Abstract
Natural killer T cell antigen receptors (NKT TCRs) recognize lipid-based antigens (Ags) presented by CD1d. Although the TCR α-chain is invariant, NKT TCR Vβ exhibits greater diversity, with one (Vβ11) and three (Vβ8, Vβ7, and Vβ2) Vβ chains in humans and mice, respectively. With the exception of the Vβ2 NKT TCR, NKT TCRs possess canonical tyrosine residues within complementarity determining region (CDR) 2β that are critical for CD1d binding. Thus, how Vβ2 NKT TCR docks with CD1d-Ag was unclear. Despite the absence of the CDR2β-encoded tyrosine residues, we show that the Vβ2 NKT TCR engaged CD1d-Ag in a similar manner and with a comparable affinity and energetic footprint to the manner observed for the Vβ8.2 and Vβ7 NKT TCRs. Accordingly, the germline–encoded regions of the TCR β-chain do not exclusively dictate the innate NKT TCR-CD1d-Ag docking mode. Nevertheless, clear fine specificity differences for the CD1d-Ag existed between the Vβ2 NKT TCR and the Vβ8.2 and Vβ7 NKT TCRs, with the Vβ2 NKT TCR exhibiting greater sensitivity to modifications to the glycolipid Ag. Furthermore, within the Vβ2 NKT TCR-CD1d-αGalCer complex, the CDR2β loop mediated fewer contacts with CD1d, whereas the CDR1β and CDR3β loops contacted CD1d to a much greater extent compared with most Vβ11, Vβ8.2, and Vβ7 NKT TCRs. Accordingly, there is a greater interplay between the germline– and nongermline–encoded loops within the TCR β-chain of the Vβ2 NKT TCR that enables CD1d-Ag ligation.
Keywords: T cell repertoire, conserved docking
Natural killer T (NKT) cells are lipid antigen (Ag) -reactive, CD1d -restricted T cells present in mice and humans (1). These cells influence the outcome in a broad range of diseases, including microbial immunity, tumor immunity, autoimmunity, and allergy (2–4). Type I NKT cells (herein referred to as NKT cells) are defined by an invariant NKT cell antigen receptor (TCR) α-chain (Vα14-Jα18 in mice and Vα24-Jα18 in humans) and specifically recognize α-galactosylceramide (α-GalCer) and related analogs of this glycolipid (reviewed in ref. 1). α-GalCer is the most extensively studied glycolipid Ag for activating NKT cells and is widely used experimentally and in translational studies as a potent NKT cell agonist (5).
NKT cells are stimulated by an array of lipid-based Ag (reviewed in refs. 3, 5, and 6), including bacteria-derived lipid Ag (7–10) and self-glycolipid Ag (11). Notably, with the exception of α-GalCer, most other glycolipid Ags seem to be differentially recognized by subsets of NKT cells (9, 12). The presence of an invariant NKT TCR α-chain suggests that the TCR β-chain, which includes the hypervariable complementarity determining region (CDR) 3β loop, determines thresholds of Ag reactivity (12–14). Interestingly, mouse NKT cells frequently use three Vβ genes (Vβ8, Vβ7, and Vβ2) and thus, possess a more diverse TCR-Vβ repertoire than human NKT cells, which mostly express Vβ11. Mouse Vβ2 NKT TCRs represent ∼5–10% of the NKT cell repertoire, although the basis of Vβ2 NKT TCR use is unclear.
The crystal structures of human Vβ11 and mouse Vβ8.2 and Vβ7 NKT TCRs in complex with CD1d-α-GalCer have provided insight into the basis of NKT recognition and some clues into the role of differential Vβ usage (14–16). Furthermore, the structures of NKT TCRs in complex with α-GalCer analogs as well as α-galactosyldiacylglycerol, the self-Ag phosphatidyl inositol, and some β-linked Ags have been determined (17–23). In all NKT TCR-CD1d-Ag complexes determined to date, a conserved, tilted, and parallel docking mode with respect to the CD1d Ag-binding cleft was observed. Within this common framework, the NKT TCR α-chain dominated the interaction (14, 15, 17, 18, 20, 22, 24). The binding of the human Vβ11 and the mouse Vβ8.2 and Vβ7 chains was largely attributable to the CDR2β-mediated contacts with CD1d. In particular, within the Vβ11 and Vβ8.2 NKT TCRs, two canonical tyrosine residues (Tyr 48β and Tyr 50β) made a conserved set of interactions with the α1-helix of CD1d. The Vβ7 NKT TCR also possessed one of these tyrosine residues (Tyr 50β) and recognized CD1d in a homologous fashion to the Vβ11 and Vβ8.2 NKT TCRs (14). Vβ11, Vβ8.2, and Vβ7 NKT TCR mutagenesis experiments highlighted the importance of the CDR1α, CDR3α, and CDR2β loops in interacting with CD1d (13–15, 25–27). Although the CDR3β loop can play little, if any, role in NKT TCR-CD1d-α-GalCer binding (14, 15), CDR3β diversity can contribute to CD1d-Ag recognition. For example, CDR3β influences binding of Vβ6+ NKT TCRs (13), and a greater role for the CDR3β loop seems to be important for autoreactivity (12, 19, 21, 22).
