Abstract
The FUT2 (Secretor) gene is responsible for the presence of ABO histo-blood group antigens on the gastrointestinal mucosa and in bodily secretions. Individuals lacking a functional copy of FUT2 are known as “nonsecretors” and display an array of differences in susceptibility to infection and disease, including Crohn disease. To determine whether variation in resident microbial communities with respect to FUT2 genotype is a potential factor contributing to susceptibility, we performed 454-based community profiling of the intestinal microbiota in a panel of healthy subjects and Crohn disease patients and determined their genotype for the primary nonsecretor allele in Caucasian populations, W143X (G428A). Consistent with previous studies, we observe significant deviations in the microbial communities of individuals with Crohn disease. Furthermore, the FUT2 genotype explains substantial differences in community composition, diversity, and structure, and we identified several bacterial species displaying disease-by-genotype associations. These findings indicate that alterations in resident microbial communities may in part explain the variety of host susceptibilities surrounding nonsecretor status and that FUT2 is an important genetic factor influencing host–microbial diversity.
Crohn disease (CD) is one of the two major forms of inflammatory bowel disease (IBD) characterized by chronic relapsing/remitting inflammation of the gastrointestinal (GI) tract (1, 2). It is posited that an abnormal immune response to microbes inhabiting the GI tract contributes to CD etiopathogenesis, which may arise from perturbed gene-by-environment interactions and include host genetic and immune factors, environmental triggers, and GI microbes. The importance of the GI microbiota is emphasized by numerous observations of alterations or imbalances of the GI microbiota in IBD patients (3–7) as well as by examples of animal models failing to manifest disease after germ-free rederivation (8). Although considerable advances in identifying genetic susceptibility loci in CD have been made, with more than 71 loci known to date (9), still very little is known regarding the details of interactions between individual susceptibility variants, environmental factors, and the GI microbiota (10, 11).
The FUT2 (Secretor) gene encodes an α-1,2-fucosyltransferase responsible for the expression of ABO histo-blood group antigens on the GI mucosa and in bodily secretions. Individuals bearing at least one functional allele are known as “secretors,” whereas those homozygous for loss-of-function mutations display a “nonsecretor” phenotype. It was recently shown that nonsecretor status is associated with CD susceptibility (12), and the wealth of information on the population genetics of FUT2 and its role in numerous host–microbe interactions make it an ideal candidate for describing the possible interactions between a genetic susceptibility variant and the endogenous microbiota.
Several mutations leading to the nonsecretor phenotype exist in human populations and display evidence of being maintained by strong selective pressure (13–16). A large body of evidence suggests that this maintenance may be because of numerous tradeoffs surrounding host–microbe interactions. For example, nonsecretors are resistant to infection with the Norwalk (Noro) (17) and respiratory viruses (18) but are more susceptible to duodenal ulcers (19), rheumatic fever (20), and cholera (21). Furthermore, the breast milk of secreting mothers provides protection against Campylobacter jejuni to their offspring by exploiting the binding affinity of the bacterium to fucosyloligosaccharides (22).
Despite variability at FUT2 mediating susceptibility to numerous pathogens, its overall conservation in mammals (23) indicates an important functional role. Experiments in mice have shown that the resident microbiota induce expression of FUT2 in the GI tract upon weaning, suggesting a role in the maintenance of homeostasis between mammalian hosts and their microbiota (24, 25). However, to date, no study of the effect of FUT2 on the overall composition and structure of the adult intestinal microbiota has been published. In this study, we have performed a survey of the colonic mucosa-attached microbiota in a panel of CD patients and controls and analyzed microbial community composition and structure with respect to disease and FUT2 genotype. As previously reported, we observe significant deviations in the mucosal communities of CD individuals. In addition, we demonstrate significant disease-by-genotype influences with respect to microbial community composition, diversity, and structure.
Results
To determine the influence of FUT2 expression on the colonic mucosa-associated microbial communities of healthy and CD individuals, we genotyped 47 individuals (29 CD, 18 controls) for the primary nonsecretor allele in Caucasian populations, W143X (G428A; rs601338) (13, 15), and generated sequence libraries of the bacterial 16S rRNA gene by using a multiplex barcoded pyrosequencing approach. Individuals homozygous for the functional allele “G” are denoted as SeSe (8 CD, 7 controls), those homozygous for the loss-of-function allele “A” are given as sese (6 CD, 3 controls), and heterozygotes are represented by Sese (15 CD, 8 controls) (Table S1). The allele most often associated with the nonsecretor phenotype in Asian populations (A385T; rs1047781) is also present in European populations at low frequency (0.4%) (15), but genotyping revealed all individuals to be homozygous for the functional variant at this position. A total of 46,990 16S rRNA gene sequence reads spanning variable regions 1 and 2 (V1 and V2) were analyzed after quality filtering and normalization of read number to 1,000 randomly chosen sequences per individual, on which all subsequent analyses are based (Materials and Methods).
