Abstract
The control of biochemical fluxes is distributed and to perturb complex intracellular networks effectively it is often necessary to modulate several steps simultaneously. However, the number of possible permutations leads to a combinatorial explosion in the number of experiments that would have to be performed in a complete analysis. We used a multi-objective evolutionary algorithm (EA) to optimize reagent combinations from a dynamic chemical library of 33 compounds with established or predicted targets in the regulatory network controlling IL-1β expression. The EA converged on excellent solutions within 11 generations during which we studied just 550 combinations out of the potential search space of ~ 9 billion. The top five reagents with the greatest contribution to combinatorial effects throughout the EA were then optimized pairwise. A p38 MAPK inhibitor with either an inhibitor of IκB kinase or a chelator of poorly liganded iron yielded synergistic inhibition of macrophage IL-1β expression. Evolutionary searches provide a powerful and general approach to the discovery of novel combinations of pharmacological agents with potentially greater therapeutic indices than those of single drugs.
Acute or chronic (non-resolving) inflammation is a well established mediator of major diseases including vascular disease (e.g. atherosclerosis, stroke), chronic obstructive pulmonary disease (COPD), cancer, diabetes, obesity, rheumatoid arthritis, psoriasis and inflammatory bowel disease 1,2 . Each of these conditions exhibits an elevated expression of the potent and pleiotropic, pro-inflammatory cytokine interleukin-1β (IL-1β) 2,3. Pharmacological therapies targeted against IL-1β are largely focused on biologics that are only likely to act extracellularly 4 (e.g. Anakinra; an IL-1 receptor antagonist), which have shortcomings such as poor CNS penetration. Small molecule modulation of IL-1β may offer benefits in certain conditions such as in cerebrovascular injury where IL-1β mediates significant cerebral damage during acute ischaemia and excitotoxic insult 5.
The lipopolysaccharide- (LPS-) induced Toll-like receptor 4 (TLR4) stimulation of macrophages is a widely used experimental model that mimics key aspects of inflammation including IL-1β expression 6. Signal transduction induced by TLR4 stimulation proceeds through the activation of a complex array of multi-protein signalling networks (e.g. MyD88, TRAF6, p38 MAPK, NF-κB), ultimately resulting in the expression of the IL-1B gene. Possibly based on the ‘one gene, one drug, one disease paradigm’ 7 a plethora of reagents has been developed to modulate individual proteins within these signalling networks. However, it is well known that multiple steps must be modulated simultaneously to have significant effects on biochemical fluxes 8. Thus, a single-target approach is unlikely to be optimal in inhibiting the expression of a pro-inflammatory cytokine such as IL-1β, where there is both inherent degeneracy and considerable complexity within the signal transduction network 9. More generally, there is an increasing recognition of the need to target multiple steps within signalling networks for their effective pharmacological modulation 7. Combinatorial chemical genetics 10 uses combinations of small molecules that allow dissection of cellular phenomena via their selective modulation of individual biological targets. Despite progress made in high-throughput screening technologies 11, the analysis of even modestly sized chemical libraries is prohibitive due to the combinatorial explosion that occurs in pharmacological space 12 (233 or ~ 9 billion combinations for all possible combinations of the chemical library explored here). Thus we sought heuristic solutions (i.e. reagent combinations) that are good but not provably globally optimal.
The terms ‘evolutionary computing’ and ‘evolutionary algorithms’ describe a set of approaches based loosely on Darwinian evolution by the natural selection of individuals and populations. In this case the population consists of individuals that each encode a candidate solution to the problem at hand. The ‘fitness’ of each solution is reflected in the objective function(s) designed by the experimenter, but normally includes the concept that fitter individuals provide more accurate solutions. There may be multiple fitness functions. For instance a simpler solution may be deemed to be a fitter solution, and algorithms with multiple objectives (multiple fitnesses), as in this work, are known as multi-objective evolutionary algorithms. Multi-objective evolutionary algorithms (EA) allow for the specification of multiple and distinct optimization objectives and their simultaneous handling 13-15. Based on the fitness(es), a selection step determines which individuals will be allowed to remain under consideration for the next generation. Some of these individuals are retained by simply being copied unchanged into the subsequent generation(s), but new diversity, based on the parents selected, is then produced from them by processes analogous to mutation and recombination. The fitnesses of these new individuals are then evaluated as above, and the algorithm continues cycling through the steps of selection, breeding and fitness evaluation until an acceptable solution is found. Decades of research within the field of evolutionary computing (e.g. 16,17) have revealed that optimization of multivariate problems can be highly effective using small numbers of experimental tests. Although the present study used an EA and an adaptive dose matrix search strategy, we recognize that other kinds of combinatorial optimization approaches might also prove effective.
Using a ‘reverse’ combination chemical genetic approach 10 our aim was to optimize combinations of reagents that minimize LPS-induced macrophage IL-1β expression, and simultaneously minimize the number of component reagents in the combination and their propensity to induce macrophage cell death. Subsequently we sought to optimize reagents to inhibit IL-1β expression at concentrations lower than the component reagents used in isolation. This was achieved by application of an EA-directed, semi-automated robotic assay of IL-1β expression to a dynamic chemical library of a total of 33 reagents (see Methods, and also 18-20). The specific algorithm used here was the Indicator Based Evolutionary Algorithm (IBEA) 21, as in preliminary simulations 22 this proved superior to a variety of other multi-objective optimization algorithms. This was followed by a dose matrix search of top-ranked reagents resulting from the EA-directed search. We demonstrate that the EA converges efficiently on good solutions, and that dual p38 MAPK inhibition and either IκB kinase inhibition or iron chelation yields synergistic and biologically-relevant inhibition of macrophage IL-1β expression.