Given the variability in the Vβ repertoire of NKT cells, it could be considered that the invariant TCR α-chain dictates the conserved NKT TCR-CD1d docking mode. Such considerations have resonances with TCR-peptide-MHC (pMHC) recognition, where the CDR2β loop of Vβ8.2-containing TCRs is considered to define the basis for MHC bias for this slice of the T cell repertoire (28–30). In the context of what determines CD1d restriction, we recently showed the existence of a population of semi-invariant NKT cells that expresses a canonical Vα10-Jα50 TCR α-chain paired with either Vβ8 or Vβ7, which are also α-GalCer–reactive (23). Nevertheless, this Vα10 NKT TCR adopted the same docking mode to CD1d-Ag as the method observed for Type I NKT cells (23). Given that a similar TCR β-chain bias was observed for these Vα10 NKT cells, this finding suggests a dominant role for the TCR β-chain in determining the conserved docking mode exhibited by NKT cells. However, mouse Vβ2 NKT TCRs lack both of the key contact residues (Tyr 48β and Tyr 50β) that underpin mouse Vβ8.2 and human Vβ11 NKT TCR binding. Additionally, rat NKT cells also possess a divergent CDR2β sequence (31) and only function with syngeneic rat CD1d as a restriction element, suggesting that the CDR2β loop plays a critical role in the specificity of the interaction. Thus, a priori, it is unclear whether the Vβ2+ NKT TCR will bind in an analogous way and with similar affinities to Vβ11, Vβ8.2, and Vβ7 NKT TCRs. It was also unclear whether the fine specificity requirements of the Vβ2 NKT TCR would be distinct from the Vβ8.2 and Vβ7 NKT TCRs. Thus, we examined the structural and functional basis of Vβ2-mediated NKT TCR recognition.
Results
Vβ2 NKT TCR Affinity Measurements.
To address the role of Vβ2 use in mouse NKT cells, we expressed and refolded soluble mouse Vα14Jα18-Vβ2, Vα14Jα18-Vβ8.2, and Vα14Jα18-Vβ7 NKT TCRs and compared their affinity for CD1d-α-GalCer and four α-GalCer analogs (Fig. 1 and Table S1). The α-GalCer analogs differed in the composition of the glycosyl head group (3′,4″-deoxy-α-GalCer, 4′,4″-deoxy-α-GalCer, and glucosylceramide (α-GlcCer) and the sphingosine chain (OCH; truncated from C18 to C9) (32–34). Accordingly, these modifications of the glycolipid Ag enabled us to address their impact on Vβ2-mediated NKT TCR recognition.
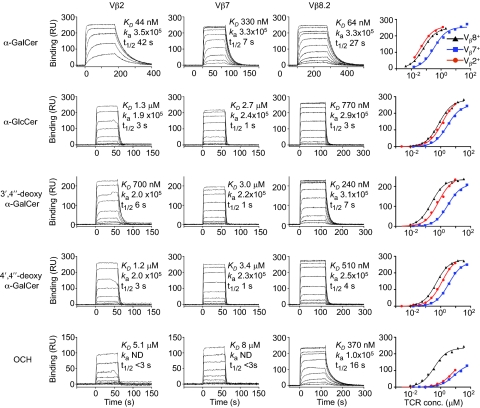
Fig. 1.
