexpression of plant genes for PSTAIRE CDKs, such as cdc2aAt of Arabidopsis (Ferreira et al., 1991; Hirayama et al., 1991), cdc2Os1 of rice (Hashimoto et al., 1992), cdc2MsA and cdc2MsB of alfalfa (Hirt et al., 1991, 1993), Amcdc2a and Amcdc2b of snapdragon (Fobert et al., 1996), cdc2ZmA of maize (Colasanti et al., 1991), and cdc2-S5 and cdc2-S6 of soybean (Miao et al., 1993). No plant CDK with an altered PSTAIRE sequence was able to rescue such yeast mutants. We overexpressed the rice cdc2Os3 gene in a mutant of Schizosaccharomyces pombe, cdc2–33 (Carr et al., 1989), and several mutants of Saccharomyces cerevisiae, such as cdc28–1N (Surana et al., 1991), cdc28–4, and cdc28–13 (Reed, 1980). However, the temperature sensitivity of each strain was not rescued by the overexpression of cdc2Os3, which was expressed from either a single-copy or a multicopy vector (data not shown). Therefore, non-PSTAIRE CDKs might have distinct functions in the cell cycle and form active complexes with particular cyclins specific to plants. Indeed, plants have several specific subclasses of cyclins (for review, see Renaudin et al., 1996), and it is likely that a mitotic cyclin that is expressed preferentially from the G2 to the M phase controls the activity of Cdc2Os3.
ACKNOWLEDGMENTS
The authors thank Dr. Hiroshi Kouchi for his helpful suggestions related to in situ hybridization. We also thank Dr. Shingo Hata for providing us with the R2 cDNA.
Abbreviations:
- CDK
cyclin-dependent protein kinase
- DAPI
4′,6-diamidino-2-phenylindole
- GST
glutathione S-transferase
Footnotes
This work was supported by a grant-in-aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan (to M.U., H.U.), and by a grant from the Rockefeller Foundation (to H.U.).
LITERATURE CITED
- Bergounioux C, Perennes C, Hemerly AS, Qin LX, Sarda C, Inzé D, Gadal P. A cdc2 gene of Petunia hybrida is differentially expressed in leaves, protoplasts and during various cell cycle phases. Plant Mol Biol. 1992;20:1121–1130. doi: 10.1007/BF00028898. [DOI] [PubMed] [Google Scholar]
- Carr AM, MacNeill SA, Hayles J, Nurse P. Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 protein kinase gene: implications for cdc2+ protein structure and function. Mol Gen Genet. 1989;218:41–49. doi: 10.1007/BF00330563. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Tyers M, Sundaresan V. Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc Natl Acad Sci USA. 1991;88:3377–3381. doi: 10.1073/pnas.88.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner PW. Cell cycle regulation in plants. Plant Physiol. 1994;106:823–827. doi: 10.1104/pp.106.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J, Fobert P. Conserved and novel regulators of the plant cell cycle. Curr Opin Cell Biol. 1997;9:824–830. doi: 10.1016/s0955-0674(97)80083-x. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F trans-activation by cyclin/cdk2 complexes. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- Ferreira P, Hemerly AS, Van Montagu M, Inzé D. Control of cell proliferation during plant development. Plant Mol Biol. 1994;26:1289–1303. doi: 10.1007/BF00016475. [DOI] [PubMed] [Google Scholar]
- Ferreira P, Hemerly AS, Villarroel R, Van Montagu M, Inzé D. The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell. 1991;3:531–540. doi: 10.1105/tpc.3.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP. CDKs and cyclins in transition(s) Curr Opin Genet Dev. 1997;7:32–38. doi: 10.1016/s0959-437x(97)80106-2. [DOI] [PubMed] [Google Scholar]
- Fobert PR, Coen ES, Murphy GJP, Doonan JH. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fobert PR, Gaudin V, Lunness P, Coen ES, Doonan JH. Distinct classes of cdc2-related genes are differentially expressed during the cell division cycle in plants. Plant Cell. 1996;8:1465–1476. doi: 10.1105/tpc.8.9.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glab N, Labidi B, Qin L-X, Trehin C, Bergounioux C, Meijer L. Olomoucine, an inhibitor of the cdc2/cdk2 kinases activity, blocks plant cells at the G1 to S and G2 to M cell cycle transitions. FEBS Lett. 1994;353:207–211. doi: 10.1016/0014-5793(94)01035-8. [DOI] [PubMed] [Google Scholar]
- Hashimoto J, Hirabayashi T, Hayano Y, Hata S, Ohashi Y, Suzuka I, Utsugi T, Toh-E A, Kikuchi Y. Isolation and characterization of cDNA clones encoding cdc2 homologues from Oryza sativa: a functional homologue and cognate variants. Mol Gen Genet. 1992;233:10–16. doi: 10.1007/BF00587555. [DOI] [PubMed] [Google Scholar]
- Hata S. cDNA cloning of a novel cdc2+/CDC28-related protein kinase from rice. FEBS Lett. 1991;279:149–152. doi: 10.1016/0014-5793(91)80271-4. [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. CABIOS. 1992;2:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hihara Y, Hara C, Uchimiya H. Isolation and characterization of two cDNA clones for mRNAs that are abundantly expressed in immature anthers of rice (Oryza sativa L.) Plant Mol Biol. 1996;30:1181–1193. doi: 10.1007/BF00019551. [DOI] [PubMed] [Google Scholar]
- Hindley J, Phear GA. Sequence of the cell division gene CDC2 from Schizosaccharomyces pombe: patterns of splicing and homology to protein kinases. Gene. 1984;31:129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Imajuku Y, Anai T, Matsui M, Oka A. Identification of two cell-cycle-controlling cdc2 gene homologs in Arabidopsis thaliana. Gene. 1991;105:159–165. doi: 10.1016/0378-1119(91)90146-3. [DOI] [PubMed] [Google Scholar]
- Hirt H, Páy A, Bögre L, Meskiene I, Heberle-Bors E. cdc2MsB, a cognate cdc2 gene from alfalfa, complements the G1/S but not the G2/M transition of budding yeast cdc28 mutants. Plant J. 1993;4:61–69. doi: 10.1046/j.1365-313x.1993.04010061.x. [DOI] [PubMed] [Google Scholar]
- Hirt H, Páy A, Györgyey J, Bakó L, Németh K, Bögre L, Schweyen RJ, Heberle-Bors E, Dudits D. Complementation of a yeast cell cycle mutant by an alfalfa cDNA encoding a protein kinase homologous to p34cdc2. Proc Natl Acad Sci USA. 1991;88:1636–1640. doi: 10.1073/pnas.88.5.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 1993;12:53–63. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imajuku Y, Hirayama T, Endoh H, Oka A. Exon-intron organization of the Arabidopsis thaliana protein kinase genes CDC2a and CDC2b. FEBS Lett. 1992;304:73–77. doi: 10.1016/0014-5793(92)80592-5. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Kidou S, Umeda M, Uchimiya H. Nucleotide sequence of rice (Oryza sativa L.) cDNA homologous to cdc2 gene. DNA Sequence. 1994;5:125–129. doi: 10.3109/10425179409039714. [DOI] [PubMed] [Google Scholar]
- King RW, Jackson PK, Kirschner MW. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Sekine M, Hata S. Distinct classes of mitotic cyclins are differentially expressed in the soybean shoot apex during the cell cycle. Plant Cell. 1995;7:1143–1155. doi: 10.1105/tpc.7.8.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees E. Cyclin dependent kinase regulation. Curr Opin Cell Biol. 1995;7:773–780. doi: 10.1016/0955-0674(95)80060-3. [DOI] [PubMed] [Google Scholar]
- Lörincz AT, Reed SI. Primary structure homology between the product of yeast cell division control gene CDC28 and vertebrate oncogenes. Nature. 1984;307:183–185. doi: 10.1038/307183a0. [DOI] [PubMed] [Google Scholar]
- Magyar Z, Mészáros T, Miskolczi P, Deák M, Fehér A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M and others. Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell. 1997;9:223–235. doi: 10.1105/tpc.9.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MC, Jørgensen J-E, Lawton MA, Lamb CJ, Doerner PW. Spatial pattern of cdc2 expression in relation to meristem activity and cell proliferation during plant development. Proc Natl Acad Sci USA. 1992;89:7360–7364. doi: 10.1073/pnas.89.16.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G-H, Hong Z, Verma DPS. Two functional soybean genes encoding p34cdc2 protein kinases are regulated by different plant developmental pathways. Proc Natl Acad Sci USA. 1993;90:943–947. doi: 10.1073/pnas.90.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Morgan DO. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–772. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- Murray JAH. Plant cell division: the beginning of START. Plant Mol Biol. 1994;26:1–3. doi: 10.1007/BF00039513. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Ohtsubo N, Nakayama T, Terada R, Shimamoto K, Iwabuchi M. Proximal promoter region of the wheat histone H3 gene confers S phase-specific gene expression in transformed rice cells. Plant Mol Biol. 1993;23:553–565. doi: 10.1007/BF00019303. [DOI] [PubMed] [Google Scholar]
- Peeper DS, Parker LL, Ewen ME, Toebes M, Hall FL, Xu M, Zantema A, van der Eb AJ, Piwnica-Worms H. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SI. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J-P, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, Jacobs T, Kouchi H, Rouzé P, Sauter M and others. Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- Sauter M. Differential expression of a CAK (cdc2-activating kinase)-like protein kinase, cyclins and cdc2 genes from rice during the cell cycle and in response to gibberellin. Plant J. 1997;11:181–190. doi: 10.1046/j.1365-313x.1997.11020181.x. [DOI] [PubMed] [Google Scholar]
- Segers G, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, Van Montagu M, Inzé D. The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 1996;10:601–612. doi: 10.1046/j.1365-313x.1996.10040601.x. [DOI] [PubMed] [Google Scholar]
- Solomon MJ. Activation of the various cyclin/cdc2 protein kinases. Curr Opin Cell Biol. 1993;5:180–186. doi: 10.1016/0955-0674(93)90100-5. [DOI] [PubMed] [Google Scholar]
- Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Uchimiya H, Kidou S, Shimazaki T, Aotsuka S, Takamatsu S, Nishi R, Hashimoto H, Matsubayashi Y, Kidou N, Umeda M and others. ) Plant J. 1992;2:1005–1009. [Google Scholar]
- Umeda M, Bhalerao RP, Schell J, Uchimiya H, Koncz C. Proc Natl Acad Sci USA. 1998;95:5021–5026. doi: 10.1073/pnas.95.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Umeda M, Uchimiya H (1999) A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J (in press) [DOI] [PubMed]