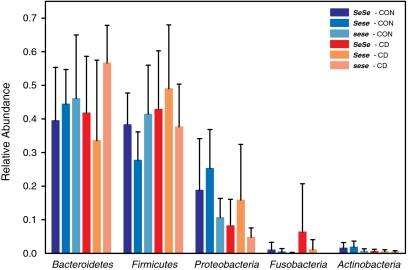
To provide an initial overview of the sequence reads associated with FUT2 genotype and disease status, we classified sequences at the phyla level by using Ribosomal Database Project (RDP) classifier (26, 27). A single Sese control individual displayed sequences belonging almost entirely (91%) to the phylum Spirochaetes, whereas this phylum was absent from all other individuals (Fig. S1). This individual was most likely displaying asymptomatic human intestinal spirochaetosis at the time of sampling and was removed because of its unclear clinical significance (28) and severe effect on bacterial community structure. All other individuals displayed communities dominated by the Firmicutes, Bacteroidetes, and Proteobacteria along with other less abundant phyla (Fig. 1). Overall, a significant increase of Firmicutes (ANOVA, P = 0.034) and corresponding decrease in Proteobacteria (ANOVA, P = 0.02) and Actinobacteria (ANOVA, P = 0.002) is apparent among CD individuals compared with controls (Statistical Analysis in Materials and Methods and Table S2). Differences were also observed with respect to secretor status, with a significant increase in Bacteroidetes among nonsecretors (ANOVA, P = 0.036). The abundance of Proteobacteria differed with respect to not only disease status but also genotype, with a significant increase in Sese compared with sese individuals, regardless of disease status (ANOVA, P = 0.047). Interestingly, several comparisons between control and CD individuals at the phyla level revealed differences only among secretors. Proteobacteria and Actinobacteria were significantly lower in abundance among CD individuals (post hoc Mann–Whitney test, P = 0.012 and 0.006, respectively), whereas Firmicutes were significantly higher in abundance (post hoc Mann–Whitney test, P = 0.005). These differences, however, were only observed among secretors, and no significant difference was present among nonsecretors (Table S2), indicating that, at the level of the relative abundance of the major phyla, control nonsecretors display more similarity to their diseased counterparts than do secretors.
Fig. 1.
Distribution of the major phyla with respect to disease status and genotype (error bars indicate SD). The corresponding results of the statistical analyses are presented in Table S2. CON, control.
Bacterial Diversity Within and Between Individuals.
To evaluate aspects of bacterial diversity that may be influenced by genotype and disease status, we first applied measures of alpha diversity, which describe species composition in one specific habitat of interest and can be informative of community functioning (29). Because microbial communities are highly diverse and are often poorly amenable to the diversity measures commonly used in community ecology (30), we used several different measures focusing on different aspects of community assembly, including species richness, evenness, and abundance based on operational taxonomic units (OTUs) at a 97% sequence similarity (species level) threshold, in addition to phylogenetic distance (Fig. S2 A–D). Phylogenetic diversity is a measure of alpha diversity that takes phylogenetic divergence into account (31). Compared with controls, a significant reduction in phylogenetic diversity was apparent among CD individuals (ANOVA, P = 0.0009; Statistical Analysis in Materials and Methods) (Fig. S2A). However, the same model also revealed a significant influence of FUT2 genotype (ANOVA, P = 0.024). The Chao1 index of estimated species richness displayed no difference with respect to disease status, but marginal differences were apparent between genotypes (ANOVA, P = 0.076, Fig. S2B). Interestingly, heterozygotes (Sese) displayed a general reduction in diversity compared with both homozygous genotypes, although only comparisons with sese genotypes were significant (post hoc Mann–Whitney test, phylogenetic diversity, P = 0.046; Chao1, P = 0.05). The modified Shannon entropy and evenness by Jost (32) displayed a similar pattern, but no differences were significant (Fig. S2 C and D). These results indicate that a reduction in bacterial diversity within CD individuals compared with controls was observable when taking the phylogenetic relationship of the present species into account, whereas measures based solely on observed species number and abundance failed to reveal differences with respect to disease status. Thus, a similar number of species is present in CD individuals compared with controls, but they are on average more closely related. Because of the significant influence of FUT2 genotype, we focused the remainder of the analysis on genotype rather than on secretor status.
