Abstract
Iron acquisition is an absolute requirement by most microorganisms for host survival. In this work, we investigated the Campylobacter jejuni iron binding Dps protein for a potential role in virulence. In vitro assays using J774A.1 macrophage-like cells demonstrated a 2.5 log reduction in C. jejuni survival of the Dps mutant and a reduction of four logs in invasion of HEp-2 epithelial cells compared to the wild-type strain. To examine the role of the dps gene in host pathogenesis, the piglet model was used in C. jejuni challenge studies. In vivo inoculation studies of newborn piglets with wild-type C. jejuni demonstrated an 11-fold upregulation of the dps gene and intestinal lesion production typical of campylobacteriosis in humans. In contrast, piglets inoculated with the dps mutant were not colonized and remained normal throughout the study period. Mucosal lesion production was restored in piglets inoculated with the complemented Dps mutant strain. Based on these results, we conclude that the C. jejuni Dps homolog is a virulence factor in the production of campylobacteriosis, and warrants further investigation.
Introduction
Campylobacteriosis caused by Campylobacter jejuni is the leading cause of bacterial gastroenteritis in the United States, causing an estimated 2.4 million cases annually (Altekruse et al., 1999; Mead et al., 1999; Samuel et al., 2004). The infectious dose is highly variable, ranging from 500 to 106 organisms (Steele and McDermott, 1978). Once ingested, a short incubation period of 24–72 h occurs, followed by an onset of symptoms marked with severe acute watery diarrhea occasionally with blood, variable fever, myalgia, and headache.
Studies on the Dps protein of Helicobacter pylori, a close relative of C. jejuni, have shown many functions linked to stress survival both in vitro and in vivo. These functions include iron binding (Tonello et al., 1999), DNA binding (Ceci et al., 2007), oxidative stress survival (Cooksley et al., 2003), and immune modulation (Amedei et al., 2006; Codolo et al., 2008; Del Prete et al., 2008).
In a previous study, a Dps homolog in C. jejuni was shown to bind iron up to 40 atoms per Dps monomer (Ishikawa et al., 2003). Additionally, the authors showed a role for C. jejuni Dps in hydrogen peroxide stress resistance, as well as evidence for constitutive expression under several in vitro conditions. Lastly, researchers have shown that C. jejuni Dps can also bind to the myelin sheath and nodes of Ranvier of rat nerves, resulting in paranodal myelin detachment and axonal degeneration, suggesting a possible role for this protein in the development of Guillain-Barre syndrome in individuals (Piao et al., 2009, 2010).
In this work, we expand the initial investigations on the C. jejuni Dps gene, and provide evidence that the Dps protein is a significant factor in C. jejuni colonization and survival. In vitro work using a J774A.1 macrophage-like and a HEp-2 epithelial cell line indicate a role for the Dps protein in extended intra-macrophage survival and invasion of epithelial cells. Additionally, we show transcriptional upregulation of dps during infection of piglets, and a loss of virulence in the piglet model after challenge with a Dps deficient mutant.
Materials and Methods
Culture of bacterial strains
All strains used in the study are provided in Table 1. Unless stated otherwise, all Campylobacter strains were routinely cultured on Mueller Hinton agar (BD, Sparks, MD) supplemented with 5% citrated bovine blood (Cleveland Scientific, Bath, OH) (MHB) and incubated under a normal atmosphere supplemented with 10% CO2 at 42°C. Escherichia coli DH5α was routinely cultured on Luria Bertani agar (BD) under a normal atmosphere at 37°C. As appropriate, the medium was supplemented with antibiotics at the following concentrations: chloramphenicol 30 μg/mL, kanamycin 50 μg/mL, and ampicillin 100 μg/mL.
Table 1.
Strains and Plasmids Used in This Study
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | endA1 hsdR17 (rk- mk-) supE44 thi-1 | Invitrogen |
| recA1 gryA relA1 Δ(lacZYA-argF)U169 | ||
| deoR [(80dlacΔ(lacZΔ M15)] | ||
| Campylobacter jejuni | ||
| NCTC11168 | C. jejuni NCTC 11168 | NCTC |
| JRT10 | NCTC 11168 1534c::cmr | This study |
| JRT101 | NCTC 11168 1534c::cmr (pJRT101) | This study |
| Plasmids | ||
| pHSS19 | Nonreplicating suicide vector derived from pUC19 | Nickoloff and Reynolds (1991) |
| pSJB49 | pHSS19 carrying cmr | This study |
| pJRT10 | pSJB49 carrying Cj1534c::cmr | This study |
| pRY107 | Complementation vector; kanr | Yao et al. (1993) |
| pJRT101 | pRY107 carrying Cj1534c; kanr | This study |
cmr, chloramphenicol resistance gene; kanr, kanamycin resistance gene.