Vβ2 NKT TCR affinity measurements. Sensorgrams illustrate binding of graded concentrations of Vα14-Vβ2+ (column 1; 1.3–0.016 μM for α-GalCer and 9.3–0.002 μM for other ligands), Vα14-Vβ7+ (column 2; 56–0.015 μM), and Vα14-Vβ8.2+ (column 3; 2.1–0.004 μM for α-GalCer and 35–0.009 μM for other ligands) soluble NKT TCRs to CD1d-α-GalCer, CD1d-α-GlcCer, CD1d-3′,4″-deoxy α-GalCer, CD1d-4′,4″-deoxy α-GalCer, and CD1d–OCH after subtraction from a control (CD1d-endogenous) flow cell. Saturation plots (column 4) show equilibrium binding. The dissociation constant (KD) derived by equilibrium analysis, association rate (ka; M−1⋅s−1), and half-life (t1/2) is shown for each interaction. The data shown is from one experiment and is representative of three separate experiments for CD1d-α-GalCer and CD1d-α-GlcCer and two separate experiments for the other ligands.
The affinity of the Vβ2 NKT TCR for CD1d-α-GalCer as determined by surface plasmon resonance (SPR) was 50 ± 10 nM compared with values of 60 ± 10 nM for Vβ8.2 and 280 ± 30 nM for Vβ7 NKT TCR, similar to the values published previously (Fig. 1 and Table S1) (14). This indicated that, despite lacking the two key Tyr motifs within the CDR2β loop, this particular Vβ2-containing TCR retained a high affinity for CD1d-Ag, suggesting that some Vβ2 NKT TCRs need not necessarily exhibit a lower affinity for CD1d-Ag than Vβ7 and Vβ8.2 NKT TCRs. The high affinity of the Vβ2 TCR-CD1d-α-GalCer interaction was primarily because of a prolonged half-life (36 ± 3 s) compared with Vβ8.2 (24 ± 2 s) and Vβ7 (6.8 ± 0.2 s) NKT TCRs (Fig. 1). Modifications at the 3′-OH (3′,4″-deoxy-α-GalCer) moiety of the glycosyl headgroup reduced Vβ2 and Vβ7 NKT TCR affinity dramatically, whereas Vβ8.2 NKT TCR was less affected (fourfold reduction). Additionally, modifications to the 4′-OH (α-GlcCer and 4′,4″-deoxy-α-GalCer) moiety had a greater impact for all NKT TCRs. The α-GalCer analog with a truncated sphingosine chain (OCH) impacted most notably on Vβ2 and Vβ7 NKT TCRs (Kd > 6 μM), whereas in comparison, it had a smaller impact on the Vβ8.2 interaction (310 ± 50 nM) (Fig. 1) (18). Collectively, these results indicate that, although the affinity of the Vβ2 NKT TCR interaction for CD1d-α-GalCer is comparable with the affinity of Vβ8.2 NKT TCRs, both Vβ2 and Vβ7 NKT TCRs are more sensitive to structural modifications of the glycolipid Ag.
Structure of the Vβ2 NKT TCR-CD1d-α-GalCer Complex.
To gain additional insight into Vβ2 NKT TCR-mediated recognition of CD1d-Ag, we formed and crystallized the complex with CD1d-α-GalCer. The structure of the Vα14Jα18-Vβ2 NKT TCR-CD1d-α-GalCer complex was subsequently determined at 3.1 Å resolution to an Rfac and Rfree of 21.7% and 26.8%, respectively (Table S2). The initial experimental phases clearly showed unbiased electron density for the α-GalCer (Fig. S1).