Although alpha diversity measures describe aspects of community structure within a given individual, they do not reveal the similarities or differences in communities between individuals (i.e., beta diversity). Thus, to reveal the relationships of the microbial communities hosted by individuals differing by FUT2 genotype and disease status, we performed several analyses describing the differences in bacterial community composition and structure between individuals, also taking age and sex as potential confounding factors into account. UniFrac is a phylogenetic-based beta diversity measure that represents the genetic distance between communities by comparing the shared versus unique branch lengths underlying different communities (33, 34). To analyze the distance between the communities, we used multivariate analysis of variance [analysis of dissimilarity or “adonis” (Materials and Methods) (35)] of the unweighted UniFrac metric, which ignores taxon abundance and thus reduces it to binary presence/absence data. This analysis revealed a highly significant distinction between the microbial communities of control and CD subjects (adonis: R2 = 0.053, P < 0.0001). For further analysis, we analyzed UniFrac distances via principal coordinate analysis (PCoA) (Fig. S3 A–C). Disease status and the interaction between disease status and genotype were strongly correlated with the ordination of the unweighted UniFrac values on all three axes (goodness of fit for disease status R2 = 0.186, P < 0.0001; genotype–disease interaction R2 = 0.266, P = 0.0005), indicating a significant contribution of these factors to variation in community composition between individuals. This finding indicates that a significant amount of the variation in bacterial community composition between individuals can be explained not only by disease status but also by disease-by-genotype interactions. We also observed a significant correlation of age (goodness of fit R2 = 0.257, P = 0.006). Additional analysis of the axis scores with a linear model framework revealed a strong correlation of axes one and two with disease status (PCo1: P = 0.013; PCo2: P < 0.0001) and a gradient among the genotypes on axis three (P = 0.053) (Table S3).
In addition to phylogenetic-based measures of beta diversity, we used OTUs (97% threshold) to analyze the similarity in species distributions between communities. To measure community similarity, we used the Jaccard and Bray-Curtis indices, which are classical ecological beta diversity measures based on the ratio of shared and unique species relative to the total number of species present between two communities, respectively. This analysis revealed a significant impact of disease status (adonis: Bray-Curtis, R2 = 0.039, P = 0.004; Jaccard, R2 = 0.032, P = 0.004). Furthermore, a significantly higher proportion of variance in community structure could be explained by adding the interaction between disease and genotype to this model (adonis: Bray-Curtis, disease status R2 = 0.039, P = 0.003; genotype–disease interaction R2 = 0.103, P = 0.037; Jaccard, disease status R2 = 0.032, P = 0.003; genotype–disease interaction R2 = 0. 097, P = 0.04). Differences in bacterial community structure are also apparent in PCoA analyses of these beta diversity indices that reveal strong differences with respect to disease status, sex, and genotype (Fig. S3 D–I and Table S3). To compare the ordinations of beta diversity measures based on OTUs and those that consider phylogeny, we conducted a Procrustes analysis (36) using the Procrustean randomization test (PROTEST). Jaccard and Bray-Curtis displayed a strong overlap with each other (PROTEST: M = 0.345, r = 0.809, P < 0.001) as well as to unweighted UniFrac (PROTEST: Jaccard, M = 0.649, r = 0.592, P < 0.001; Bray-Curtis, M = 0.390, r = 0.781, P < 0.001). Thus, despite measuring different aspects of community structure, metrics based on shared abundance or phylogenetic distance appear to converge to related solutions.