Mutation and complementation
C. jejuni chromosomal DNA was extracted using the Wizard genomic DNA purification kit (Promega, Madison, WI). Primers Cj1534KOF1 and Cj1534KOR1 (Table 2) were used to amplify a 982-base pair (bp) fragment containing the initial 96 bp of the dps gene and the flanking DNA. Primers Cj1534KOF2 and Cj1534KOR2 (Table 2) were used to amplify a second 843-bp fragment containing the terminal 110 bp of the dps gene and flanking DNA. The resultant inserts were digested with PstI–XbaI and BamHI–EcoRI, respectively. The two fragments were consecutively cloned into pSJB49, a pHSS19 derivative that contains the Campylobacter chloramphenicol acetyltransferase (cat) gene cloned between the BamHI and XbaI sites (Nickoloff and Reynolds, 1991). Polymerase chain reaction (PCR) and sequencing of the cloned products were used to confirm the construct. Once confirmed, the construct was introduced into C. jejuni via electroporation (1.25 kV, 600Ω and 25 μF). Electroporated cells were plated on MHB and incubated for 16h. After incubation, cells were harvested and mutants were selected from growth on MHB supplemented with chloramphenicol (30 μg/mL), and confirmed via PCR.
Table 2.
Primers Used in This Study
| Primer | DNA sequence (5′ to 3′) |
|---|---|
| Cj1534KOF1 | GGATTCAAAACTGCAGCAAGAAGGTG |
| Cj1534KOR1 | TTGCAAACCTTCTAGATTCCAGTGAT |
| Cj1534KOF2 | AAAAAGAAAGGGATCCTACAACAGCT |
| Cj1534KOR2 | TTTTAAGGTAGAATTCACATAAGTAT |
| Cj1534TCF1 | TTCTTAATCAGAATTCATTAAAAAAG |
| Cj1534TCR1 | TTTCAATTTTATCGATTAAAAAAGGA |
| Cj0402RTP1 | CGATGGAACGGATAATCACC |
| Cj0402RTP2 | AATACCTGCATTTCCAAGAGC |
| Cj1534RTP1 | AAAAAGAAAGTGATACTACAACAGCT |
| Cj1534RTP2 | AAGCACCTTGTAAAGTAGCGCCTATC |
| IpxAC.jejuni | ACAACTTGGTGACGATGTTGTA |
| IpxARKK2m | CAATCATGDGCDATATGASAATAHGCCAT |
The mutation was complemented in trans using the replicative plasmid pRY107 (Yao et al., 1993). Briefly, primers Cj1534TCF1 and Cj1534TCF2 (Table 2) were used to amplify the entire dps gene, plus 126 bases upstream and 60 bases downstream of the gene. The insert was digested with EcoRI–ClaI, and cloned into pRY107, creating pJRT101. The insertion was confirmed by PCR and sequencing. This plasmid was then introduced into C. jejuni via electroporation using previously described settings. Electroporated cells were plated on MHB for 16 h at 42°C. After incubation, cells were harvested and transformants were selected from growth on MHB supplemented with kanamycin (50 μg/mL). PCR was used to confirm the presence of the plasmid.
RNA isolation
For plate growth, C. jejuni was grown on MHB plates for 72 h under routine culture conditions. Postincubation, cells were harvested in phosphate-buffered saline (PBS) and combined with an equal volume of RNA protect (Qiagen, Valencia, CA). For pig samples, total colonic contents, including mucosal scrapings and fecal material, were harvested from piglets 4 days postchallenge with wild-type C. jejuni NCTC11168 and resuspended in a 50/50 PBS-RNA protect solution. Colon contents were then centrifuged (1000 g, 10 min) to pellet large debris. The supernatant was then filtered through a 0.8-μm filter, and centrifuged (10,000 g, 15 min) to pellet the filtered C. jejuni. Pelleted material containing C. jejuni was retained and stored on ice until RNA extraction was performed. RNA from harvested samples was isolated using RNeasy Mini Kits (Qiagen) and E.Z.N.A. RNase Free DNase I kit (Omega, Norcross, GA) as per manufacturers' instructions. Total RNA collected was quantified spectrophotomically and screened by PCR to ensure that no contaminating DNA was present in the extracted material.