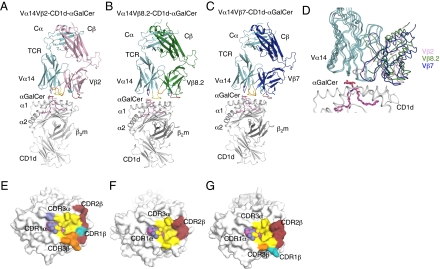
The Vβ2 NKT TCR adopted a parallel docking mode above the F′ pocket of the CD1d-α-GalCer binding cleft and thus, adopted a docking topology similar to the topology previously observed (14, 15, 17, 18) with the Vβ11, Vβ8.2, and Vβ7 NKT TCRs (Fig. 2 A–C). However, there was a slight difference in the Vα-Vβ juxtapositioning between the three mouse NKT TCRs (∼8–14° rotation between the Vβ8.2 vs. Vβ2 and Vβ7 vs. Vβ2 complexes, respectively) (Fig. 2D). The Vβ2 NKT TCR contacted CD1d, spanning residues 72–87 and 145–152 of the α1- and α2-helices, respectively (Table S3). The buried surface area on ligation was ∼920 Å2, a value higher than the corresponding Vβ8.2 and Vβ7 NKT TCR-CD1d-α-GalCer complexes (buried surface area ∼ 760–860 Å2). The higher buried surface area was attributable to the increased interactions made by the TCR Vβ2 chain contacting CD1d and more specifically, the CDR3β loop (Fig. 2 E–G). Within the invariant NKT TCR α-chain, which contributed 58% of the buried surface area, the CDR1α and CDR3α loops contacted CD1d-α-GalCer and were observed to be very similar to the loops previously described for the Vβ8.2 and Vβ7 NKT TCR-CD1d-α-GalCer complex structures. Namely, in the Vβ2 complex, the CDR3α loop dominated the interactions by contributing 43% of buried surface area, whereas CDR1α contributed 15% of buried surface area. The CDR3α interactions mediated by Asp94α, Arg95α, and Arg103α were electrostatic in nature but also included some van der Waals (vdw) -mediated contacts by Gly96α, Ser97α, Leu99α, and Gly100α (Fig. 3A and Table S3). The mode of Vβ2 NKT TCR docking enabled the CDR1α loop to contact CD1d, which was observed recently in Vβ8.2 NKT TCR-CD1d-α-GalDAG and CD1d-α-GalCer analog structures, further showing the role of CDR1α in mediating interactions not only with the Ag but also with CD1d (17, 18).
Fig. 2.
Structure of mouse NKT TCRs in complex with mouse CD1d-αGalCer. (A) Vα14Vβ2 NKT TCR-CD1d-α-GalCer. Magenta, α-GalCer; gray, CD1d heterodimer; cyan, Vα14; pink, Vβ2; purple, CDR1α; yellow, CDR3α; teal, CDR1β; ruby, CDR2β; orange, CDR3β. (B) Vα14Vβ8.2 NKT TCR-CD1d-α-GalCer (14) [Protein Data Bank (PDB) reference ID 3HE6]. Green, Vβ8.2. α-GalCer, CD1d, Vα14, and CDR loops color coding as in A. (C) Vα14Vβ7 NKT TCR-CD1d-α-GalCer (14) (PDB reference ID 3HE7). Dark blue, Vβ7; α-GalCer, CD1d, Vα14, and CDR loops color coding as in A. (D) Superposition of Vβ2, Vβ8.2, and Vβ7 NKT TCR-CD1d-α-GalCer. Color coding as in A to C. (E) Footprint of Vα14Vβ2 on the surface of CD1d-α-GalCer. α-GalCer is shown in spheres. (F) Footprint of Vα14Vβ8.2 on the surface of CD1d-α-GalCer. (G) Footprint of Vα14Vβ7 on the surface of CD1d-α-GalCer. E to G color coding is the same as in A.
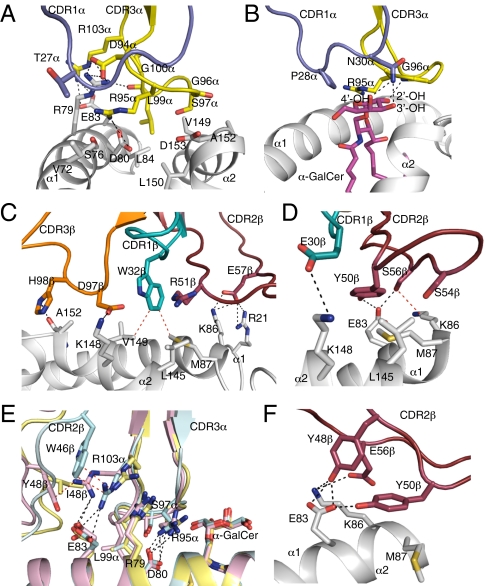
Fig. 3.