Results
Rapid convergence of Multi-objective IBEA to near-optimal solutions
Concentration-effect curves for a selection of reagents with known or predicted targets in the IL-1β expression network were determined in order to identify the most appropriate concentration for use in the EA (Supplementary Results, Supplementary Fig. 2). On the basis of these data we selected a sub-optimal dose (3μM) which would provide scope for observing combinatorial synergy.
The Indicator Based Evolutionary Algorithm (IBEA) 21 directed a semi-automated robotic assay utilizing chemical combinations to inhibit IL-1β expression was initialized (Fig. 1, Loop 1). As described in Methods (Implementation of the Indicator Based Evolutionary Algorithm (IBEA)), the IBEA generates subsets of combinations from the library and then assesses the performance of these subsets with respect to inhibition of IL-1β expression, decreases in LDH release (a marker for cell death) and the number of member reagents within the combination. In the present case, we confined the number of experiments in each of the first and subsequent generations to 50 and those for generation 1 were selected randomly by the EA from the first-generation library of chemicals. Superior combinations are retained and recombined with other library components in successive subsets and assessed iteratively until satisfactory combinations are found. Assay of successive generations of chemical combinations from a dynamic chemical library (a total of 33 reagents, each at a concentration of 3 μM, see supplementary methods) revealed their convergence towards a set of highly effective cocktails (Fig 2; see also Supplementary Fig. 3 and Supplementary Table 3 and 4 (top) for data on the individual generations). In addition, Supplementary Spreadsheet 1 gives all of the data on both reagent combinations and experimental measurements in tabular form.
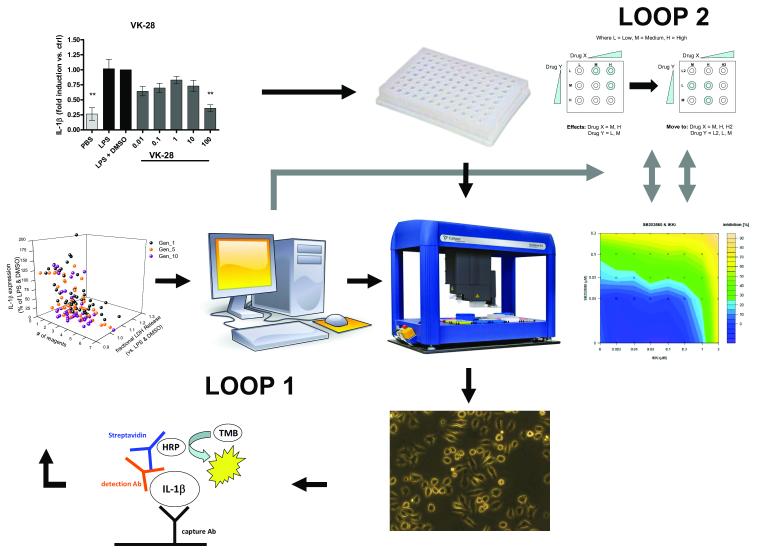
Figure 1.
Combinatorial evolutionary inhibition of IL-1β expression (Loop 1, clockwise, black arrows). Known drugs were tested alone before being used at a single concentration (3μM) in a chemical library. Initialization of the Indicator Based Evolutionary Algorithm (IBEA) creates a random selection of combinations that are incubated with stimulated cells before measurement of cell death (LDH release) and IL-1β expression. Evaluation of these data against the number of compounds in the combination (All data n = 3) is performed by IBEA prior to a new generation of combinations being computed and tested. After 11 generations, concentration-dependent optimization (Loop 2) of five top-ranked reagents was undertaken. Synergy was detected in novel dual-combinations.
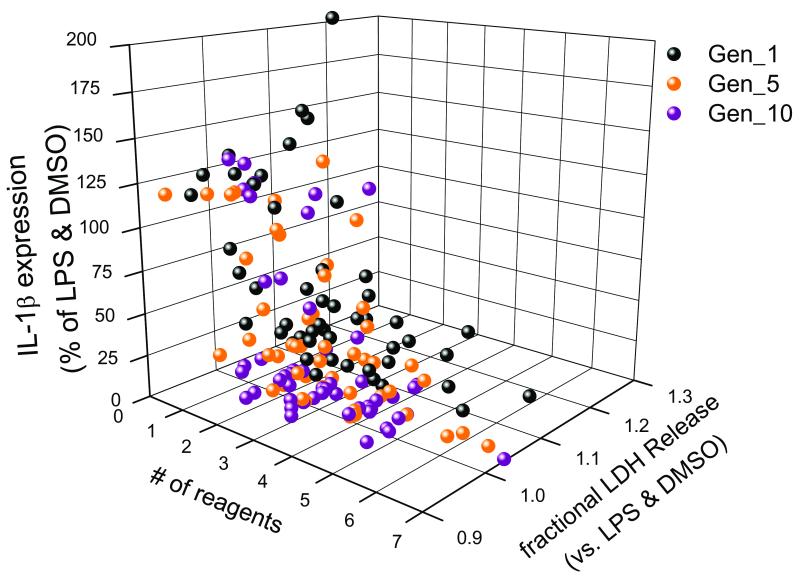
Figure 2.
Analysis of successive generations (Generations 1; initialization, 5 and 10) of reagent combinations reveals their convergence to a subset of highly effective combinations reflecting the inhibition of IL-1β expression with concomitant decreases in LDH release and the number of member reagents. All data presented are the means of 3 determinations. Data points appearing as zero on the number of (#) reagents axis were reflective of positive control responses (LPS (1 μg / mL) and DMSO (0.5 % v/v).
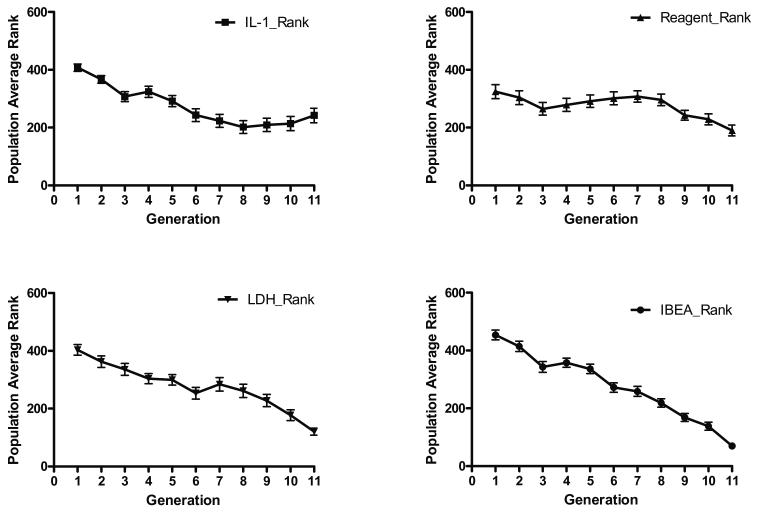
Convergence of solutions derived from the IBEA’s search of the chemical library was assessed by measuring the population average rank of the IBEA hypervolume (see supplementary methods) and the three objective functions (IL-1β expression, number of component reagents within combinations and LDH release (Fig. 3). The observed plateau in IL-1β expression between generations 9 and 10 and very marginal decreases in LDH decrease (Fig 3 top and bottom left respectively) were triggers to halt the IBEA-directed search. By this stage, although not provably globally optimal, almost all IL-1β expression had been ablated by some combinations, with negligible toxicity.
Figure 3.
Population average rank for inhibition of IL-1β expression (top left), number of component reagents in combinations (top right), LDH release (bottom left) and overall IBEA hypervolume (bottom right) respectively. Error bars are the standard errors. The IBEA hypervolume is a composite (see Methods; Implementation of the Indicator Based Evolutionary Algorithm (IBEA)) of the performance of the different generations with regard to the three objectives, a smaller number being better.
A particular strength of this algorithm is the ability to add and remove reagents to/from the library during the evolution of the combinations 23 (see supplementary methods – reagent removal / addition and data analysis), and generations 10 and 11 explored these a little further. We also noted that many of the more successful reagent combinations contained the p38 MAPK inhibitor SB203580.
Post-hoc analysis of the IBEA search of combinatorial chemical space
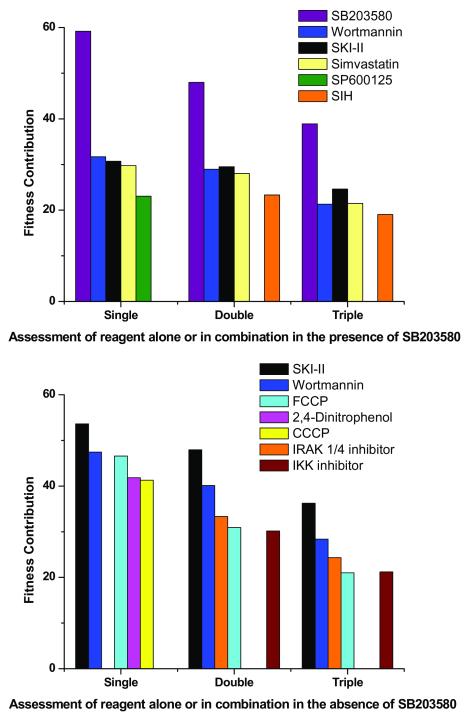
The search of combinatorial chemical space yielded 51 and 188 reagent combinations that exhibited an inhibition of IL-1β expression that was greater than or equal to either 95 % or 70 % of the control response, respectively (Supplementary Table 3 and 4, (Top)) across all generations. We chose to explore the inhibitory activities of other reagents independently of SB203580 because these effects might have been ‘masked’ in the EA by the dominance of SB203580 (70% (35/50)) of sampled combinations at generation 10 contained SB203580). Thus, a ‘post hoc’ analysis was conducted of all data to assess the fitness contributions of single reagents (their overall score against our defined objectives) alone and in two- and three- component combinations (in the presence and absence of SB203580) for all reagents (Fig. 4 and see Methods). These component reagents included the p38 MAPK inhibitor (SB203580), a sphingosine kinase inhibitor, (SKI-II), a statin (simvastatin), an iron chelator (SIH) and an inhibitor (BMS-345541) of the inhibitory κB kinase (IKKi). Inhibitors of p38 MAPK and IKK are well established as inhibitors of IL-1β expression, and (as well as its effects on HMG-CoA reductase) simvastatin is a known anti-inflammatory and an abundance of evidence implicates poorly liganded iron in inflammatory processes 24,25. However, the appearance of SKI-II may have been unexpected, albeit that there is evidence for the involvement of at least one sphingosine kinase in inflammation 26. Despite the appearance of both wortmannin and the mitochondrial uncouplers in the post-hoc analysis, these compounds were not pursued further owing to their lack of specificity and potential toxicity, respectively. The ability to observe substantial inhibition of IL-1β expression with just pairs of inhibitors (Supplementary Table 3 and 4 (bottom)) led us to study the concentration-dependent pair-wise optimization of the top-ranked reagents.
Figure 4.
Analysis of all EA generations (1 – 11) in the presence (top) and absence (bottom) of SB203580 yielded a rank order for the fitness contribution (see Methods; Post-hoc analysis of IBEA search and calculation of reagent fitness) of each reagent within the library. Only five top ranked reagents are displayed here either alone (i.e. single) or in double or triple combinations in the presence and absence of SB203580.
Concentration-dependent search reveals combinatorial synergism
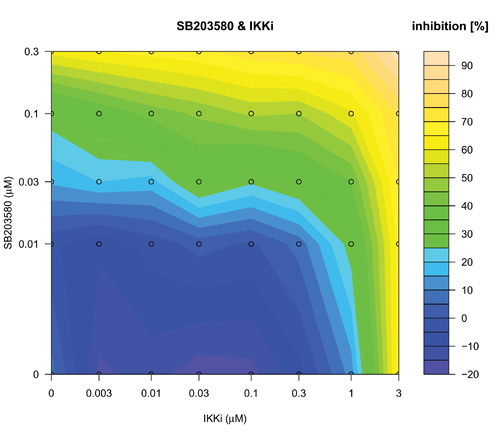
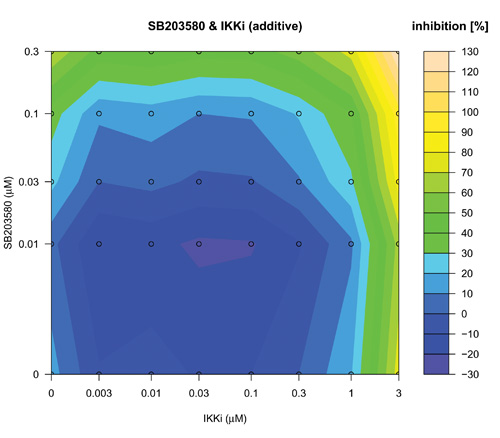
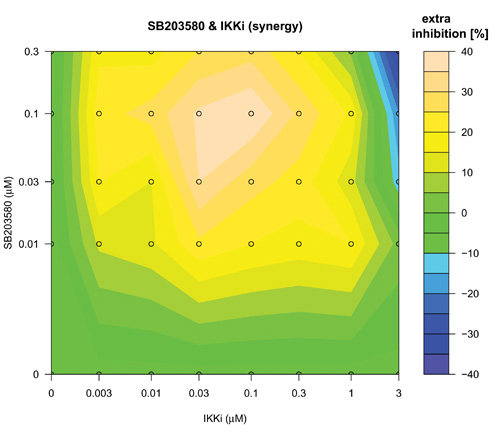
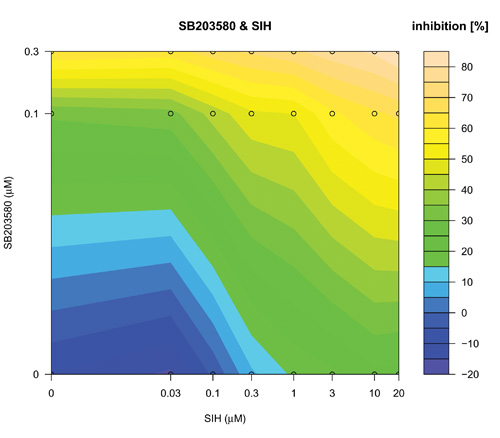
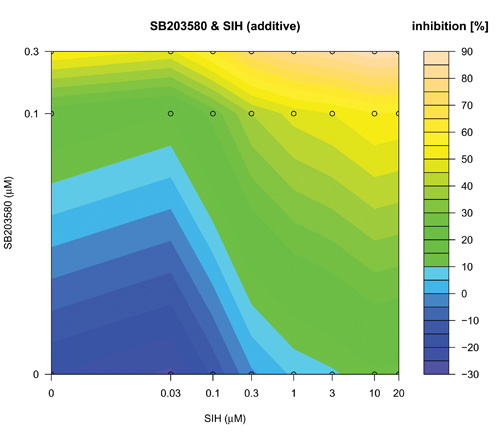
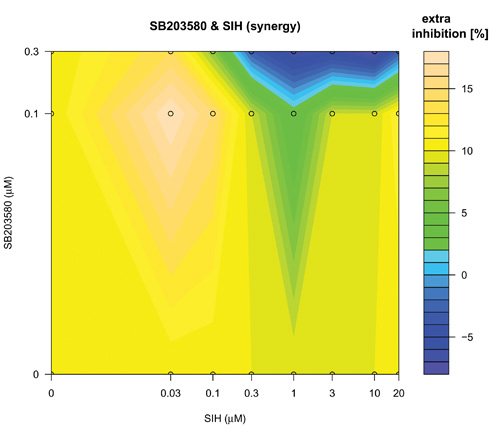
The search over defined concentration ranges of all pairs of five top-ranked reagents was assessed using an adaptive dose matrix search protocol (Figure 1, Loop 2 and Supplementary Fig. 4). Briefly, this protocol adaptively changes the concentrations of chosen reagents (See methods and compare SB203580 and SIH (Supplementary Fig. 4) versus the same combination presented here (Fig. 5)). Reagents were assessed alone and in pairs (Fig 5). Modes of pharmacological effect driven by reagent combinations have their own nomenclature 27-29. Hence, additivity is the linear superposition of two different reagent effects and synergy a non-linear (excess) inhibition from a reagent combination beyond that expected for simple additivity 27. Evidence of synergy (Fig 5(IKKi; c) or 5(SIH; f)) was determined by subtraction of predicted additive effects (Fig 5(IKKi; b) or 5(SIH; e)) of each combination (based on single reagent efficacy) from the actual experimental data (Fig 5(IKKi; a) or 5(SIH; d)). Synergy was observed for most combinations; however, we noted in some instances that this was in fact attributable to a loss of a potentiating effect on IL-1β expression that occurred with either of the reagents when used alone. In these cases, the net (i.e. resulting) magnitude of IL-1β inhibition was minimal and therefore we focus here on examples of combinatorial synergy generating biologically-relevant levels of IL-1β inhibition. SB203580 (0.1 μM) and IKKi (1 μM) alone inhibited IL-1β expression by 28 ± 7 % and 12 ± 9 % respectively (mean ± SEM, n=7) and SB203580 (0.1 μM) and IKKi (1 μM) in combination achieved significantly greater inhibition than did either drug alone (59 ± 5 %, n=7, p< 0.02 and p< 0.0005, one way ANOVA with Tukey’s multiple test correction versus SB203580 or IKKi alone respectively). The inhibitory effect of SB203580 (0.1 μM) and IKKi (1 μM) in combination was 19% greater than that predicted for a purely additive effect thus demonstrating a marked synergistic interaction at these concentrations (Fig. 5(c)). Similarly, SB203580 (0.1 μM) and SIH (3 μM) alone inhibited IL-1β expression by 31 ± 10 % and 19 ± 8 % respectively (n=7 plates) and combinatorially inhibited IL-1β expression by a significantly greater magnitude (59 ± 4 %, n=7 plates, p<0.04 and p< 0.004, one way ANOVA with Tukey’s multiple test correction versus SB203580 and SIH alone, respectively). This combinatorial inhibitory effect was 9% greater than that predicted for pure additivity therefore indicating a synergistic interaction (Fig. 5(f)). We also observed marked synergistic effects for combinations of IKKi and SKI-II, IKKi and SIH and SB203580 and SIH (Supplementary Fig. 4), To assess whether a triple combination of SB203580, IKKi and SIH could provide an inhibition of IL-1β expression beyond the synergy already observed for both paired combinations (i.e SB203580 with either IKKi or SIH) we superimposed increasing concentrations of SIH (0, 0.1, 0.3, 1 and 3 μM) onto SB203580 and IKKi dose matrices (Supplementary Fig. 5). We did not observe any further synergy with the triple-combination.
Figure 5.
Concentration-dependent adaptive dose matrix optimization of paired reagent combinations was achieved by adaptively changing the concentrations of reagents after assessing the inhibition of IL-1β expression. A p38 MAPK inhibitor (SB203580) and an Iκ Kinase inhibitor (IKKi) (a) or an iron chelator (SIH) (d) were assessed alone and as paired combinations. Potential synergy of the SB203580 & IKKi (c) and SB203580 & SIH (f) combinations were revealed by subtraction of simple additive effects ((b) and (e); calculated from single reagent data in the absence of the other reagent respectively) of the respective combinations from the experimental data (a) & (d). Synergistic inhibition of IL-1β expression was revealed with the combinations of SB203580 & IKKi (c) and SB203580 & SIH (f) as an ‘extra’ inhibition additional to the additive inhibitions of the individual reagents taken together.
Discussion
There is a growing recognition that drugs, to be effective whether singly or in combination, must affect multiple steps simultaneously 7,10,30,31. This, however, leads immediately to a combinatorial explosion of experimental possibilities that limits the number of drugs that can reasonably be tested exhaustively. We have applied a multi-objective evolutionary algorithm (EA) to the optimization of reagent combinations, using a panel of candidate reagents selected from our own studies and the literature 32 as targeting the pro-inflammatory IL-1β expression network. The objective assessment of reagent combinations arising from the IBEA-directed search utilizing the ‘post hoc’ analysis of reagent fitness contributions is useful as it removes a layer of decision making 20, reducing bias and potentially enhancing ‘hidden’ phenomena (e.g. off target effects of reagents) that may have beneficial effects on the system output (i.e. IL-1β expression). In this regard, it is worth mentioning the increasingly effective use of adaptive dosing regimes in clinical trials of pharmaceutical drugs both singly and in combinations 33.
Combination therapy is now returning to the fore with a greater understanding of pharmacological mechanism being uncovered by advances in parallel measurements of biological endpoints 11,34,35 The use of the Gur-game stochastic search algorithm has been reported 36,37 in elucidating the anti-viral and NF-κB-activating efficacy of drug and cytokine combinations, respectively. Briefly, this algorithm functions by generating a random number (e.g. one representing a specified anti-viral activity) and switching the concentration of component drugs if their efficacy is below this value. In contrast, stochastic and deterministic elements of our search were based on experimental data output and recombination of reagents in new cocktails that had not yet been evaluated. In addition, the multi-objective nature of the EA-driven search presented here allows assessment of a number of biological endpoints. Following this approach with an adaptive dose-matrix driven search enabled a search of pharmacological space using just top-ranked ‘hits’. Applications of search algorithms and the use of machine learning in the optimization of combinatorial therapies have recently been reviewed 38, and within their categorization our method would fall into “E – Model-free Biological Search”.
Recently, two papers have cited the pairing of reagent combinations as indicative / predictive of higher order effects 39,40. The latter 40 used a series of chemotherapeutic reagents to monitor for additivity, antagonism or synergy of combinations on YFP-tagged protein dynamics in the H1299 cell line. The authors propose that a linear superposition of weighted sums from the effect single drugs have on protein dynamics can predict higher order (i.e. combinatorial) effects for these reagents, although they were unable to demonstrate this for Wortmannin (a PI3K inhibitor). The former 39 looked for an enhancement in the [Ca2+]i signal of platelets using a pairwise agonist scanning approach of six reagents at three different concentrations. Using these data the authors trained a neural network model to predict higher order effects and were successful in doing so. However, the rapid and non-transcriptional signal transduction required for Ca2+ mobilization may not be entirely reflective of multi-protein signalling networks, where there is extensive cross-talk and feedback loops that modulate responses on timescales ranging in minutes to hours and beyond. In the present case, we found a substantial inhibition of IL-1β production by the p38 MAPK inhibitor SB203580, that was enhanced synergistically by either the IKK inhibitor BMS 345541 or the iron chelator SIH 41. Perhaps surprisingly, the triple combination of SB203580, IKKi and SIH did not reveal additional effects beyond those observed for pairwise combinations of SB203580 and either of the other two reagents. The effects of iron chelation are of especial interest here, as there is abundant but widespread evidence for the role of unliganded iron in a variety of inflammatory disorders 24,25,42.
p38 inhibitors have proved disappointing in clinical trials, but this is not so much because they are not active in vivo but because (at the doses used) they lose specificity for the α isoform of p38 and are toxic 43. The particular benefit of the synergy when combining a p38 inhibitor such as SB203580 with BMS345541 or SIH observed here is that we can use them at lower concentrations in combinations than those at which they would be used alone (Fig 5). We note too that such inhibitors seem not to have been designed to exploit the specificity of pharmaceutical drug transporters 44. Finally, it was noted too the efficacy of some combinations that did not involve the p38 inhibitor.
In conclusion, the application of an EA in conjunction with semi-automated assay of a dynamic chemical library enables a rapid scanning of reagent combinations without the need of initial hypotheses 45 about likely higher-order effects. Our results show that synergistic combinations can be revealed quickly, and that these combinations survived further experimental scrutiny, leading to pairwise combinations that seem promising to use in practice. Synergism of the SB203580 and either IKKi or SIH combination presented here, in contrast to the comparatively marginal effect of the individual reagents at the same concentrations, shows that pharmacological modification of biological targets and processes may be effected at concentrations that are more likely to avoid toxicities. This has particular relevance to the treatment of chronic inflammatory conditions such as irritable bowel syndrome46 and chronic obstructive pulmonary disease 47 where treatment may be maintained for extensive periods. Our approach is essentially generic, and the time required per generation is determined by the time needed for setting up and running the assays (typically 3 days, one each for cell preparation, combination preparation and ELISA analyses), since the time needed for the algorithm to analyse the results and then to choose the cocktails for the next generation was negligible in comparison. Overall, our new method substantially decreases the time taken for the triage of pharmacologically useful chemical diversity within chemical libraries 48. Additionally, we demonstrate how combinations of known drugs or reagents could allow them to be repurposed 49 and could provide an elegant adjunct to existing therapeutic strategies in chronic inflammatory conditions.
Methods
All procedures, protocols and methods were carried out under aseptic conditions where deemed necessary.
Construction and composition of the chemical library
The choice of reagents with which to populate the chemical library searched here was guided in part by Oda and Kitano’s TLR signalling network 32 and via identification of suitable ligands from single reagent studies in peritoneal macrophages. The following pharmacological classes of reagents were used: Iron chelators, TPEN; a zinc chelator, anti / pro-oxidants, NADPH oxidase inhibitors, PI3Kinase inhibitors, MAPK pathway inhibitors, NF-κB pathway inhibitors, Genistein; a tyrosine kinase inhibitor, mitochondrial uncouplers (removed after generation 3), statins and small-GTPase inhibitors. In the evolutionary optimization process each of these reagents corresponds to a single binary variable indicating whether the reagent is included in a combination or not; the combination itself represents a candidate solution to the problem. Detailed information regarding the construction, storage, maintenance and removal and replacement of reagents within the chemical library can be found in the supplementary methods (Reagent removal / addition and data analysis).
Implementation of the Indicator Based Evolutionary Algorithm (IBEA)
In order to select a suitable multi-objective evolutionary algorithm (EA) for addressing the search of the fitness landscape of the chemical library, a comparison of four EAs; IBEA, SPEA2 and NSGA2 with binary tournament and NSGA2 with probabilistic selection 50 was undertaken 23. All EAs were assessed on a family of test problems used to simulate the reagent combination problem. The test problems model a scenario where pharmacological interactions among reagents can be described by single, binary, and ternary effects only. A reagent combination can effect minimal IL-1β expression by killing cells; measured as a large release of LDH. To de-risk the detection of lethal versus effective and benign combinations respectively, positive, negative, and no correlations were considered between these two objectives. The different levels of correlation were realized by assigning certain probabilities to effect values; first for IL-1β expression and dependent on this value a subsequent probability determining an effect value for the LDH release. Depending on the correlation level, the effect values were drawn uniformly from the interval [−1,0) and/or (0,1].
All EAs tested were capable of locating combinations of compounds of similar quality in the presence of 80 % and 10 % variability in IL-1β expression and LDH release measurements respectively. However, IBEA had the best performance at finding effective compound combinations that contained only a few compounds (although its search was unrestricted and could have used any number of compounds). This was the only EA tested that was not based on Pareto ranking; rather IBEA searches for those solutions that maximize their hypervolume within objective space. Initialization of the 1st generation of reagent compounds in IBEA was conducted by fixing the probability of compound selection to 3 / 33 to ensure a random selection of compounds from across the library, with on average three compounds in a cocktail. See the Supplementary methods (IBEA directed evolutionary search of combinations inhibiting IL-1β expression) and results (Supplementary table 2) for other details of the algorithm and its parameters.
Production and assay of reagent combinations
A Sciclone ALH3000 laboratory robot (Caliper Life Sciences) under the indirect control of an Indicator Based Evolutionary Algorithm (IBEA) enabled the semi-automated assay of chemical combinations (Supplementary methods – Production of combinatorial chemical cocktails) in LPS stimulated J774.A1 macrophages. The iterative searching and analysis of incremented generations of combinations was conducted via measurements of an IL-1β expression ELISA (R&D Systems; DY401) and LDH release (Promega) (see Supplementary methods – Measurement of LDH and IL-1β expression).
Treatment of Peritoneal and J774.A1 macrophages with single reagents and combinations
Peritoneal and J774.A1 macrophages were prepared and cultured (see supplementary methods) for either single reagent or combinatorial and dose matrix studies respectively. Peritoneal macrophages were exposed to either single reagents (0.01 μM - 100 μM) or DMSO (0.5 % v/v) for 0.5 h prior to stimulation with LPS (1 μg / mL). Similarly, J774.A1 macrophages were treated with chemical combinations (3 μM or varying concentration) or DMSO (0.5 % v/v or 0.1% v/v) during the EA-directed and adaptive dose-matrix search respectively for 10 min prior to stimulation with LPS (1 μg / mL). After 4 h (Peritoneal) or 2 h (J774.A1) aliquots of well supernatants were taken for the measurement of LDH during single-reagent and EA driven combinatorial assessment respectively (see supplementary methods) prior to disposal of remaining supernatant, lysis of cells and freezing before measurement of IL-1β expression (see Supplementary methods – Measurement of LDH and IL-1β expression).
Post-hoc analysis of IBEA search and calculation of reagent fitness
Calculation of the fitness contribution of a single reagent within a combination was assigned as follows (1):
| (1) |
Where; Fi is the fitness contribution of any given single reagent (i), and where and are the mean IL-1β expression values of all combinations where this single reagent (i) was present or absent respectively. Thus, a larger fitness contribution Fi indicates that a reagent is more efficient in decreasing IL-1β expression.,
Concentration-dependent optimization of paired reagent combinations using an adaptive dose matrix search protocol
Upon completion and post-hoc prioritization of reagent combinations from the IBEA search a concentration-dependent optimization step was implemented. Briefly, to assess the potentially synergistic effects of paired combinations on IL-1β expression we serially and logarithmically decreased the test concentrations of reagents from those used during the EA-directed search. Similarly, after this initial optimization step, we extended the scanned concentration ranges of promising combinations by adding in test concentrations of reagents at approximate 0.5 log10 spacings within the dose-matrix. This allowed effect (i.e. IL-1β expression) comparisons at multiple doses of paired reagents. Pseudocolor mappings were performed by linear interpolation between samples; those mappings that move away from the blue end of the spectrum within combination response shape plots are indicative of synergistic inhibition of IL-1β expression between two reagents. (Figure 5, Supplementary Fig 4).
Supplementary Material
Acknowledgements
We would like to thank the BBSRC for funding this project (BB/F018398/1) and the BBSRC/EPSRC and Wellcome Trust for the funding of studentships and fellowships. We thank the Manchester Centre for Integrative Systems Biology for providing access to the Sciclone instrument. BGS would like to thank Dr. Susan Wasley and Dr. Darren Stubbs of Caliper Life Sciences for their help and assistance in commissioning and their image of the Sciclone instrument (Figure 1) respectively. The images of the computer and 96-well plate (Figure 1) were used under the creative commons CC0 public domain dedication and obtained from Clker.com.
Footnotes
Competing Financial Interests Statement DBK transferred his responsibility as Principal Investigator of BBSRC Grant BB/F018398/1 to NJR and PM prior to taking up his appointment as Chief Executive of the BBSRC on 1st October 2008. NJR is a non-executive director of AstraZeneca. BGS as a former employee of AstraZeneca and Eli Lilly and Company is a shareholder of stock from both companies.
References
- 1.Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–82. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–147. [PubMed] [Google Scholar]
- 4.Luheshi NM, Rothwell NJ, Brough D. Dual functionality of interleukin-1 family cytokines: implications for anti-interleukin-1 therapy. Brit J Pharmacol. 2009;157:1318–29. doi: 10.1111/j.1476-5381.2009.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relton JK, Rothwell NJ. Interleukin-1 receptor antagonist inhibits ischaemic and excitotoxic neuronal damage in the rat. Brain Res Bull. 1992;29:243–6. doi: 10.1016/0361-9230(92)90033-t. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y-C, Yeh W-C, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–51. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 8.Fell DA, Thomas S. Physiological control of metabolic flux: the requirement for multisite modulation. Biochem J. 1995;311(Pt 1):35–9. doi: 10.1042/bj3110035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon YJ, et al. Dexamethasone inhibits IL-1beta gene expression in LPS-stimulated RAW 264.7 cells by blocking NF-kappaB/Rel and AP-1 activation. Immunopharmacology. 2000;48:173–183. doi: 10.1016/s0162-3109(00)00199-5. [DOI] [PubMed] [Google Scholar]
- 10.Léhar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–81. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8:567–78. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 12.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24:805–15. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 13.Coello CAC, Lamont GB, Veldhuizen DAV. Evolutionary algorithms for solving multi-objective problems. Springer; 2007. [Google Scholar]
- 14.Handl J, Kell DB, Knowles J. Multiobjective optimization in bioinformatics and computational biology. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:279–92. doi: 10.1109/TCBB.2007.070203. [DOI] [PubMed] [Google Scholar]
- 15.Knowles J, Corne D, Deb K. Multiobjective problem solving from nature: from concepts to applications. Springer; Heidelberg: 2008. [Google Scholar]
- 16.Bäck T, Fogel DB, Michalewicz Z. Handbook of evolutionary computation. Institute of Physics Pub.; 1997. [Google Scholar]
- 17.Goldberg DE. The design of innovation: lessons from and for competent genetic algorithms. Kluwer Academic Publishers; 2002. [Google Scholar]
- 18.Knight CG, et al. Array-based evolution of DNA aptamers allows modelling of an explicit sequence-fitness landscape. Nucleic Acids Res. 2008;37 doi: 10.1093/nar/gkn899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Hagan S, Dunn WB, Brown M, Knowles JD, Kell DB. Closed-loop, multiobjective optimization of analytical instrumentation: gas chromatography/time-of-flight mass spectrometry of the metabolomes of human serum and of yeast fermentations. Anal Chem. 2005;77:290–303. doi: 10.1021/ac049146x. [DOI] [PubMed] [Google Scholar]
- 20.Knowles J. Closed-Loop Evolutionary Multiobjective Optimization. IEEE Comput Intell M. 2009;4:77–91. [Google Scholar]
- 21.Zitzler E, Künzli S. Indicator based selection in multi-objective search. In: Yao X, editor. Parallel problem solving from nature - PPSN VIII : 8th international conference, Birmingham, UK, September 18-22, 2004 : proceedings; Berlin; [London]: Springer; 2004. pp. 832–842. [Google Scholar]
- 22.Allmendinger R, Knowles J. Analysis of Several Evolutionary Algorithms on the Noisy Three-Objective Chemical Mixture Optimization Problem. 2009:1–9. Technical report MLO-12009. http://www.cs.man.ac.uk/~allmendr/publications.html.
- 23.Allmendinger R, Knowles J. Evolutionary Optimization on Problems Subject to Changes of Variables. In: Schaefer R, Cotta C, Kolodziej J, Rudolph G, editors. Parallel Problem Solving from Nature – PPSN XI. Springer; Berlin / Heidelberg: 2010. pp. 151–160. Vol. 6239. [Google Scholar]
- 24.Kell D. Iron Behaving Badly: Inappropriate Iron Chelation as a Major Contributor to the Aetiology of Vascular and Other Progressive Inflammatory and Degenerative Diseases. BMC Med Genomics. 2009;2:2. doi: 10.1186/1755-8794-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kell DB. Towards a unifying, systems biology understanding of large-scale cellular death and destruction caused by poorly liganded iron: Parkinson’s, Huntington’s, Alzheimer’s, prions, bactericides, chemical toxicology and others as examples. Arch Toxicol. 2010;84:825–889. doi: 10.1007/s00204-010-0577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puneet P, et al. SphK1 regulates proinflammatory responses associated with endotoxin and polymicrobial sepsis. Science. 2010;328:1290–4. doi: 10.1126/science.1188635. [DOI] [PubMed] [Google Scholar]
- 27.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacological reviews. 1995;47:331–85. [PubMed] [Google Scholar]
- 28.Léhar J, et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat Biotechnol. 2009;27:659–66. doi: 10.1038/nbt.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimmermann GR, Léhar J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Ejim L, et al. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol. 2011;7:348–50. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]
- 31.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100057. 2006 0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bornkamp B, et al. Innovative approaches for designing and analyzing adaptive dose-ranging trials. J Biopharm Stat. 2007;17:965–95. doi: 10.1080/10543400701643848. [DOI] [PubMed] [Google Scholar]
- 34.Jia J, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8:111–128. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 35.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 36.Sun C-P, et al. Integrative systems control approach for reactivating Kaposi’s sarcoma-associated herpesvirus (KSHV) with combinatory drugs. Integrative Biology. 2009;1:123–130. doi: 10.1039/b815225j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong PK, et al. Closed-loop control of cellular functions using combinatory drugs guided by a stochastic search algorithm. Proc Natl Acad Sci USA. 2008;105:5105–10. doi: 10.1073/pnas.0800823105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feala JD, et al. Systems approaches and algorithms for discovery of combinatorial therapies. Wiley interdisciplinary reviews. Systems biology and medicine. 2010;2:181–93. doi: 10.1002/wsbm.51. [DOI] [PubMed] [Google Scholar]
- 39.Chatterjee MS, Purvis JE, Brass LF, Diamond SL. Pairwise agonist scanning predicts cellular signaling responses to combinatorial stimuli. Nat Biotech. 2010;28:727–32. doi: 10.1038/nbt.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geva-Zatorsky N, et al. Protein dynamics in drug combinations: a linear superposition of individual-drug responses. Cell. 2010;140:643–51. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Horackova M, Ponka P, Byczko Z. The antioxidant effects of a novel iron chelator salicylaldehyde isonicotinoyl hydrazone in the prevention of H2O2 injury in adult cardiomyocytes. Cardiovas Res. 2000;47:529–36. doi: 10.1016/s0008-6363(00)00088-2. [DOI] [PubMed] [Google Scholar]
- 42.Sindrilaru A, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammaker D, Firestein GS. “Go upstream, young man”: lessons learned from the p38 saga. Ann Rheum Dis. 2010;69:77–82. doi: 10.1136/ard.2009.119479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobson PD, Kell DB. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nat Rev Drug Discov. 2008;7:205–20. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 45.Kell DB, Oliver SG. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays. 2004;26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 46.Scully P, et al. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am J Gastroenterol. 2010;105:2235–43. doi: 10.1038/ajg.2010.159. [DOI] [PubMed] [Google Scholar]
- 47.Löfdahl C-G. COPD and co-morbidities, with special emphasis on cardiovascular conditions. Clin Respir J. 2008;2(Suppl 1):59–63. doi: 10.1111/j.1752-699X.2008.00085.x. [DOI] [PubMed] [Google Scholar]
- 48.Akella LB, DeCaprio D. Cheminformatics approaches to analyze diversity in compound screening libraries. Curr Op Chem Biol. 2010;14:325–30. doi: 10.1016/j.cbpa.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 49.Tobinick EL. The value of drug repositioning in the current pharmaceutical market. Drug News Perspect. 2009;22:119–25. doi: 10.1358/dnp.2009.22.2.1303818. [DOI] [PubMed] [Google Scholar]
- 50.Zitzler E, et al. Evolutionary Multi-Criterion Optimization. Springer; Berlin / Heidelberg: 2001. Evolutionary Multi-objective Ranking with Uncertainty and Noise; pp. 329–343-343. Vol. 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.