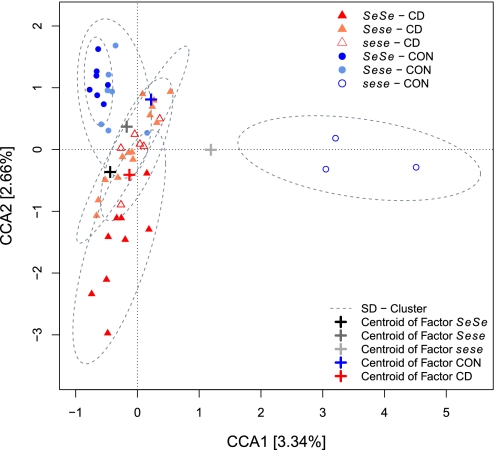
To validate the factors responsible for community clustering and gain deeper insight into the actual determinants explaining this community clustering, it is advisable to use ordination methods such as canonical correspondence analysis (CCA) because these methods use the full range of OTU data and not merely the relationships among the individuals, allowing specific hypothesis testing. CCA models assume a unimodal distribution of species around an optimum (niche) to calculate the relationships among individuals via an iterative process. To avoid a potential confounding influence of age and sex, we performed partial CCA with these variables as conditioning factors. With this approach, we identified significant clusters of individuals by using artificial gradients based on disease status and the interaction with genotype (Fig. 2). Model optimization for CCA was carried out for maximizing the explained variance using a minimum of variables as well as a strong contribution of these variables to the model (total inertia = 17.739, constrained inertia = 2.195, conditioned inertia = 0.828, unconstrained inertia = 14.716 (82.96%), and explained variance by constraints and conditions = 17.04%). By this procedure, we obtained two highly significant axes explaining 5.99% of the variance in the data. Disease status (F1, 38 = 1.202, χ2 = 0.466, P = 0.008), genotype (F2, 38 = 1.120, χ2 = 0.867, P = 0.023), and genotype–disease status interaction (F2, 38 = 1.113, χ2 = 0.862, P = 0.06) contributed to the fit of the ordination as assessed via permutation tests (105 permutations). An additional related unconstrained method [detrended correspondence analysis (DCA)] also revealed a strong influence of genotype and disease status (Table S3 and Fig. S3 J–L), thus verifying that disease status and FUT2 genotype significantly contribute to the differences in community structure observed between individuals. Although these analyses describe factors that significantly contribute to differences in community structure, they explain a relatively small proportion of the total interindividual variability. Other factors such as diet, lifestyle, additional genetic factors, and differences in disease manifestation likely contribute to the remaining unexplained variation in intestinal communities. However, additional analysis of different disease subphenotypes (i.e., colitis, ileitis, and ileocolitis) did not reveal any significant patterns in Dataset S1.
Fig. 2.
Ordination of individuals using CCA of disease status and its interaction with genotype, conditioned by the factors age and sex. The ordination includes only nonredundant factors that were validated by variance inflation scores ranging between 1.1 and 3.6. Only axes that significantly contribute to the explained variance (expl. var.) are displayed (CCA1: expl. var. = 3.34%, P = 0.007; CCA2: expl. var. = 2.66%, P = 0.03; CCA3: expl. var. = 2.44%, P = 0.171; CCA4: expl. var. = 2.19%, P = 0.476; CCA5: expl. var. = 1.76%, P = 0.944).
Indicator Species.
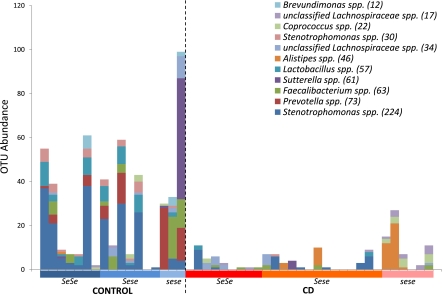
To identify individual bacterial taxa that contribute to the patterns identified by multivariate analysis, we analyzed OTUs at both the species (≥97% identity) and genus (≥95% identity) levels by using a common ecological measure of species habitat association, i.e., “indicator species” analysis (37). We defined our “habitats” by disease status, genotype, and genotype within the disease status. Using this analysis, we identified numerous species-level OTUs distributed across the three major phyla that were more frequently present and/or abundant in a given habitat(s) (Table S4). The analysis with respect to disease status identified 10 species-level OTUs restricted to healthy individuals belonging to Prevotella, Lactobacillus, Coprobacillus, Clostridium, Faecalibacterium, and Stenotrophomonas. The analysis with respect to genotype revealed only two OTUs belonging to Coprococcus and unclassified Lachnospiraceae. However, extending the analysis to genotypes within disease status revealed numerous interesting associations (Fig. 3). An OTU belonging to Lactobacillus and two OTUs belonging to Stenotrophomonas were associated with healthy secretor genotypes (SeSe and Sese). Likewise, five OTUs were identified among healthy sese individuals and belonged to Prevotella, Brevundimonas, unclassified Lachnospiraceae, Sutterella, and Faecalibacterium. Interestingly, three OTUs belonging to Alistipes, unclassified Lachnospiraceae, and Coprococcus were found to be associated with sese individuals with CD, suggesting that they may contribute to a bacterial community structure that is a subphenotype among CD patients and depends on FUT2 genotype. Performing this analysis at the genus level identified a single, but strong, association of an unclassified genus belonging to the family Lachnospiraceae that is more abundant among CD individuals and an additional 15 genera associated with healthy individuals (Fig. S4 and Table S4).
Fig. 3.
Abundance of species-level OTUs identified by indicator species analysis with respect to genotype within disease status. Numbers in parentheses indicate the total read number in the normalized dataset (1,000 reads per individual).
Discussion
In this study, we sought to determine the influence of the nonsense SNP W143X at FUT2, a major susceptibility variant for CD, on the composition and structure of the intestinal mucosa-associated microbiota in both healthy and CD individuals. By using high-throughput sequencing of bacterial 16S rRNA gene sequences at a single location along the GI tract, we revealed several unique aspects of bacterial communities with respect to both disease status and W143X genotype. Our analysis of bacterial diversity both within and between individuals revealed significant influences of disease and disease-by-genotype interactions. Furthermore, through the application of an ecological measure of species habitat association to disease and genotype habitats, we identified important candidate bacterial species and genera contributing to these patterns.
The distribution of reads according to bacterial phyla was concordant with previously published analyses of the intestinal microbiota (38) but deviated somewhat from previous investigations of CD patients (e.g., a decrease in the relative abundance of Proteobacteria was found as opposed to an increase) (5, 39). This difference may be attributable to the fact that samples in these studies were taken from acutely inflamed tissue or from multiple locations throughout the gut. Cloning bias and/or overlooked diversity because of different sequencing technologies also could contribute to this discrepancy (40) as well as account for the general lack of decreased alpha diversity in CD patients observed in our study. Another explanation for the latter could be a loss of specific taxa and corresponding replacement by other members of the bowel community. This scenario may maintain the number of observed species but result in a different phylogenetic relationship among them, thus impacting phylogenetic- rather than OTU-based diversity indices, which was observed in Dataset S1. Our observation of a significant increase in prevalence and abundance of a single bacterial genus in the context of CD compared with 15 such genera in healthy individuals (Fig. S4 and Table S4) also supports this explanation.
The contribution of W143X genotype to variation in bacterial communities between individuals is, in some respect, overshadowed by the effects of CD. In CD, the architecture of the intestinal mucosal lining undergoes a drastic remodeling process. Apart from the increased influx of inflammatory cells, even in remission phases without obvious inflammation, studies have shown an increased intestinal permeability in CD patients and defects in the amount and structure of the dense carbohydrate-rich layer of mucus covering the intestinal epithelium (41, 42). However, combining the effects of CD and the underlying factor of W143X genotype explained significantly more variation between microbial communities, supporting the hypothesis of an interaction of these three elements. Interestingly, some differences in community composition and structure, i.e., alpha diversity and the relative abundance of the major phyla, were less apparent between control and CD nonsecretors than in the other genotypes. Given the overall conservation of FUT2 among mammals (23), these observations associated with a lack of functional enzyme activity in the GI tract could be indicative of an altered mucosal constitution that may contribute to the manifestation and progression of CD.
The intestinal mucosal barrier has a high regenerative capacity and builds up a pivotal barrier not only for mechanical or chemical stressors but also for those induced by the microbiota. The immunological interface of the intestines is composed of a multilayered structure from the luminal-secreted mucus, the intestinal epithelial cells, and the underlying mucosa-associated migratory immune cells. Bacteria are mostly restricted to the outer layer of the mucus and interact directly with the underlying epithelia only in pathological conditions (43). These adjacent bacteria are able to trigger the secretion of specific glycans, such as blood groups antigens, which in return can serve as nutrients or attachment sites for the bacteria, highlighting the close interaction of resident bacteria with the gut (24, 25, 44). Thus, the clustering we observed according to W143X genotype might be explained by the common presence/absence of preferred attachment site(s), which are, in several cases, fucosylated glycoconjugates (45). We hypothesize that the glycoconjugate profile of the mucosa of heterozygotes (Sese) may differ from each respective homozygote, for example, via competition for substrates with other glycosyltransferases expressed in the GI tract, resulting in an influence on the availability of bacterial attachment sites or nutritional resources (46). Variation in α-1,2-fucosyltransferase activity was observed in fucosylated milk proteins, and it is hypothesized that variation not only at the level of secretor status but also at the level of genotype plays an important role in determining which substrates are glycosylated by the same set of enzymes (46). Experimental evidence also points toward a dose effect of FUT2, with significant differences among SeSe and Sese individuals (47).
To shed light on the groups contributing to differences in beta diversity at higher taxonomic levels, we performed an indicator species analysis at both the genus and species levels. At the genus level, we identified among CD patients only a single taxa belonging to the family Lachnospiraceae. The family Lachnospiraceae has been recovered in numerous metagenomic surveys and belongs to the core gut microbiota (48) but was also previously demonstrated to be increased in ileal CD (49) and in mice with dextran sodium sulfate (DSS)-induced colitis (50).
At the species level, most OTUs displayed a strong association with control individuals, stressing the compositional difference with respect to disease status. Importantly, several of these OTUs are considered to be probiotic [e.g., Lactobacillus (51) and Faecalibacterium (52)] and/or were previously observed to be reduced in the context of CD (e.g., Faecalibacterium prausnitzii). Our observation that OTUs belonging to the Lactobacillus genus are particularly associated with healthy secretors is also consistent with the fact that they possess adhesins specifically targeting ABO blood group antigens in the mucosa (53), which may contribute to the probiotic function of Lactobacillus by blocking the attachment of potential pathogens to mucosal surfaces (54).
Interestingly, although Stenotrophomonas is becoming increasingly important as a nosocomial pathogen—associated with airway infections, weak bacteremia in immunocompromised subjects, resistance to antibiotics, and even life-threatening chronic enteritis (55, 56)—several species’ OTUs belonging to this genus were identified among our controls. Notable features of this genus are its weak invasiveness but its variety of colonization mechanisms and strong ability to form biofilms, making it a successful colonizer of various hosts (56). This genus was more prevalent among SeSe and Sese controls, which might indicate a preference for mucosal substrates containing blood group antigens.
Finally, we identified several species significantly associated with sese individuals, in both control and CD groups. Unclassified species belonging to the family Lachnospiraceae were identified as indicator species in both healthy and CD nonsecretors. As mentioned above, this group becomes more abundant upon experimental induction of colitis in mice by the chemical irritant DSS (50). Because DSS treatment changes the characteristics of the inner and outer mucosal barriers, facilitating bacterial penetration by reduced thickness and permeability (42), it is possible that this group may also display increased invasiveness in undisturbed mucosa lacking protective ABO antigens. Furthermore, flagellins of this bacterial group serve as elicitors of inflammation in CD (57). Another noteworthy observation is an OTU belonging to Prevotella that is associated with sese controls. Because these bacteria are able to digest mucins and are of increasing clinical relevance for chronic infections, it is possible that they contribute to mucosa impairment (58, 59).
Our results offer important insight into both the host genetic basis of diversity in the intestinal microbiota and the potential means through which alternative FUT2 alleles contribute to disease susceptibility. Although several individual interactions between microbes and FUT2-dependent antigens in the GI tract were known before this study, our results indicate that differences in the composition and structure of bacterial communities according to FUT2 genotype may contribute to CD susceptibility. Because of their long-term maintenance and repeated evolutionary origin (14, 15), loss-of-function mutations at FUT2 are extremely common among human populations. Given the association of genetic variants at FUT2 with multiple immune phenotypes, further understanding of the role of FUT2 in maintaining homeostasis between mammalian hosts and their complex associated microbial communities may considerably contribute to future improvements in preventative and therapeutic patient care in acute infectious and chronic inflammatory diseases.
Materials and Methods
Human Samples.
The biopsy bank of the outpatient clinic of the Department of General Internal Medicine of University Hospital Schleswig-Holstein was screened for individuals with a diagnosis of CD and for healthy controls of Caucasian (northern European) ancestry. Symptoms were in remission at the time of sampling, and all biopsies were taken from noninflamed tissue (for details, see SI Materials and Methods). All procedures related to patients and healthy subjects were approved by the University Hospital Schleswig-Holstein ethics committee (B231/98 and A154/06) and follow the guidelines of the Declaration of Helsinki.
Genotyping.
Functionally tested TaqMan SNP Genotyping Assays (Applied Biosystems) were used to genotype the primary nonsecretor allele in Caucasian populations, W143X (G428A; rs601338) (13, 15), and A385T (rs1047781) on an automated platform (60). All process data were written to and administered by a previously described database-driven laboratory information management system (LIMS) (61).
DNA Extraction and 16S rRNA Gene Pyrosequencing.
DNA from sigmoid colonic biopsies was extracted with the AllPrep DNA/RNA Mini Kit (Qiagen) following the manufacturer's instructions with the addition of a bead-beating step after the addition of the RLT buffer to enhance cell lysis. The 27F–338R region of the 16S rRNA gene was amplified and sequenced on the 454 Life Sciences GS-FLX platform using Titanium sequencing chemistry (SI Materials and Methods).
Sequence Processing and Quality Control.
Raw sequences were trimmed by a Perl script using a Smith–Waterman alignment algorithm to identify multiplex identifier primer sequences. A mean quality score of ≥20 and a minimum length of 200 nt for the coupled V1–V2 region was required. Sequences were then aligned to the highly curated seed database from SILVA (62) using a k-mer alignment procedure as implemented in mothur 1.12 (63). Sequences that did not match the defined core region of the seed alignment were manually removed. Chimeric sequences were removed by using the UCHIME function in UCLUST 3.0 (64) with the SILVA gold database as a reference. For all subsequent analyses, we used a random subset of 1,000 sequences per individual to normalize the read distribution (a single individual was included with 990 sequences), which was previously suggested as a good balance between sample number and coverage (65). This sampling depth corresponded to an average Good's coverage value of 0.86 ± 0.07 SD. Sequences were confirmed as bacterial by using the Ribosomal Database Project (RDP) classifier with an 80% bootstrap threshold (26, 27). Aligned sequences were used to compute a distance matrix and group-related sequences into OTUs using mothur (OTU abundances are provided as Dataset S1). Phylogenetic tree construction was carried out by using FastTree 2.0 with a generalized time-reversible (GTR) substitution model (66).
Statistical Analysis.
Alpha diversity indices were calculated in R (67). Phylogenetic diversity was calculated according to Faith et al. (31). Analyses of phyla abundances and alpha diversity were performed with an ANOVA framework. Alternative models were tested sequentially by using all possible combinations of predictor variables (i.e., genotype, disease status, age, and sex), and the best model was chosen according to the Akaike information criterion. FAST UniFrac was used to calculate the unweighted UniFrac metric (33, 34). For statistical analysis of beta diversity indices, we performed nonparametric matrix-based analysis of variance by using adonis implemented in the vegan package for R (35, 68). PCoAs were performed in R without constraints. Goodness of fit was assessed with 105 permutations on all three axes. A correlation of each single axis with specific factors (i.e., genotype, disease status, age, and sex) was assessed by using linear models with model selection procedures as described above. For the comparison of ordinations, we used Procrustes analysis with 105 permutations (36). CCA was carried out with the least number of variables to assess the highest explained variation (69).
Indicator species analysis, as described by De Cáceres et al. (37), was implemented in the R package indicspecies and based on 105 permutations. To reduce the number of candidate OTUs, we applied the sample discrimination (SIMPER) method implemented in PRIMER 6 (70). We limited the analysis to OTUs with a minimum contribution of 0.5% to the overall similarity within each cluster as assessed by the Bray-Curtis distance. The thresholds for P values of OTU association were adjusted for multiple testing by the method of Benjamini and Hochberg (71). We set the threshold for significance after correction at the 5% level and that for trends in the data at 10%.
Supplementary Material
Acknowledgments
We thank all study participants and Katja Cloppenborg-Schmidt, Tanja Wesse, and Manuela Kramp for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft Excellence Cluster “Inflammation at Interfaces,” the National Genome Research Network (NGFN) “Systematic Genomics of Chronic Inflammatory Barrier Diseases” (Subprojects GP1 and 10), and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive, www.ebi.ac.uk/ena/data/view/ERP000888 (accession no. ERP000888).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106408108/-/DCSupplemental.
References
- 1.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Ott SJ, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lepage P, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm Bowel Dis. 2005;11:473–480. doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 5.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, Lay C, Seksik P, Tannock GW. Analysis of bacterial bowel communities of IBD patients: What has it revealed? Inflamm Bowel Dis. 2008;14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 7.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taurog JD, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DN, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. doi: 10.1002/ibd.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehman A, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354–1362. doi: 10.1136/gut.2010.216259. [DOI] [PubMed] [Google Scholar]
- 12.McGovern DPB, et al. International IBD Genetics Consortium Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum Mol Genet. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group α(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270:4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 14.Koda Y, Tachida H, Soejima M, Takenaka O, Kimura H. Ancient origin of the null allele se428 of the human ABO-secretor locus (FUT2) J Mol Evol. 2000;50:243–248. doi: 10.1007/s002399910028. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer-Admetlla A, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. 2009;26:1993–2003. doi: 10.1093/molbev/msp108. [DOI] [PubMed] [Google Scholar]
- 16.Andrés AM, et al. Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 18.Raza MW, et al. Association between secretor status and respiratory viral illness. BMJ. 1991;303:815–818. doi: 10.1136/bmj.303.6806.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans DA, Horwich L, McConnell RB, Bullen MF. Influence of the ABO blood groups and secretor status on bleeding and on perforation of duodenal ulcer. Gut. 1968;9:319–322. doi: 10.1136/gut.9.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haverkorn MJ, Goslings WR. Streptococci, ABO blood groups, and secretor status. Am J Hum Genet. 1969;21:360–375. [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri A, DasAdhikary CR. Possible role of blood-group secretory substances in the aetiology of cholera. Trans R Soc Trop Med Hyg. 1978;72:664–665. doi: 10.1016/0035-9203(78)90031-7. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 23.Abrantes J, Posada D, Guillon P, Esteves PJ, Le Pendu J. Widespread gene conversion of α-2-fucosyltransferase genes in mammals. J Mol Evol. 2009;69:22–31. doi: 10.1007/s00239-009-9239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 25.Meng D, et al. Bacterial symbionts induce a FUT2-dependent fucosylated niche on colonic epithelium via ERK and JNK signaling. Am J Physiol Gastrointest Liver Physiol. 2007;293:G780–G787. doi: 10.1152/ajpgi.00010.2007. [DOI] [PubMed] [Google Scholar]
- 26.Cole JR, et al. Ribosomal Database Project The Ribosomal Database Project (RDP-II): Previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmiedel D, et al. Rapid and accurate diagnosis of human intestinal spirochetosis by fluorescence in situ hybridization. J Clin Microbiol. 2009;47:1393–1401. doi: 10.1128/JCM.02469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peter H, et al. Function-specific response to depletion of microbial diversity. ISME J. 2011;5:351–361. doi: 10.1038/ismej.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 32.Jost L. Partitioning diversity into independent α and β components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- 33.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 36.Peres-Neto P, Jackson D. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia. 2001;129:169–178. doi: 10.1007/s004420100720. [DOI] [PubMed] [Google Scholar]
- 37.De Cáceres M, Legendre P, Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. [Google Scholar]
- 38.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temperton B, et al. Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing. ISME J. 2009;3:792–796. doi: 10.1038/ismej.2009.32. [DOI] [PubMed] [Google Scholar]
- 41.Pullan RD, et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut. 1994;35:353–359. doi: 10.1136/gut.35.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson MEV, et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoskins LC, Boulding ET. Degradation of blood group antigens in human colon ecosystems. I. In vitro production of ABH blood group-degrading enzymes by enteric bacteria. J Clin Invest. 1976;57:63–73. doi: 10.1172/JCI108270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magalhães A, et al. Fut2-null mice display an altered glycosylation profile and impaired BabA-mediated Helicobacter pylori adhesion to gastric mucosa. Glycobiology. 2009;19:1525–1536. doi: 10.1093/glycob/cwp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erney RM, et al. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr. 2000;30:181–192. doi: 10.1097/00005176-200002000-00016. [DOI] [PubMed] [Google Scholar]
- 47.Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoën-Clouet N. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J Infect Dis. 2005;192:1071–1077. doi: 10.1086/432546. [DOI] [PubMed] [Google Scholar]
- 48.Sekelja M, Berget I, Næs T, Rudi K. Unveiling an abundant core microbiota in the human adult colon by a phylogroup-independent searching approach. ISME J. 2011;5:519–531. doi: 10.1038/ismej.2010.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854, e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 50.Nagalingam NA, Kao JY, Young VB. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 2011;17:917–926. doi: 10.1002/ibd.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72:728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe M, et al. Identification of a new adhesin-like protein from Lactobacillus mucosae ME-340 with specific affinity to the human blood group A and B antigens. J Appl Microbiol. 2010;109:927–935. doi: 10.1111/j.1365-2672.2010.04719.x. [DOI] [PubMed] [Google Scholar]
- 54.Chan RC, Reid G, Irvin RT, Bruce AW, Costerton JW. Competitive exclusion of uropathogens from human uroepithelial cells by Lactobacillus whole cells and cell wall fragments. Infect Immun. 1985;47:84–89. doi: 10.1128/iai.47.1.84-89.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hellmig S, et al. Life-threatening chronic enteritis due to colonization of the small bowel with Stenotrophomonas maltophilia. Gastroenterology. 2005;129:706–712. doi: 10.1016/j.gastro.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Ryan RP, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 57.Duck LW, et al. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 58.Rho JH, et al. A novel mechanism for desulfation of mucin: Identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J Bacteriol. 2005;187:1543–1551. doi: 10.1128/JB.187.5.1543-1551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucke K, Miehlke S, Jacobs E, Schuppler M. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol. 2006;55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 60.Hampe J, et al. An integrated system for high throughput TaqMan based SNP genotyping. Bioinformatics. 2001;17:654–655. doi: 10.1093/bioinformatics/17.7.654. [DOI] [PubMed] [Google Scholar]
- 61.Teuber M, et al. Improving quality control and workflow management in high-throughput single-nucleotide polymorphism genotyping environments. J Assoc Lab Autom. 2005;10:43–47. [Google Scholar]
- 62.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 65.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price MN, Dehal PS, Arkin AP. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 68.Oksanen J, et al. vegan: Community Ecology Package. 2011. (Comprehensive R Archive Network), R package Version 1.17-6, http://cran.r-project.org/web/packages/vegan/index.html.
- 69.ter Braak CJF. Canonical correspondence analysis: A new eigenvector technique for multivariate direct gradient analysis. Ecology. 1986;67:1167–1179. [Google Scholar]
- 70.Clarke K, Gorley R. PRIMER v6: User Manual/Tutorial. Plymouth, UK: PRIMER-E Ltd; 2006. [Google Scholar]
- 71.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.