Vα14Vβ2 NKT TCR-mediated interactions with mouse CD1d and α-GalCer. (A) Vα14Vβ2 NKT TCR CDR1α- and CDR3α-mediated contacts with CD1d and (B) α-GalCer. Gray, CD1d; purple, CDR1α; yellow, CDR3α. (C) Vα14Vβ2 NKT TCR CDR1β-, CDR2β-, and CDR3β-mediated contacts with CD1d. Gray, CD1d; teal, CDR1β; ruby, CDR2β; orange, CDR3β. (D) Vα14Vβ7 NKT TCR CDR1β- and CDR2β-mediated contacts with CD1d. CD1d, CDR1β, and CDR2β color coding as in C. (E) Superposition of Vβ2, Vβ8.2, and Vβ7 NKT TCR-CD1d-α-GalCer complexes. Blue, Vβ2 complex; pink, Vβ8.2 complex; yellow, Vβ7 complex. (F) Vα14Vβ8.2 NKT TCR CDR2β-mediated interaction with CD1d. CD1d and CDR2β color coding as in C. H-bond or salt bridge interactions are shown in black dashed lines, and some vdw interactions are shown in red dashed lines.
The galactose head group of α-GalCer protruded out of the binding cleft and made contact solely with the CDR1α and CDR3α loops of the Vβ2 NKT TCR, similar to the action observed for the Vβ8.2 and Vβ7 NKT TCR-CD1d-α-GalCer complexes (14, 18) (Fig. 3B and Table S3). Namely, Asn30α H-bonded to the 3′-OH and 4′-OH of the galactose head group and along with Pro28α, made various vdw interactions involving C-1 to C-4 carbons of the sugar moiety. Within the CDR3α loop, Gly96α H-bonds with the 2′-OH of galactose and Arg95α and Gly96α made vdw interactions with the galactose head group, and additionally, Arg95α made vdw contacts with the 3′-OH of the sphingosine chain.
The TCR β-chain of Vβ2 shares 28% and 27% sequence identity with Vβ8.2 and Vβ7, respectively, which is notably less than the 54% sequence identity shared by Vβ8.2 and Vβ7 (Fig. S2). As such, sequence differences in the TCR β-chain resulted in altered contacts between the Vβ2, Vβ8.2, and Vβ7 TCRs (Table S3) (14). The TCR β-chain of Vβ2 contributed 42% of the buried surface area, with the CDR1β, CDR2β, and CDR3β loops comprising 4%, 23%, and 14% of buried surface area, respectively. Although no CDR1β–CD1d interactions were present in the Vβ8.2 complex, they were observed in the Vβ7 and Vβ2 ternary complexes. Within the Vβ2 complex, the CDR1β interactions are dominated by Trp 32β, which made vdw contacts with Met87 and Val149 of the α1- and α2-helices of CD1d, respectively (Fig. 3C), whereas in the Vβ7 NKT TCR (14), Glu 30β salt-bridged to Lys148 from CD1d (Fig. 3D).
When comparing the Vβ8.2 and Vβ7 NKT TCR complex structures, sequence differences in the TCR β-chain altered the Ag-binding interface and indirectly impacted contacts made by the invariant α-chain at the tip of the CDR3α loop (14). Similar perturbations were also observed at the Vα14-Jα18-Vβ2 interface. Namely, Arg103α of the CDR3α loop from the Vβ2 NKT TCR occupied an intermediate position with respect to Arg103α in the Vβ8.2 and Vβ7 NKT TCRs (Fig. 3E). This finding was attributable to the presence of Trp46β in the Vβ2 chain that made extensive vdw interactions with Arg103α, thus allowing it to retain the polar interactions with Glu83, which was observed in the Vβ8.2 TCR. This variation at the interface, combined with altered Vα-Vβ juxtapositioning, resulted in slight changes in the interactions made by the invariant TCR α-chain of the Vβ2 NKT TCR, specifically Arg95α, Ser97α, and Leu99α compared with the Vβ8.2 and Vβ7 NKT TCR complex structures (Fig. 3E and Table S3).
The CDR3β loop-mediated interactions with residues from the α2-helix of CD1d, namely Asp97β, salt-bridged to Lys148 as well as made vdw interactions with Val149 and Ala152, the latter of which also interacted with His98β (Fig. 3C and Table S3). These CDR3β interactions were notably more extensive compared with the Vβ11, Vβ7, and Vβ8.2 NKT TCR-CD1d-α-GalCer complexes (14, 15), where minimal and no contacts were observed with CD1d, respectively. The site of the CDR3β-mediated contacts with CD1d was analogous to the interactions made by the CDR3β loop of an autoreactive Vα14 NKT TCR as well as a Vα14Vβ8.2 NKT TCR that exhibits relatively high affinity for a range of CD1d-Ags (17, 19–22).
The interactions made by germline-encoded residues Tyr 48β in Vβ8.2 NKT TCR and Tyr 50β in both Vβ8.2 and Vβ7 NKT TCRs are energetically important for the NKT TCR–CD1dAg interaction (13, 14, 25). The Vβ2 NKT TCR, however, does not possess these tyrosine residues in the CDR2β loop, and the buried surface area contribution of the Vβ2 CDR2β loop (23% buried surface area) was slightly lower compared with the Vβ8.2 and Vβ7 counterparts (26% and 27%, respectively). Within the Vβ2 NKT TCR CDR2β, Arg51β and Glu57β mediated interactions with CD1d. Namely, the side chain of Arg51β made vdw contacts with Leu145, whereas Glu 57β salt-bridged to Lys86 and Arg21 of the α1 helix of CD1d (Fig. 3C and Table S3). Thus, although the CDR2β loop of the Vβ2 NKT TCR docked in a similar location to that of Vβ8.2 and the Vβ7 NKT TCRs, the interatomic contacts at the respective CDR2β-CD1d interfaces were markedly different (Fig. 3 C–F and Table S3). Thus, despite the lack of the canonical tyrosine residues in the CDR2β loop, the Vβ2 NKT TCR docked in a conserved manner similar to the manner of the Vβ11, Vβ8.2, and Vβ7 NKT TCRs.
Mutagenesis at the Vβ2 NKT TCR-CD1d-α-GalCer Interface.
Next, we aimed to establish the Vβ2 and CD1d residues that were energetically important in the interaction. The structure of the Vβ2 NKT TCR-CD1d-α-GalCer complex allowed us to undertake precise structural correlates of the alanine-scanning mutagenesis study previously conducted (13). Namely, the marked effect of the Tyr30βAla Vβ2 NKT TCR mutant in interacting with CD1d-α-GalCer was indirect, because Tyr30β does not contact CD1d-α-GalCer, and its mutation to Ala would impact on the conformation of the CDR1β loop. The marked effect of the Trp32βAla mutant underscored the importance of this residue in mediating contacts, although because the aromatic ring of Trp 32β packed against Arg51β of CDR2β and the CDR3α loop, the Trp32βAla mutation may also affect the structure of the local environment. Within the CDR2β loop, no mutation abrogated Vβ2 NKT TCR-CD1d-α-GalCer recognition, which contrasted the central role of this CDR2β loop in the Vβ11, Vβ8.2, and Vβ7 NKT TCR-mediated interaction (13, 14, 27). Arg51βAla and Glu57βAla had the greatest impact on the affinity (∼50% and 80% reduction, respectively) (13), which was consistent with their role in contacting CD1d. Interestingly, the Asp55βAla mutation improved the affinity of the interaction, indicating that this polar-based residue at the periphery of the interface does not contribute energetically to the complexation. Similar heteroclytic effects have been observed in TCR–pMHC complexes (35, 36) and also, the Vβ7 NKT TCR-CD1d-α-GalCer interaction (14), and our findings also highlight the suboptimal nature of the Vβ2 CDR2β loop in contacting CD1d.
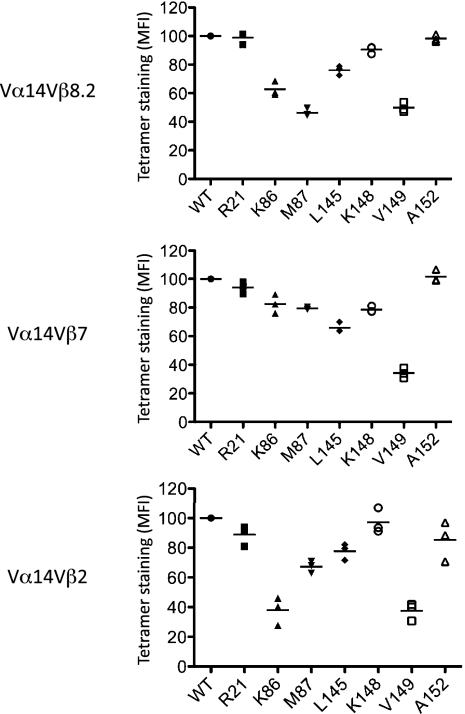
To further evaluate the individual role of residues within the Vβ2 NKT TCR-CD1d interface, we mutated the CD1d contact residues, namely Arg21, Lys86, Met87, