Abstract
Histone deacetylases (HDACs) are a family of enzymes which regulate the acetylation state of nucleosomal histones, as well as non-histone proteins. By altering local chromatin architecture, HDACs play important roles in shaping cell differentiation and morphogenesis. Expression of class I HDACs during early chick development has so far not been analyzed. Here, we report the expression profile of chick class I HDACs from the onset of gastrulation (HH2) to day 4 of development and compare it to relevant stages during mouse development. Visualized by in situ hybridization to whole mount embryos and tissue sections, we found tissue-specific overlapping temporal and spatial expression domains for all four class I HDACs in chick and mouse, although species-specific differences could be identified. All class I HDACs in both species are highly expressed in the developing brain. In particular, HDAC1 is expressed at sites of anterior and posterior neural tube closure most obvious in the hot spot-like expression of HDAC1 in HH12 chicken embryos. A significant species-specific spatio-temporal expression pattern was observed for HDAC8. Whereas HDAC8 is exclusively found in fore- and midbrain regions during early mouse embryogenesis, the chick ortholog shows an expanded expression pattern, suggesting a more diversified role of HDAC8 in the chick system. Our results present a basis for further functional analysis of class I HDACs in chick development.
Keywords: histone deacetylase, chromatin, development, chick, mouse
Transcriptional regulation in eukaryotes occurs within a chromatin setting and is strongly influenced by the posttranslational modification (PTM) of histones, the building blocks of chromatin. PTMs, such as methylation, phosphorylation and acetylation largely influence the readout of gene expression and represent an interface for binding factors and chromatin modifying enzymes (Strahl and Allis, 2000). A “histone code” has been suggested for the combination of distinct histone modifications that dramatically escalates the functional plasticity of the genome beyond the one-dimensional nature of the primary DNA sequence (Jenuwein and Allis, 2001). Reversible histone acetylation is a dynamic process controlled by the antagonistic actions of two large families of enzymes - the histone acetyltransferases (HATs) and the histone deacetylases (HDACs). HDACs remove acetyl moieties from histone tails, resulting in chromatin condensation and an overall reduction in transcriptional potential, while HAT activity results in local opening of chromatin thereby setting the stage for transcription. By controlling dynamic acetylation patterns at specific chromatin regions, HATs and HDACs also modulate the affinity of acetyl-histone binding factors. The balance between the actions of these enzymes serves as a key regulatory mechanism for gene expression and plays important roles in the programming of multicellular development. Furthermore, it has been demonstrated that also nonhistone proteins such as transcription factors (e.g. p53, STAT1 or STAT3), cytoskeleton proteins (e.g. α-tubulin) and other cellular proteins (e.g. HSP90 or KU70) are targets for reversible acetylation. Differential acetylation of target proteins has been suggested to influence protein stability, activity, localization and binding efficiency with co-factors (reviewed in Brunmeir et al., 2009).
The HDAC superfamily is vast and ancient, dating back to prokaryotes, which differ in structure, enzymatic function, subcellular localization and expression patterns. Based on sequence similarities HDACs have been divided into four classes: classic HDACs comprise class I (HDAC1, HDAC2, HDAC3, and HDAC8), class II ((HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10) and class IV (HDAC11-like) enzymes. Class III consists of NAD-dependent, functionally unrelated Saccharomyces cerevisiae Sir2-like deacetylases named sirtuins (Gregoretti et al., 2004). In all model organisms studied, class I HDACs are expressed in many different cell types during most stages of development.
With the exception of HDAC8, where no complex has been described so far, all class I members can function as the catalytic subunits of multiprotein complexes (Yang and Seto, 2008). HDAC1 and HDAC2 share more than 80% identity on the protein level and are generally found together in repressive complexes such as the Sin3, NuRD, CoREST, NODE and SHIP complexes (Brunmeir et al., 2009). HDAC3 is found in distinct complexes, such as the nuclear hormone receptor N-CoR/SMRT complex (Yang and Seto, 2008). One common feature of these complexes is that they interact with DNA sequence specific transcription factors to repress transcription and cooperate with other chromatin modifiers to shape epigenetic programs (Grozinger and Schreiber, 2002). The ubiquitous expression, high deacetylase activity towards common substrates and high homology between class I HDACs suggests functional redundancy among these HDACs in vivo. However, deletion of each member of the class I HDAC family in mice leads to lethality in all cases, demonstrating the unique roles of each HDAC in the control of specific gene expression programs.
Hdac1-null mice die before embryonic day 10.5 (E10.5) due to severe proliferation defects and general growth retardation (Lagger et al., 2002). Conflicting reports exist regarding the global deletion of Hdac2 in vivo. One study found that mice lacking Hdac2 die within the first 24 hours after birth as a result of severe cardiac defects associated with uncontrolled proliferation of ventricular cardiomyocytes, which leads to obliteration of the right ventricular chamber (Montgomery et al., 2007). The neonatal lethal phenotype observed contrasts with that of another recent study in which nearly 50% of the offspring homozygous for a lacZ insertion in the Hdac2 gene were viable, whereas the other half of affected animals revealed postnatal lethality within the first 25 days after birth (Trivedi et al., 2007). A possible explanation for these varying phenotypes could be different genetic backgrounds of the mice tested. Alternatively, the mutation described by Trivedi might be a hypomorphic allele, rather than a true null, allowing adequate expression of Hdac2 for viability. Deletion of Hdac3 in the germ line of mice causes embryonic lethality by E9.5 owing to defects in gastrulation (Knutson et al., 2008; Montgomery et al., 2008). The in vivo functions of Hdac8 have been described just recently. Global deletion of Hdac8 in mice leads to a highly specific deficiency of cranial neural crest cells, resulting in perinatal death due to skull instability (Haberland et al., 2009).
Although certain aspects of class I HDAC expression have been reported in the context of knock-out studies, surprisingly no detailed transcription pattern during mouse embryogenesis has been published. Furthermore, to understand the specific functions of individual class I HDACs during chick embryogenesis, a detailed knowledge of their expression at relevant stages of development is required. Here, we describe for the first time the expression pattern of class I HDACs in chick embryos and compare it to the temporal and spatial expression profile during mouse embryonic development.
Our results show a distinct spatio-temporal expression pattern of class I HDACs during mouse and chicken development. Most notably, all class I HDACs in both species are highly expressed in the developing brain with a general tendency for higher expression levels in forebrain regions. Class I HDACs, in particular Hdac1, are expressed at sites of anterior and posterior neural tube closure most obvious in a hot spot-like expression of Hdac1 in HH12 chicken embryos. Other sites of robust class I HDAC expression are the developing inner ears, eyes, and limbs as well as neural crest. These expression profiles may serve as useful primer for further analyzing the individual biological functions of the different class I HDACs in chicken development.