Reverse transcription and real-time PCR
Reverse transcription (RT) of 250 ng total isolated RNA was carried out in 10 μL reactions using qScript cDNA SuperMix (Quanta, Gaithersberg, MD) as per manufacturer's instructions. Reaction conditions were as follows: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. All cDNA samples were stored at 4°C until processed. Gene amplification and real-time analysis were carried out using a Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA). One microliter of cDNA and 800 nM concentrations of primers Cj1534RTP1 and Cj1534RTP2 (Table 2) were used in 20 μL reactions using PerfeCta SYBR Green FastMix (Quanta). Reactions were carried out as follows: 5 min at 95°C, followed by 40 cycles of 30 sec at 95°C, and 1 min at 57.5°C. To determine PCR efficiencies, standard curves were generated using 10-fold serial dilutions of genomic DNA and their respective threshold cycles. To account for variances in total RNA used and RT efficiency, the gene Cj0402 (primers Cj402RTP1 and Cj402RTP2) (Table 2) was used as an internal control, as its expression was found to be constant during piglet colonization (Joens, unpublished data). All results were analyzed using the Pfaffl method (Pfaffl, 2001). Each sample was analyzed in triplicate, with the averages presented.
C. jejuni growth curves
Mueller Hinton broth was inoculated with C. jejuni to a final OD600 of 0.01, and incubated under standard conditions with aeration (150 RPM). Enumeration was performed at 3 h intervals, by plating 10-fold serial dilutions. Titers obtained were used to generate growth curves, and subsequently the doubling time for each strain in this study. Each strain was tested in three independent experiments.
Intra-macrophage survival assays
Intra-macrophage survival assays were performed using a modification of a method described elsewhere (Day et al., 2000). Briefly, 24-well polystyrene plates were seeded with 2×105 J774A.1 murine macrophage-like cells and incubated for 24 h in 5% CO2 at 37°C. The resulting monolayers were washed twice with Dulbecco's modified Eagle's medium (DMEM; Cellgro, Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Atlas Biologicals, Fort Collins, CO) (DMEM+10% FBS) and inoculated with 1×107 colony forming units (CFU) C. jejuni in DMEM+10% FBS. Cells were incubated as above for 21, 45, or 69 h. After incubation, cells were rinsed once with DMEM+10% FBS and incubated as above an additional 3 h in DMEM+10% FBS and 250 μg/mL gentamicin. Postincubation, cells were rinsed three times with PBS, and lysed with 0.5% sodium deoxycholate. Ten-fold serial dilutions were performed on the lysates and the lysates plated to determine surviving bacteria. Assays were repeated in three independent experiments.
HEp-2 epithelial cell attachment and invasion assay
The attachment assay was performed as described by Monteville et al. (2003). Briefly, 24-well polystyrene plates were seeded with 2×105 HEp-2 cells and incubated for 24 h in 5% CO2 at 37°C. The resulting monolayers were washed twice with Eagle's minimum essential media (EMEM; Cellgro, Herndon, VA) supplemented with 10% FBS (EMEM+10% FBS) and inoculated with 1×107 CFU C. jejuni in EMEM+10% FBS. Cells were incubated 3 h as above, rinsed three times with EMEM+10% FBS, and incubated an additional 3 h in EMEM+10% FBS. For invasion assays, cells were incubated 3 h, rinsed three times with EMEM+10% FBS, and incubated an additional 3 h in EMEM+10% FBS and 250 μg/mL gentamicin. After incubation, cells were rinsed three times with PBS, and then lysed with 0.5% sodium deoxycholate. Lysates were serially diluted, plated on MHB agar, and incubated under standard conditions. The assay was repeated in three independent experiments.