Results
Class I HDAC expression during early chick development
To identify class I HDAC chick orthologs we analyzed the recently sequenced genome of chick Gallus gallus (International Chicken Genome Sequencing Consortium, 2004). Search analysis in the database determined that the chick genome comprises four genes that are highly related to mouse and human class I HDACs, Hdac1 (accession number AF_039751), Hdac2 (NM_204831), Hdac3 (NM_204747), and Hdac8 (XM_420178.2). Comparison of the full length proteins between chick and mouse showed that each chick class I HDAC has a strong homology with its mouse counterpart: HDAC1 93,1%, HDAC2 97,8%, HDAC3 96,9% and HDAC8 87,8%. Furthermore, individual class I HDAC protein sequence similarities between species are higher than the homologies observed between class I HDACs within the same species.
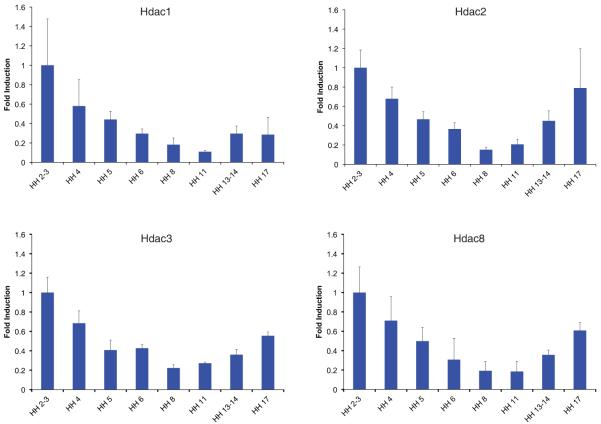
Class I HDACs are generally believed to be ubiquitously expressed, although varying mRNA and protein levels between species and embryonic stages have been reported (for a recent review see (Brunmeir et al., 2009). Therefore, we first examined gene expression profiles of class I HDACs during early stages of chick development (HH2-HH17) by quantitative Real-time PCR. We observed a similar temporal expression pattern for all four HDACs with peak levels during gastrulation, followed by a continuous decrease in transcription reaching lowest amounts at HH8, from which on expression levels increased again (Fig. 1). These data are in accordance with recent studies showing that mouse and bovine preimplantation embryos express most class I HDACs with increased levels at the blastocyte stage (Ma and Schultz, 2008, McGraw et al., 2003). Whereas transcript profiling of mouse preimplantation embryos did not detect a convincing signal for Hdac8 (Zeng et al., 2004), in this study Hdac8 transcripts could clearly be observed in the chick embryo at the earliest stage examined (HH2). Furthermore, the Hdac8 expression pattern nearly perfectly mirrored that of the other three class I HDAC members at early stages of chick development.
Fig. 1. Class I HDAC mRNA levels during early stages of chick development.
Real time PCR from RNA extracts of whole embryos between stages HH2/3–17 showing Hdac1, Hdac2, Hdac3 and Hdac8 levels. Measurements were done in duplicates. Error bars represent the standard deviation of 3 individual experiments, with the range indicated. The values were normalized to 28S levels.
We next analyzed the temporal and spatial expression pattern of Hdac1, Hdac2, Hdac3 and Hdac8 during chick embryogenesis by whole mount in situ hybridization.
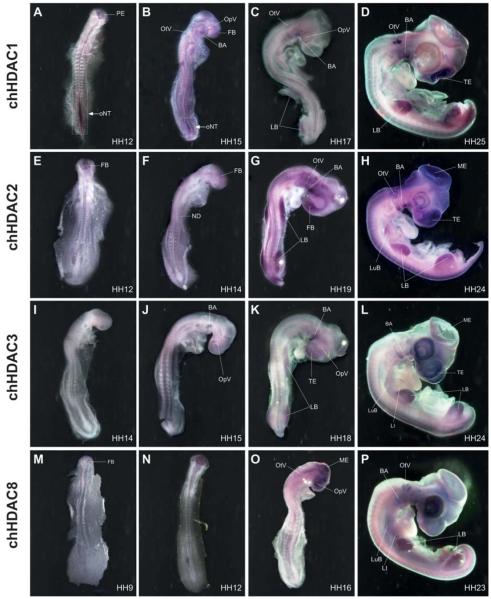
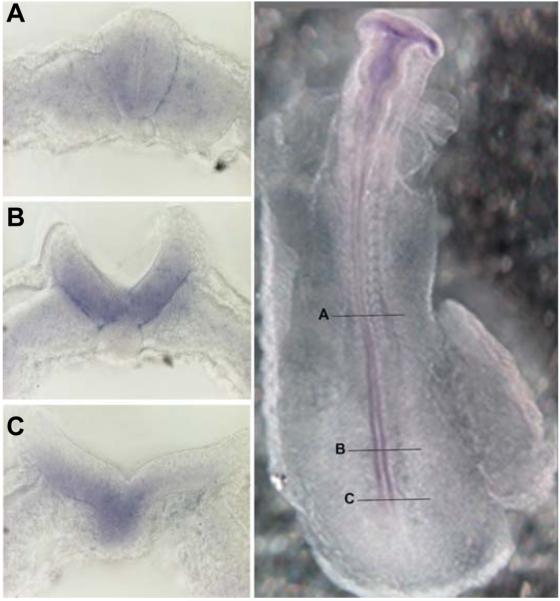
Transcripts of Hdac1 can be detected as early as stage HH10 as faint labelling in all three primary brain vesicles and in the developing neural tube (data not shown). Between HH10 and HH13 the overall staining in the brain increases and is most prominent in the prosencephalon with decreasing levels towards the rhombencephalon. Hdac1 expression extends into the neural tube at low levels. However, a prominent hot spot of Hdac1 expression can be found in the posterior part of the neural tube. This hot spot correlates with neural tube closure and is stronger in the open region diminishing where neural tube closure proceeds (Fig. 2A). This observation is also evident in transverse vibratome sections of a HH11 embryo as shown in Fig. 3. At the level of the last somite formed, the neural tube has closed and Hdac1 expression is strongly reduced (Fig. 3A), whereas more caudal sections from the same embryo clearly detect Hdac1 transcripts in both neural folds. Interestingly, the expression is excluded from the most dorsal tip (Fig. 3B). Further caudally, where neurulation is less advanced, also the notochord is positive for Hdac1 (Fig. 3C). Between stage HH14 and 15, the Hdac1 expression pattern in the developing brain persists as in previous stages although at reduced intensity. Within the brain, elevated transcript levels are found in the forebrain. In the posterior open neural tube Hdac1 expression at the hot spot has strongly decreased but is still visible. At this stage, Hdac1 expression is detectable in tissues other than the central nervous system (CNS), with increased intensities in the optic and otic vesicles, the branchial arches and the somites (Fig. 2B). At stage HH17 and 18, the general Hdac1 expression in the CNS is low and transcripts can only be detected in the tel- di- and mesencephalon after prolonged staining. A hot spot staining in the posterior neural tube can no longer be detected. As in the previous stages, Hdac1 expression is found in optic and otic vesicles, branchial arches and somites. In addition, the appearing limb anlagen are strongly labelled and with lower intensities the neural crest, the lung buds and the liver anlage (Fig. 2C). After 4 days of development (HH25), robust expression reappears in the telencephalon and to a lower degree in the other parts of the brain. Concomitantly, expression in the spinal cord increases as individual cells in both ependymal and mantle layer become positively labelled (data not shown). As in previous stages, strong Hdac1 expression is visible in otic vesicles, branchial arches and in limb buds, whereas reduced levels are found in developing eyes. The pattern of Hdac1 expression appears rather homogenously distributed in limb buds of previous stages, whereas now staining becomes progressively lower in the distal part of limbs and stronger in the more proximal parts, concomitant with developing musculo-skeletal elements. Hdac1 expression is also observable in the somites, lung buds, liver anlage, faintly in the heart, and in dorsal root ganglia and head mesenchyme, indicating neural crest cells (Fig. 2D).