Piglet model
Piglets were challenged with C. jejuni as described previously (Babakhani et al., 1993). Briefly, colostrum-deprived piglets were obtained at parturition and housed in Campylobacter-free facilities. Piglets were fed a diet of 40 mL Similac® (Abbott Laboratories, Abbott Park, IL) every 4 h. At 3 days of age, piglets were challenged via oral gavage with ∼3×1010 CFU C. jejuni resuspended in Similac. Four days postchallenge, piglets were euthanized and subjected to postmortem examination. Lesions were recorded and tissue samples taken and prepared for histological examination as described by Babakhani et al. (1993).
Detection of fecal shedding of C. jejuni
To determine shedding patterns of the various strains used, daily fecal swabs were collected from all pigs postchallenge at 24 h intervals throughout the study and assayed for C. jejuni. Briefly, fecal swabs were transported to the laboratory in Cary-Blair transport media (Copan, Murrieta, CA). Swabs were placed in 20 mL of Bolton broth supplemented with 20 μg/mL sodium cefoperazone, 26.4 μg/mL trimethoprim, 20 μg/mL vancomycin, and 10 μg/mL amphotericin B for 24 h at 42°C under standard conditions. Postincubation, enrichments were assayed for the presence of C. jejuni via PCR using the C. jejuni primers IpxA and IpxARKK2m (Table 2) (Klena et al., 2004).
Statistical analysis
Attachment and invasion data were analyzed using univariable factorial analysis: strain vs. repeats. Macrophage survival data were analyzed using repeated measure analysis using strain and repeats as between factors, and the time variable served as the within factor. Macrophage data were further analyzed with the Wilks lambda test.
Animal care and use
All animal work was approved and overseen by the Institutional Animal Care and Use Committee at the University of Arizona, under protocol number 06-037.
Results
Construction and analysis of a dps mutant in C. jejuni
To determine the role of the Dps protein in pathogenesis, a dps mutant was constructed by allelic exchange in the sequenced strain NCTC11168 of C. jejuni. The C. jejuni NCTC11168 Dps mutant was complemented in trans using the vector pRY107. PCR and sequencing of the insertion product confirmed the genetic manipulations. Additionally, to demonstrate that observed effects were the result of dps mutation and not changes in growth rates from the mutation, growth curves were generated and the doubling time of the cells calculated. No difference in doubling times was observed.
Dps effects on in vitro intra-macrophage survival
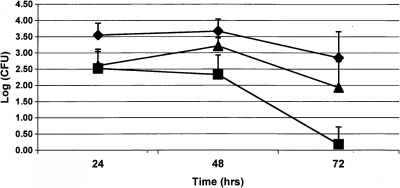
To assess the effects of Dps loss for Campylobacter survival in macrophages, in vitro J774A.1 intra-macrophage survival assays were performed using C. jejuni strain NCTC11168, NCTC11168Δdps mutant, and complemented NCTC11168Δdps (pJRT101). When compared to the wild type, the loss of the Dps protein resulted in a significant decrease (p≤0.05) at all three time points (24, 48, 72 h). Complementation of the mutant partially restored wild-type function (Fig. 1).
FIG. 1.
Intra-macrophage survival assay. Macrophage survival assays were performed using J774A.1 murine macrophage cell line and Campylobacter jejuni strains NCTC11168 (diamond), NCTC11168Δdps (square), or NCTC11168Δdps (pJRT101) (triangle) at 24, 48, and 72 h time points. All studies were repeated in three independent experiments, with the average log of viable bacteria that survived within cultured J774A.1 cells (CFU/well of 24-well plate) presented. Error bars indicate 1 standard deviation. CFU, colony forming units.
Dps effects on in vitro attachment and invasion
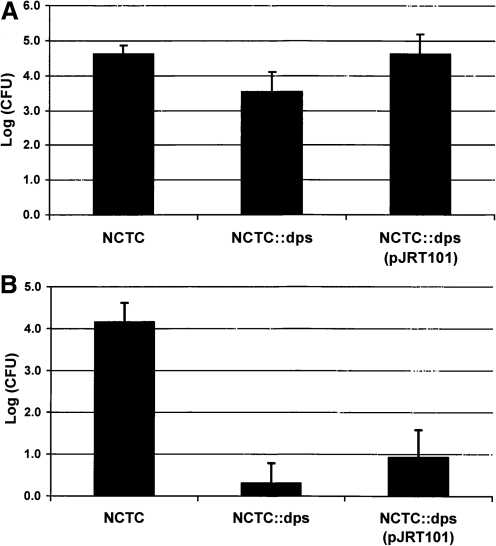
To determine if the Dps protein is involved in attachment and invasion of epithelial cells, assays were performed with HEp-2 cells and a wild-type NCTC11168, NCTC11168Δdps mutant, or complemented NCTC11168Δdps (pJRT101) C. jejuni. Results demonstrated a significant (p≤0.05) one-log decrease in attachment to the HEp-2 cells by the Dps mutant compared to the wild-type NCTC11168 strain. Complementation of the Dps mutant restored wild-type levels of attachment (Fig. 2). Examination of viable C. jejuni after exposure to gentamicin demonstrated a significant (p≤0.05) decrease in cell invasion by 4 logs with the Dps mutant compared to the wild-type strain. However, the complemented mutant failed to restore the invasive ability of the strain (Fig. 2).
FIG. 2.
Attachment and invasion assay. Effects of the loss of Dps on attachment and invasion of cultured Hep-2 cells. Attachment (A) and invasion (B) assays were repeated in three independent experiments with the average log of viable bacteria that attached or invaded cells (CFU/well of 24-well plate) presented. Error bars indicate 1 standard deviation.
Dps-deficient C. jejuni is avirulent in the porcine model of infection
To assess the phenotypic effect of the Dps mutation on pathogenesis, piglets were intragastrically inoculated with wild-type NCTC11168, NCTC11168Δ dps, or NCTC11168Δ dps (pJRT101) C. jejuni and monitored daily for fecal shedding of the bacterium. All piglets infected with the wild-type strain NCTC11168 C. jejuni were positive for shedding at all time points throughout the study, whereas shedding of the C. jejuni Dps mutant was not detected at any time points during the study. Piglets infected with the Dps complemented strain showed sporadic shedding, positive in half of the samples taken.
At necropsy, all piglets infected with C. jejuni strain NCTC11168 demonstrated mucosal lesions of catarrhal inflammation accompanied by hyperemia and petechial hemorrhage. Piglets infected with the NCTC11168 Dps mutant were normal at necropsy. Piglets infected with the NCTC11168 complemented Dps mutant had intestinal lesions that were indistinguishable from piglets infected with the wild-type strain.
Histologically, piglets infected with the wild-type and complemented strains had marked blunting of intestinal villi and mildly increased lymphocytes plus scattered neutrophils infiltrating into the lamina propria. Additionally, some fibrin was present on the intestinal mucosal surface with focal areas of epithelial erosion (Table 3). Piglets infected with the Dps mutant or negative controls lacked microscopic lesions.
Table 3.
Microscopic Lesion Development in Piglets Infected with Campylobacter jejuni
| Strain | Congested mucosa | Epithelial cell erosion | Villous degeneration |
|---|---|---|---|
| NCTC11168 | 2/4 | 3/4 | 2/4 |
| NCTC11168Δdps | 0/4 | 0/4 | 0/4 |
| NCTC11168Δdps(pJRT101) | 2/3 | 2/3 | 3/3 |
Piglets were intragastrically inoculated with ∼3×1010 colony forming units of wild-type NCTC11168, NCTC11168Δdps mutant, or complemented NCTC11168Δdps (pJRT101) Campylobacter jejuni. Results indicate number of piglets with congested mucosa, epithelial cell erosion, or villous degeneration.
C. jejuni dps is transcriptionally upregulated in piglets
RNA isolated from the intestine of piglets infected with wild-type NCTC11168 was subjected to RT real-time PCR to assess if dps transcription is increased in the in vivo piglet model as compared to in vitro growth. Results of this work yielded an 11.2-fold increase in the expression of dps in the piglet model.
Discussion
In this work, we examined the role of the Dps protein in cell survival and C. jejuni piglet pathogenesis. Beyond simply binding iron, the Dps protein has been shown to have additional roles in the cell, including protection from oxidative stress. This protection is provided through utilizing hydrogen peroxide (either generated from the Fenton reaction or exposure from the environment) for oxidation of iron or reducing the formation of hydroxyl radicals (Bellapadrona et al., 2010). Ishikawa et al. (2003) partially demonstrated this occurring in C. jejuni, when they demonstrated that a Dps-deficient mutant had an increased sensitivity to hydrogen peroxide in vitro. This work provides further evidence of this function in C. jejuni, as the Dps-deficient strain was significantly attenuated in its ability to survive in J774A.1 murine-like macrophages compared to the wild-type strain.
An additional role that has been attributed to the Dps protein is the ability to function as an adhesion, where it has been specifically shown to bind sulfated carbohydrates in vitro (Namavar et al., 1998). This ability to bind select carbohydrates may explain the one log decrease in attachment observed in this work. In addition to the decrease in attachment, a 4 log decrease in invasion was also observed. These results are possibly due to a synergistic effect of the aforementioned decrease in attachment, and an overall decrease in stress tolerance, making the strain less viable intracellularly.
Genes (as well as subsequent proteins) that are biologically relevant under certain conditions are typically upregulated during exposure to those given conditions. To determine a potential role in vivo for the Dps protein, RT real-time experiments using C. jejuni RNA isolated from infected piglets were performed to determine if upregulation of the dps gene was occurring. Results of these experiments demonstrated an 11.2-fold increase in dps expression in the host, indicating a potential role in colonization and pathogenesis.
Piglet challenge studies confirmed that C. jejuni Dps has a role in piglet colonization and Campylobacter pathogenesis. Overall results of the piglet studies indicate that the C. jejuni Dps mutant is severely attenuated, causing no gross or histological lesions. Comparatively, wild-type and complemented strains produced strong inflammatory responses accompanied by intestinal cell degradation and diarrhea. The lack of lesions appears to be from an inability to effectively colonize the piglets, as no C. jejuni was detected in fecal swabs taken from piglets challenged with the Dps mutant, whereas all wild-type challenged piglets were positive at all time points. How the loss of the Dps protein is causing this has not been definitively proven, but several contributing factors may exist. First, considering the loss of attachment and invasion in vitro, the loss of Dps may prevent the organism from escaping the lumen of the intestine into the epithelial cells, resulting in a washout of the organism within the first 24 h post-challenge. Second, introduction into a host will expose an organism to numerous stressors, including prolonged exposure to acidic environments, bile salts, and reactive oxygen species, among others. The loss of the Dps protein may have an effect of rendering the strain unfit to survive these stresses, and unable to colonize the host.
In conclusion, continuing the initial investigations of Ishikawa et al. (2003), we have demonstrated a role for the Dps protein in the pathogenesis of campylobacteriosis. Considering the myriad of functions that have been attributed to the dps gene, particularly in the closely related H. pylori, further research on its role in C. jejuni infections is definitely required.
Acknowledgments
The authors wish to thank Dr. Mohammad Torabi for his help with the statistical analysis and Dr. Stephen Billington for his assistance with the genetic manipulations. These studies were supported by the Food Safety Research Response Network, a Coordinated Agricultural Project, funded through the National Research Initiative (now National Institute for Food and Agriculture, Agricultural and Food Research Initiative) of the USDA Cooperative State Research, Education and Extension Service, Grant number #2005-35212-15287.
Disclosure Statement
No competing financial interests exist.
References
- Altekruse SF. Stern NJ. Fields PI. Swerdlow DL. Campylobacter jejuni—an emerging foodborne pathogen. Emerg Infect Dis. 1999;5:28–35. doi: 10.3201/eid0501.990104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedei A. Cappon A. Codolo G. Cabrelle A. Polenghi A. Benagiano M. Tasca E. Azzurri A. D'Elios MM. Del Prete G. de Bernard M. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Investig. 2006;116:1092–1101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babakhani FK. Bradley GA. Joens LA. Newborn piglet model for campylobacteriosis. Infect Immun. 1993;61:3466–3475. doi: 10.1128/iai.61.8.3466-3475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellapadrona G. Ardini M. Ceci P. Stefanini S. Chiancone E. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic Biol Med. 2010;48:292–297. doi: 10.1016/j.freeradbiomed.2009.10.053. [DOI] [PubMed] [Google Scholar]
- Ceci P. Mangiarotti L. Rivetti C. Chiancone E. The neutrophil-activating Dps protein of Helicobacter pylori, HP-NAP, adopts a mechanism different from Escherichia coli Dps to bind and condense DNA. Nucleic Acids Res. 2007;35:2247–2256. doi: 10.1093/nar/gkm077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codolo G. Mazzi P. Amedei A. Del Prete G. Berton G. D'Elios MM. de Bernard M. The neutrophil-activating protein of Helicobacter pylori down-modulates Th2 inflammation in ovalbumin-induced allergic asthma. Cell Microbiol. 2008;10:2355–2363. doi: 10.1111/j.1462-5822.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- Cooksley C. Jenks PJ. Green A. Cockayne A. Logan RP. Hardie KR. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J Med Microbiol. 2003;52:461–469. doi: 10.1099/jmm.0.05070-0. [DOI] [PubMed] [Google Scholar]
- Day WA., Jr. Sajecki JL. Pitts TM. Joens LA. Role of catalase in Campylobacter jejuni intracellular survival. Infect Immun. 2000;68:6337–6345. doi: 10.1128/iai.68.11.6337-6345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete G. Chiumiento L. Amedei A. Piazza M. D'Elios MM. Codolo G. de Bernard M. Masetti M. Bruschi F. Immunosuppression of TH2 responses in Trichinella spiralis infection by Helicobacter pylori neutrophil-activating protein. J Allergy Clin Immunol. 2008;122:908–913. doi: 10.1016/j.jaci.2008.08.016. e905. [DOI] [PubMed] [Google Scholar]
- Ishikawa T. Mizunoe Y. Kawabata S. Takade A. Harada M. Wai SN. Yoshida S. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J Bacteriol. 2003;185:1010–1017. doi: 10.1128/JB.185.3.1010-1017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klena JD. Parker CT. Knibb K. Ibbitt JC. Devane PM. Horn ST. Miller WG. Konkel ME. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J Clin Microbiol. 2004;42:5549–5557. doi: 10.1128/JCM.42.12.5549-5557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS. Slutsker L. Dietz V. McCaig LF. Bresee JS. Shapiro C. Griffin PM. Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteville MR. Yoon JE. Konkel ME. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology. 2003;149:153–165. doi: 10.1099/mic.0.25820-0. [DOI] [PubMed] [Google Scholar]
- Namavar F. Sparrius M. Veerman EC. Appelmelk BJ. Vandenbroucke-Grauls CM. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect Immun. 1998;66:444–447. doi: 10.1128/iai.66.2.444-447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff JA. Reynolds RJ. Subcloning with new ampicillin- and kanamycin-resistant analogs of pUC19. BioTechniques. 1991;10:469–470. 472. [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H. Minohara M. Kawamura N. Li W. Matsushita T. Yamasaki R. Mizunoe Y. Kira J. Tissue binding patterns and in vitro effects of Campylobacter jejuni DNA-binding protein from starved cells. Neurochem Res. 2010;36:58–66. doi: 10.1007/s11064-010-0263-7. [DOI] [PubMed] [Google Scholar]
- Piao H. Minohara M. Kawamura N. Li W. Mizunoe Y. Umehara F. Goto Y. Kusunoki S. Matsushita T. Ikenaka K. Maejima T. Nabekura J. Yamasaki R. Kira J. Induction of paranodal myelin detachment and sodium channel loss in vivo by Campylobacter jejuni DNA-binding protein from starved cells (C-Dps) in myelinated nerve fibers. J Neurol Sci. 2009;288:54–62. doi: 10.1016/j.jns.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Samuel MC. Vugia DJ. Shallow S. Marcus R. Segler S. McGivern T. Kassenborg H. Reilly K. Kennedy M. Angulo F. Tauxe RV. Epidemiology of sporadic Campylobacter infection in the United States and declining trend in incidence, FoodNet 1996–1999. Clin Infect Dis. 2004;38(Suppl 3):S165–S174. doi: 10.1086/381583. [DOI] [PubMed] [Google Scholar]
- Steele TW. McDermott S. Campylobacter enteritis in South Australia. Med J Aust. 1978;2:404–406. doi: 10.5694/j.1326-5377.1978.tb76814.x. [DOI] [PubMed] [Google Scholar]
- Tonello F. Dundon WG. Satin B. Molinari M. Tognon G. Grandi G. Del Giudice G. Rappuoli R. Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol. 1999;34:238–246. doi: 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- Yao R. Alm RA. Trust TJ. Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]