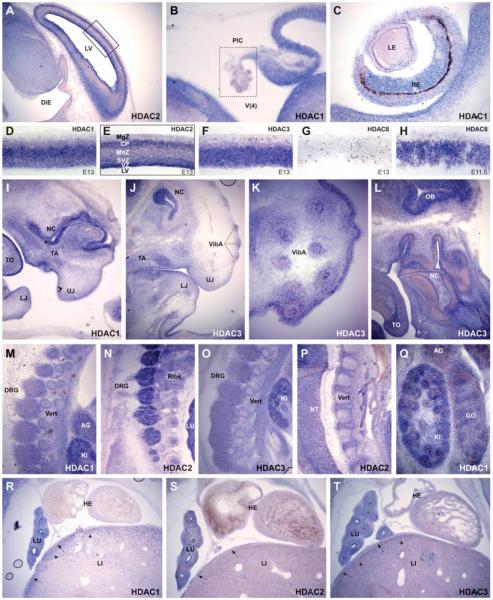
Fig. 2. Class I HDAC expression during chick development.
Whole mount in situ hybridization with probes against chick Hdac1 (A-D), chick Hdac2 (E-H), chick Hdac3 (I-L) and chick Hdac8 (M-P). Shown are embryos between 30 hours and 4 days of development with characteristic distribution of expression. A-C, E-G, I-K, M-O are dorsal views; D,H, L, P are lateral views. Hdac1 hot-spot expression in the open neural tube at HH12 (A). At HH15 only the most posterior tip of the neural tube shows strong Hdac1 expression (B). At stage HH17 overall Hdac1 levels decrease (C) but increase again at stage HH25, with most prominent staining in the face mesenchyme, otic vesicle, wing and limb buds (D). Hdac2 expression is first detected in the forebrain at stage HH12 (E). At stage HH14 brain and head mesenchyme are positive, as well as the first branchial arch, the notochord, the somites and the tail bud. (F). At stage HH19 strong Hdac2 signal is visible in fore brain, branchial arches, neural crest, wing and leg buds (G). At stage HH24 Hdac2 expression is most notably in the brain and head region (H). Between HH14-15 faint Hdac3 transcripts can be detected in the brain and neural tube, branchial arches, somites, notochord and optic vesicles (I-J). At HH18 it is more pronounced in the face, forebrain and in wing and limb buds (K). The most prominent staining in a HH24 embryo is seen in the face, forebrain and in the eyes (L). Hdac8 is visible in a HH9 embryo in the primary brain vesicle (M) and throughout all brain regions in a HH12 embryo (N). At HH16 it is expressed stronger in the brain and to a lesser extend in the spinal cord (O). At HH23 Hdac8 is found in the brain and also in the eyes, otic vesicles, wing and limb buds (P). BA; branchial arches. FB; forebrain. LB; limb buds. LI; liver. LuB; lung buds. ME; mesencephalon. ND, nephric duct. oNT; open neural tube. OpV; optic vesicle. OtV; otic vesicle. PE; prosencephalon. TE; telencephalon.
Fig. 3. HDAC1 hot spot in the chick open neural tube.
Whole mount embryo at HH11 and transverse vibratome sections at indicated regions, showing the expression of Hdac1 during the process of neural tube closure. In the closed neural tube Hdac1 is only weakly expressed (A). Elevated neural folds with strong Hdac1 expression; the signal is absent at the most dorsal tip (B). At the start of neural fold elevation the Hdac1 signal is restricted to the ventral region of the neural plate extending into the notochord (C).
Hdac2 transcripts are first detectable around stage HH12 in the forebrain region. Staining is also present in the head mesenchyme, optic and otic vesicles and in the nephric ducts (Fig. 2E). At stages HH14 and 15, Hdac2 signal in the brain shows highest intensities in the forebrain with declining levels towards the hindbrain. In contrast to Hdac1, the neural tube is only weakly stained. As in previous stages, Hdac2 expression can also be observed in otic vesicles and nephric ducts. In addition, signal now appears in the notochord, the first branchial arch, in somites and in the tail bud whereas staining in the eye is reduced (Fig. 2F). The expression pattern in the brain is similar at stages HH 18 and 19, with prominent signal in the forebrain and declining staining towards hindbrain and low signal in the neural tube. Strong Hdac2 expression can be detected in the head mesenchyme, the branchial arches and throughout the limb buds. Further sites of Hdac2 expression are optic and otic vesicles, tailbud and somites (Fig. 2G). As development proceeds, expression areas in the CNS are more intensively labelled around stage HH24 with most prominent staining in the telencephalic and mesencephalic region. Optic and otic vesicles, limb buds, lung buds, somites, and dorsal root ganglia and branchial arches are strong positively stained whereas the head mesenchyme, the notochord, the nose placodes and the lung buds express Hdac2 at lower levels (Fig. 2H).
Hdac3 expression starts relatively late compared to the other members of class I histone deacetylases. Hdac3 transcripts were first detected in the brain of stages HH14 to 15 embryos. Staining is strongest in the midbrain and to a lesser degree in the fore- and hindbrain. Staining extends into the neural tube, which is weakly positive. In addition, the optic vesicles, the branchial arches, somites and the notochord express Hdac3 only faintly (Fig. 2 I,J). At stage HH18, Hdac3 expression in the brain is strongest in the telencephalon and mesencephalon and to a lesser degree in the other brain vesicles. Strong signal can be observed in the head mesenchyme, optic vesicles, branchial arches, and throughout the limb buds. Relatively weak signal can be found in otic vesicles and in somites (Fig. 2K). The overall staining intensity is enhanced at around 4 days of development. In stage HH24 embryos, the expression pattern in the CNS is similar to the previous stages with strongest expression in tel- and mesencephalon. Strong Hdac3 expression can be observed in the eyes, throughout limb buds and in branchial arches. Further signal can be detected in the head mesenchyme, in the notochord, in somites and in the developing liver, lung and heart (Fig. 2L).
Hdac8 is expressed with the earliest onset of all class I HDACs. After in situ hybridization, transcripts can already be seen around stage HH9 in the primary brain vesicles (Fig. 2M). At stages HH11-12, Hdac8 is predominantly expressed throughout all areas of the brain (Fig 2N). Around stage HH16, Hdac8 expression in the CNS is similar compared to previous stages with prominent staining of all parts of the brain extending into the neural tube at lower levels. A prominent hybridization signal can be detected in the optic vesicles whereas weaker signal intensities can be found in the otic vesicles, the head mesenchyme, notochord, branchial arches, nephric ducts, tailbud and in the heart (Fig. 2O). Robust Hdac8 expression is visible in the 4 day old embryo at stage HH23. Hdac8 transcripts are present in all parts of the brain although at a relatively lower level of intensity than in previous stages. Specifically strong signals can be detected in the otic vesicles, branchial arches, nose placodes and throughout the limb buds. The head mesenchyme, heart, lung buds and the liver are only weakly stained (Fig. 2P).
HDAC class I expression during mouse development
Despite the growing knowledge on the mechanisms of HDAC-dependent gene repression by a panoply of biochemical analyses in vitro, mainly involving studies in cultured cells, surprisingly little is known about the distinctive patterns of gene expression of class I HDACs during development in vivo. Prior to analyzing temporal and spatial expression profile of Hdac1, 2, 3, and 8 during early mouse development by in situ hybridization, we first compared overall protein levels between corresponding developmental stages of chick and mouse by immunoblotting.
As shown in Fig.4, chick HDAC1 protein levels are low at stage HH13 and HH17, but increase at stage HH25. Interestingly, the opposite trend is seen for mouse embryos where the strongest HDAC1 signal is already detected at E10 followed by decreasing levels at later stages. Furthermore, a strong difference in HDAC1 protein levels can also be observed in tissue culture cells with high levels in the mouse embryonic fibroblast cell line Swiss 3T3 whereas HDAC1 protein is nearly undetectable in the chick embryonic fibroblast cell line CEF 32. In contrast, HDAC2 and 3 protein levels are similar in chick and mouse with an invariable pattern in the different embryonic stages examined. Remarkably, HDAC8 exhibited a dynamic protein expression pattern. Whereas the chick showed a steady increase reaching highest levels at stage HH25, the mouse already reached peak expression at E11.5 followed by a strong decrease to nearly undetectable amounts at E13. Also in fibroblast cell lines HDAC8 protein is nearly undetectable in both systems, which is in agreement with the tissue-specific expression detected during in situ hybridization experiments.
Fig. 4. Comparison of HDAC protein levels between mouse and chick embryos.
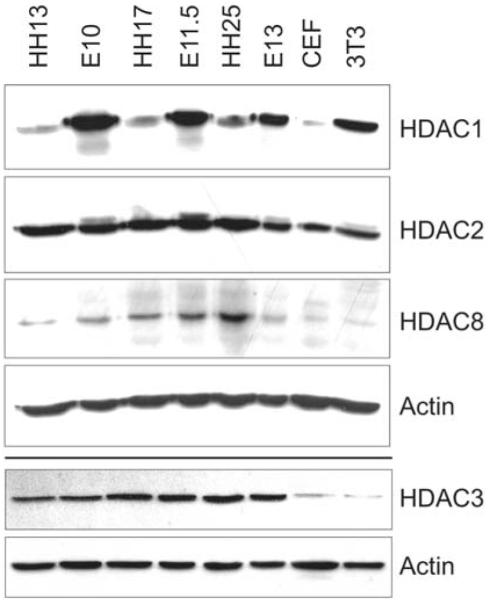
Protein levels of class I HDACs at comparable stages of mouse and chick development. Chick embryos at HH13 (lane 1), HH17 (lane 3), and HH25 (lane 5) were compared to mouse embryos at E10 (lane 2), E11.5 (lane 4), and E13 (lane 6). Chicken embryonic fibroblast cell line CEF 32 (lane 7) and mouse embryonic fibroblast cell line swiss 3T3 (lane 8) were included. One blot was probed against HDAC1, HDAC2 and HDAC8, a second blot was probed against HDAC3. Actin was used as loading control in both cases.
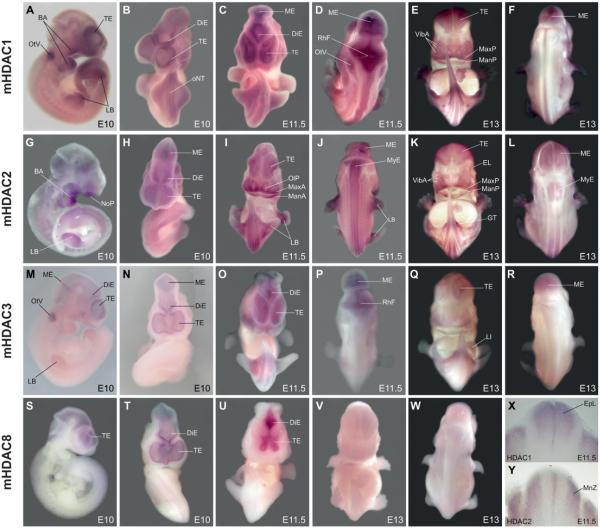
Using whole mount in situ hybridization, Hdac1 expression at E10 is strongest in telencephalic vesicles and with lower intensities in the di- and mesencephalon, rhombic fossa and only faint in the myelencephalon. Staining in the spinal cord is not detectable with the exception of the caudal regions where neural tube closure proceeds. Apart from the CNS, strong staining can be found in the otic vesicles and in branchial arches. Lower but distinct expression is visible in the head mesenchyme and throughout the limb buds (Fig. 5 A,B). At E11.5, the Hdac1 expression pattern is similar to the previous stage with strong expression in the tel-di-mes-metencepahlon and in the rhombic fossa. Faint expression can now be observed in the myelencephalon extending caudally as two faint stripes adjacent to the midline of the spinal cord. Hybridization signal is stronger in the caudal part of the neural tube, which is undergoing neural tube closure. Furthermore, branchial arches and otic vesicles are again strongly positive, as are the limb buds. The head mesenchyme, somites, dorsal root ganglia and liver anlage are faintly positive (Fig. 5 C,D). At E13, Hdac1 expression is high in the tel- di- and mesencephalon and in the developing cerebellum but is no longer detectable in the rhombic fossa (Fig. 5 E,F; Fig. 6 B,D). The choroid plexus is negative for Hdac1 (Fig. 6B) as well as for all other class I Hdacs (data not shown). Starting from the rhombic lips, two distinct stripes located adjacent to the neural tube midline extend caudally along the spinal cord (Fig. 5 E,F). Apart from the CNS, elevated Hdac1 transcript levels can be found in the mandibulary and maxillary processes of the first branchial arch, philthrum region, genital tubercle and in the limbs (Fig. 5E). In the latter, the signal appears concentrated around the forming skeletal elements, which themselves are negative. Further sites of Hdac1 expression are the developing eyes and eyelids, field of vibrissae anlagen, tooth anlagen, otic vesicles, dorsal root ganglia and the somites (Fig. 5 E,F; Fig. 6 C,I). Hybridization to sections further revealed strong Hdac1 expression in the lung and kidney. Hdac1 positive cells in the liver where only detected in the capsule and in the subcapsular region (Fig. 6 M,Q,R). Contrary to that, we could not detect any staining in the heart of E13 embryos, which is also the case for Hdac2 and 3 (Fig. 6 R-T). Noteworthy, cells positive for Hdac1, 2 and 3 are present in the outflow tract (data not shown).
Fig. 5. Class I HDAC expression during mouse development.
Mouse embryos at E10, E11.5 and E13 days of embryonic development were hybridized with probes against mouse Hdac1 (A-F, X), mouse Hdac2 (G-L,Y), mouse Hdac3 (M-R) and mouse Hdac8 (S-W). (A,G,M,S) are lateral views, (B,C,E,H,I,K,N,O,Q,T,U,V) are ventral views; (D,F,J,L,P,R,W) are dorsal views; (X, Y) are 100μm sagittal sections from the embryos shown in (C-D) and (I-J) at the level between the fore and hind limbs. Hdac1 is most prominently expressed in the developing brain, the branchial arches and the fore and hind limb regions from E10 to E13 (A-F). Hdac2 is also expressed in the brain at all stages. Branchial arches, extremities and the mouth region are also strongly positive (G-L). Across the spinal cord Hdac1 and 2 show distinct, nonoverlapping expression areas, as seen in the sections from E11.5 embryos (X-Y). Hdac3 is restricted to fore and midbrain regions and the otic vesicles at E10 (M-N). At E11.5 and E13 there is also faint expression in limb buds, face mesenchyme, liver and urogenital tubercle (O-R). Hdac8 is only expressed in fore and midbrain regions at E10 and E11.5 (S-U), whereas at E13 Hdac8 is nearly undetectable (V-W). BA; branchial arches. DiE; diencephalon. EL; eye lid. EpL; ependymal layer. GT; genital tubercle. LB; limb buds. LI; liver. MnZ; mantle zone. ManP; mandibulary process. MaxP; maxillary process. ME; mesencephalon. MyE; myelencephalon NoP; nose placode. OlP; olfactory placode. oNT; open neural tube. OpV; optic vesicle. OtV; otic vesicle. RhF; rhombic fossa. TE; telencephalon. VibA; vibrissae anlagen.
Fig. 6. In situ hybridization analysis of class I HDAC expression during mouse development.
Longitudinal sections of mouse embryos at E13 (except for (H), which represents E11.5) were hybridized with probes against mouse Hdac1 (B,C,D,I,Q,R), mouse Hdac2 (A,E,N,P,S), mouse Hdac3 (J-L,O,T) and mouse Hdac8 (G-H). Prominent expression of all four class I HDACs in the head region (A-L), particularly in the CNS (A-H) including the olfactory bulb (L). A distinct spatio-temporal expression pattern of the different class I HDACs is observable in the cortex (A, D-H), whereas no expression can be detected in the choroid plexus (B). Hdac1, 2 and 3 are expressed in the developing retina (C) and prominently in nose cavity, field of vibrissae and tooth anlage (I-L). In the trunk region (M-T) Hdac1, 2 and 3 are expressed in dorsal root ganglia (M-O), developing vertebrae and ribs (M-P), kidney (M,O,Q), adrenal gland (M, Q), gonad (Q), lung (N,R-T). In the liver expression is confined to the capsule and the subcapsular region (R-T). No expression of Hdac1, 2 and 3 is detectable in the developing heart (R-T). AG; adrenal gland. CP; cortical plate. DiE; diencephalon. DRG; dorsal root ganglia. GO; gonade. HE; heart. KI; kidney. LE; lens. LI; liver. LJ; lower jaw. LU; lung. MgZ: marginal zone. MnZ: mantle zone. NT; neural tube. LV; lateral ventricle. NC; nose cavity. PlC; plexus choroideus. RE; retina. SVZ; subventricular zone. TO; tongue. TA; tooth anlage. UJ; upper jaw. V(4); fourth ventricle. Vert; vertebrae. VibA; vibrissae anlagen. VZ; ventricular zone.
At E10, strong expression of Hdac2 in the brain extends from the telencephalic vesicles to the rhombic fossa, whereas it is weak in the myelencephalon and in the spinal cord. Apart from the CNS, a strong signal is present in the branchial arches and throughout the limb buds. Further Hdac2 expression domains can be observed in the otic vesicles, the head mesenchyme and the somites (Fig. 5 G,H). At E11.5 expression can be seen in all parts of the fore- mid- and hindbrain extending into the spinal cord as two faint stripes running from the caudal rhombic lips through the entire length of the myelencephalon and spinal cord. These bands are located in the lateral parts of the neural tube separated by unstained medio-lateral stripes, which are reminiscent of Hdac1 positive domains. Indeed, transverse vibratome sections reveal spatially separated, non-overlapping regions with Hdac1 expression confined to the ependymal layer and Hdac2 expression restricted to the mantle layer of the developing neural tube (Fig. 5 X,Y). In the head, strong Hdac2 expression can be seen in the processes around the olfactory placodes, mandibulary and maxillary processes and in the area of future vibrissae. Prominent labelling can also be observed in the limb buds, head mesenchyme and the genital tubercle whereas only moderate staining can be found in otic vesicles, somites, dorsal root ganglia and in the developing Iiver (Fig. 5 I,J). At E13, Hdac2 expression is still strong in the fore- and midbrain region. Preclusive Hdac1 and 2 expression areas became apparent in sections from the cortex of E13 embryos. Whereas Hdac2 is predominantly expressed in cells of the ventricular and subventricular zones and in the cortical plate, Hdac1 expression is highest in the mantle zone (Fig. 6 A,D-E). The two distinct mediolateral stripes extending from the myelencephalon to the caudal tip of the spinal cord are now even more pronounced than in the previous stage. Strong hybridization signal can be found in the maxillary and mandibulary processes, tooth anlage, field of vibrissae and in the limbs where the signal is concentrated around the forming skeletal elements (Fig. 5 K,L). The developing vertebrae and ribs are also positively stained for Hdac2 (Fig. 6 N,P), as well as for Hdac1 and 3 (Fig. 6 M,O). Further Hdac2 expression domains localize to the head mesenchyme, otic vesicles, somites, dorsal root ganglia and the eyelid (Fig. 5 K,L, Fig. 6 N). Hdac2 shows high levels of expression in the lung and only faint expression in the liver (Fig. 6S), resembling Hdac1 (Fig. 6R).
At E10, Hdac3 expression is restricted to the tel- diencephalon including hypophyseal pouch and the mesencephalon. Strongest Hdac3 signal can be detected in the otic vesicles and faint staining is present throughout the limb buds (Fig. 5 M,N). At E11.5, Hdac3 expression in the brain extends further caudally until the metencephalon and rhombic fossa. Apart from the CNS, staining can be found in the head mesenchyme, mandibulary processes, otic vesicles, limb buds and in the genital tubercle (Fig. 5 O,P). At E13, expression of Hdac3 within the brain is again restricted to the tel- di- and mesencephalon (Fig. 5 Q, R) extending into the olfactory bulb (Fig. 6L). Hdac3 expression in the cortex resembles the expression of Hdac1 and is strongest in subventricular and mantle zones but missing in the cortical layer (Fig. 6F). The developing eyelids, fields of vibrissae and tooth primordia express high levels of Hdac3 (Fig. 6 J-K).
Additional prominent Hdac3 hybridization signals can be detected in the genital tubercle and to lower degrees in upper and lower jaws, liver and in limbs around the forming skeletal elements (Fig. 5 Q,R; Fig. 6T). Hybridization to sections further revealed prominent expression in the lung, dorsal root ganglia, and developing vertebrae and ribs (Fig. 6 O,T), as observed for Hdac1 and 2.
Mouse Hdac8 displays the most restricted spatio-temporal expression pattern of all class I HDACs and is expressed predominately in the forebrain and to a lower degree in midbrain. At E10, Hdac8 transcripts are first observed in tel- and diencephalon, including hypophyseal pouch and with lower intensity in the mesencephalon. This signal pattern is maintained in E11.5 (Fig. 5 S,T,U; Fig. 6H), whereas transcript levels at E13 are dramatically reduced (Fig. 5 V,W), which becomes particularly evident on brain sections (Fig. 6 G-H) complementing the results of the Western blot analysis (Fig. 4).
Collectively, positive signals in head mesenchyme, branchial arches (including maxillar and mandibular processes), ectomesenchyme of tooth anlagen, field of vibrissae anlagen, dorsal root ganglia, and genital tubercle demonstrate class I HDAC expression in neural crest.
Discussion
In this study we have addressed the spatio-temporal expression pattern of class I HDACs during early development in chick and mouse embryos. We found overlapping expression profiles for all four class I HDACs in chick and mouse, although we noticed some species-specific differences suggesting also distinct roles in development.
All class I HDACs in both species are highly expressed in the developing brain. Within the brain, a general tendency for higher expression levels in forebrain regions and declining expression towards hindbrain regions can be observed. Hdac1 expression is detected at sites of anterior and posterior neural tube closure, consistent with previous observations of elevated Hdac1 levels in the head and neural folds of mouse embryos at E8.5 (Lagger et al., 2002). As a major feature of Hdac1 expression in chicken, we identified enhanced concentration in a hot spot domain in the posterior neural tube of HH12 chicken embryos. Preliminary data in our lab suggest a role of Hdac1 in neural tube closure as RNAi induced knock down targeting the hot spot resulted in neural tube closure defects. Interestingly, in mice however, starting from E11.5 spatially preclusive Hdac1 and 2 expression in the neural tube becomes evident. Hdac1 is confined to the proliferative ependymal zone whereas Hdac2 is restricted to the mantle layer. This finding is consistent with a recent study reporting the distinct expression of Hdac1 and Hdac2 during different stages of neuro-glial development in the mouse CNS. At all developmental stages, Hdac1 is expressed in progenitor cells. As progenitors commit to the neuronal lineage, expression of Hdac1 is down-regulated, whereas Hdac2 is up-regulated in neuroblasts and post-mitotic neurons. In contrast, Hdac2 is not expressed in differentiated glia where Hdac1 expression is maintained (MacDonald and Roskams, 2008). Given the fact that both paralogs evolved from a recent gene duplication event and can associate within the same transcriptional repressor complexes (Brunmeir et al., 2009) a high degree of functional redundancy has been proposed. However, the unique spatial Hdac1 and 2 expression pattern in the vertebrate CNS suggests specific cell-autonomous functions of the two enzymes. Recently, Hdac2 has been linked to memory formation and regulation of synaptic plasticity, revealing a predominant role for HDAC2 in restricting adult neuronal synapse maturation (Guan et al., 2009), whereas HDAC1 reduction was only able to prohibit early synaptogenesis (Akhtar et al., 2009). Furthermore, the nitrosylation of HDAC2 leads to derepression of neuron-specific genes, thereby promoting dendritic growth, synapse formation and neuron plasticity (Nott et al., 2008). These results are well in accordance with our expression data and demonstrate an essential function of HDAC2 in post-mitotic neurons. In this line of argumentation it is surprising that individual deletion of Hdac1 or Hdac2 in neuronal precursors and oligodendrocytes shows no obvious phenotype in mice, whereas only the loss of all four Hdac1 and 2 alleles results in postnatal lethality (Montgomery et al., 2009; Ye et al., 2009). In contrast, in zebrafish, where only one Hdac1/Hdac2 copy exists, loss of the gene results in severe neurodevelopmental associated abnormalities further underlining the fact that functional diversification of Hdac1 and Hdac2 has added higher levels of complexity and fine tuning of HDAC1 and HDAC2 functions within the CNS (Cunliffe, 2004). A mechanistic redundancy between Hdac1 and Hdac2 has already been described in mouse knockout embryos and ES cells, where both enzymes are expressed within the same cell (Lagger et al., 2002; Montgomery et al., 2007; Zupkovitz et al., 2006). However, within the CNS, displaying a preclusive Hdac1 and Hdac2 expression pattern, it is tempting to speculate that a cross-regulation also exists beyond the cell type. As no compensatory upregulation of these closely related enzymes on the transcriptional level has been observed so far, a posttranscriptional mechanism guided by micro RNAs might be involved. Alternatively, HDAC1 and HDAC2 might be cross-regulated by differential protein stability executed by the action of PTMs directly on the individual HDACs.
Interestingly, in the chick Hdac1 and 2 expression in the spinal cord does not show the same distinctive pattern as in the mouse. Instead, both paralogs are expressed at low levels in overlapping regions at embryonic day 4, a developmental stage comparable to E13 in the mouse. However, it cannot be excluded that spatial separation of chick Hdac1 and 2 occurs at later time points.
Overall, Hdac3 in chick and mouse embryos shows a similar expression pattern as Hdac1 and 2 although slight differences exist. In the chick, a generally delayed onset and a lack of expression in the inner ear could be observed. Furthermore, we noticed an absence of Hdac3 expression in hindbrain regions of mouse embryos. Conditional deletions of Hdac3 have been so far described for the liver and the heart. Consistent with the described liver specific phenotype in postnatal mouse (Knutson et al., 2008) leading to disrupted lipid and cholesterol homeostasis we could detect Hdac3 expression in the liver of E13 mouse embryos as well as in the developing liver in chick. Cardiac specific deletion of Hdac3 led to massive abnormalities in cardiac energy metabolism, resulting in 100% lethality by 16 weeks of age (Montgomery et al., 2008). Whereas we observed Hdac3 positive staining in the heart of chicken embryos, transcripts were not detectable in the myocard of the developing mouse heart. We did however find Hdac3 positive cells in the outflow tract.
In mouse development, Hdac8 displays a significantly restricted spatio-temporal expression pattern compared to the chicken ortholog. Hdac8 expression in mice is exclusively found in fore- and midbrain regions, whereas chicken Hdac8 is more widely expressed throughout embryonic development. This difference between species becomes particularly evident at later stages of development. Whereas in mouse Hdac8 expression dramatically decreases at E13 both in intensity and localization, the chick shows an expansion in expression domains to other parts, including otic vesicles, branchial arches, nose placodes and throughout the limb buds.
Interestingly, the deletion of Hdac8 in mice results in a highly specific deficiency of cranial neural crest cells, phenotypically first observed around E16.5 as ossification defects in skull skeltal elements (Haberland et al., 2009). In contrast, whole mount in situ experiments detect Hdac8 predominantly in the forebrain and to a lower degree in the midbrain at E10 and E11.5, stages of mouse embryonic development where rather Hdac1, 2 and 3 show expression in cranial neural crest cells. Explanations for this seemingly diverging findings might be that HDAC8 exerts its effects on cranial neural crest cells before developmental stage E10 perhaps by controlling neural crest cell number or that Hdac8 expression at later stages of development (i.e. older than E13.5) leads to mispatterning of anterior skull elements in knock-out mice as ossification of neural crest-derived skull osteoblast cells commences at E16.5. In contrast to the original papers, documenting Hdac8 mRNA expression in multiple human organs (Buggy et al., 2000; Hu et al., 2000; Van den Wyngaert et al., 2000) a recent study indicated that in normal human tissues Hdac8 is exclusively expressed in cells showing smooth muscle differentiation. In particular, this work reports that Hdac8 expression is completely absent from the nervous system including brain and spinal cord (Waltregny et al., 2004). Conversely, in situ hybridization experiments clearly reveal ubiquitous Hdac8 expression in P55 old mouse brains, most prominent in the Purkinje cell layer of the cerebellum and the dentate gyrus of the hippocampus (Lein et al., 2007). A detailed expression pattern of Hdac8 in the adult chick would have the potential to contribute significantly to an understanding of the biological role of Hdac8 at later stages of development and elucidate existing discrepancies throughout different species.
Finally, class I histone deacetylases have attracted significant interest as anti-tumour targets during the last decade. However, pharmacological inhibition of HDACs in tumour therapy has one major disadvantage: the inability of various inhibitors to distinguish between members of the enzyme class. At the moment the search for more specific inhibitors against single HDAC enzymes is ongoing. Therefore, the clarification of embryonic and adult HDAC expression patterns in different species may supply helpful information when administration of specific inhibitors is useful and does not lead to undesired side effects.
Materials and Methods
Chick and mouse embryos
Fertilized eggs (Gallus gallus) were obtained from a local supplier and incubated at 38°C, 50% humidity. Embryos were staged according to Hamburger and Hamilton (Hamburger and Hamilton, 1992). Mouse embryos were collected from wild type Swiss OTF mice. For staging, the appearance of a vaginal plug after overnight breeding was considered as day 0.5.
RNA isolation and real time RT-PCR analysis
Total RNA was isolated from chick embryos using TRIzol reagent (GibcoBRL) according to the manufacturer’s instructions. 1μg of RNA was used for reverse transcription with an iScript cDNA synthesis kit (Bio-Rad). Real time PCR was done with the i-Cycler iQ System (Bio-Rad) and a SYBR-green-Fluoresceine based detection system. HDAC values obtained with the i-Cycler iQ software were normalized to 28S mRNA.
Primer and hybridization probes
For detection of HDAC transcripts we designed primers to amplify a unique region about 300-400 base pairs from each transcript. For generation of the templates used to transcribe in situ hybridization probes a T7 polymerase promoter sequence was added to the antisense primer. The primers used to detect the chicken Hdac transcripts by Real time RT-PCR were the same as used for generation of the in situ hybridization probes. All transcripts were sequence verified. Labelled probes were generated with the MaxiScript kit (Ambion) and dioxigenin-UTP (Roche). Primer sequences used are:
chHDAC1 f: GGATGAAGAAGAAGAAGATCC, r: GATAACTATGCACTGACAGG (AF_039751, bp1239- 1541); chHDAC2 f: AAGGTGGACGGCGAAATG, r: GATACGGTCCATGCCAAATAG (NM_204831.1, bp 1356- 1663); chHDAC3 f: TTGGATCAGATCAGGCAGAC, r: CCCAGTTAACTGCACCAATG (NM_204747.1, bp 1185- 1500); chHDAC8 f: AGATATTGGCCTGGGGAAAG, r: CCAGTCAAGTACGTCCAGCA (XM_420178.2, bp 655- 969); ch28S f: GGTATGGGCCCGACGCT, r: CCGATGCCGACGCTCAT (BQ037787, 145- 288); mHDAC1 f: AGAGATCCCTAATGAGCTGCC, r: CTCCCTCCTCATCTGAGTC (NM_008228.2, bp 966-1282); mHDAC2 f: AGTGATGGAGATGTACCAGCCT, r: TGAACACCAGGTGCATGTGG (NM_008229.2, bp 952-1350); mHDAC3 f: GGCCATTAGTGAGGAACTTCC, r: TCCACATCACTTTCCTTGTCG (NM_010411.2, bp 951-1281); mHDAC8 f: ACCGAATCCAGCAAATCCTCA, r: ATAAAATTCTTCCCCCCAACTTGC (AK_131998, bp 1163- 1522)
In situ hybridization
Whole mount in situ hybridization was performed according to the protocol of Henrique (Henrique et al., 1995). Some embryos were subsequently embedded in 2% agarose and sectioned with a vibratome at 100μm.
For in situ hybridization to frozen sections, mouse embryos were fixed in 4% PFA, cryopreserved in sucrose and embedded in Tissue-Tek embedding medium (Gröpl Electron Microscopy). Sagittal sequential sections of 20μm were taken on a cryostat, mounted onto Superfrost Plus Gold glass slides (Roth-Lactan) and stored at −20°C. Before hybridization slides were incubated at 50°C, fixed in methanol and washed in PBS, followed by proteinase K digestion, postfixation in 4% PFA and acetylation with 0,25% acetic anhydride in Tris buffer. Sections were hybridized overnight at 65°C in a humidified chamber, followed by several high stringency washes in SSC buffer. Alkaline phosphatase conjugated anti dioxigenin antibody (Roche, 1:2000 dilution) was used to detect bound probes. Signals were developed with NBT/BCIP (Roche), and sections were embedded in Cityfluor mounting medium (Gröpl Electron Microscopy). As controls sense probes were used.
Western blot analysis
Whole cell protein extracts were separated on a 10% SDS-page and blotted onto a nitrocellulose membrane. After blocking, the membrane was sequentially incubated with the following primary antibodies at indicated dilutions: HDAC1: 2E10 monoclonal mouse 1:1000 (Millipore); HDAC2: 3F3 monoclonal mouse 1:500 (Millipore); HDAC3: polyclonal rabbit 1:2000 (Abcam); HDAC8: polyclonal rabbit 1:1000 (Abcam); beta-Actin: monoclonal mouse 1:1000 (Sigma). After washing, the membrane was incubated with horseradish conjugated secondary anti mouse or rabbit antibodies and the proteins were visualized using the Perkin Elmer ECL system.
Acknowledgements
The authors would like to thank Simon Simonovic for his expertise in scientific illustration design. We would also like to thank Professor Michael Freissmuth for generously providing funds for C.M. in critical financial circumstances. We are thankful to Pavel Uhrin for kindly providing mouse embryos and Christine Hartmann for providing protocols for in situ hybridization to tissue sections. The work in the laboratory of C.S. was supported by the Austrian Science Fund (FWF P16443 and FWF P18746) and the GEN-AU project “Epigenetic Plasticity of the Mammalian Genome” (Austrian Ministry of Science and Research (BM:WF)). S.L. was a fellow of the Vienna Biocenter international PhD program supported by the FWF.
Abbreviations used in this paper
- HDAC
histone deacetylase
- PTM
posttranslational modification
References
- AKHTAR MW, RAINGO J, NELSON ED, MONTGOMERY RL, OLSON EN, KAVALALI ET, MONTEGGIA LM. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29:8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUNMEIR R, LAGGER S, SEISER C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- BUGGY JJ, SIDERIS ML, MAK P, LORIMER DD, MCINTOSH B, CLARK JM. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 2000;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- CUNLIFFE VT. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development. 2004;131:2983–2995. doi: 10.1242/dev.01166. [DOI] [PubMed] [Google Scholar]
- GREGORETTI IV, LEE YM, GOODSON HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- GROZINGER CM, SCHREIBER SL. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- GUAN JS, HAGGARTY SJ, GIACOMETTI E, DANNENBERG JH, JOSEPH N, GAO J, NIELAND TJ, ZHOU Y, WANG X, MAZITSCHEK R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HABERLAND M, MOKALLED MH, MONTGOMERY RL, OLSON EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMBURGER V, HAMILTON HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- HENRIQUE D, ADAM J, MYAT A, CHITNIS A, LEWIS J, ISH-HOROWICZ D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- HU E, CHEN Z, FREDRICKSON T, ZHU Y, KIRKPATRICK R, ZHANG GF, JOHANSON K, SUNG CM, LIU R, WINKLER J. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- JENUWEIN T, ALLIS CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- KNUTSON SK, CHYLA BJ, AMANN JM, BHASKARA S, HUPPERT SS, HIEBERT SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAGGER G, O’CARROLL D, REMBOLD M, KHIER H, TISCHLER J, WEITZER G, SCHUETTENGRUBER B, HAUSER C, BRUNMEIR R, JENUWEIN T, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEIN ES, HAWRYLYCZ MJ, AO N, AYRES M, BENSINGER A, BERNARD A, BOE AF, BOGUSKI MS, BROCKWAY KS, BYRNES EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- MA P, SCHULTZ RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACDONALD JL, ROSKAMS AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev Dyn. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- MCGRAW S, ROBERT C, MASSICOTTE L, SIRARD MA. Quantification of histone acetyltransferase and histone deacetylase transcripts during early bovine embryo development. Biol Reprod. 2003;68:383–389. doi: 10.1095/biolreprod.102.005991. [DOI] [PubMed] [Google Scholar]
- MONTGOMERY RL, DAVIS CA, POTTHOFF MJ, HABERLAND M, FIELITZ J, QI X, HILL JA, RICHARDSON JA, OLSON EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONTGOMERY RL, HSIEH J, BARBOSA AC, RICHARDSON JA, OLSON EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONTGOMERY RL, POTTHOFF MJ, HABERLAND M, QI X, MATSUZAKI S, HUMPHRIES KM, RICHARDSON JA, BASSEL-DUBY R, OLSON EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOTT A, WATSON PM, ROBINSON JD, CREPALDI L, RICCIO A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- STRAHL BD, ALLIS CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- TRIVEDI CM, LUO Y, YIN Z, ZHANG M, ZHU W, WANG T, FLOSS T, GOETTLICHER M, NOPPINGER PR, WURST W, et al. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- VAN DEN WYNGAERT I, DE VRIES W, KREMER A, NEEFS J, VERHASSELT P, LUYTEN WH, KASS SU. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 2000;478:77–83. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- WALTREGNY D, DE LEVAL L, GLENISSON W, LY TRAN S, NORTH BJ, BELLAHCENE A, WEIDLE U, VERDIN E, CASTRONOVO V. Expression of histone deacetylase 8, a class I histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. Am J Pathol. 2004;165:553–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG XJ, SETO E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YE F, CHEN Y, HOANG T, MONTGOMERY RL, ZHAO XH, BU H, HU T, TAKETO MM, VAN ES JH, CLEVERS H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG F, BALDWIN DA, SCHULTZ RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- ZUPKOVITZ G, TISCHLER J, POSCH M, SADZAK I, RAMSAUER K, EGGER G, GRAUSENBURGER R, SCHWEIFER N, CHIOCCA S, DECKER T, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]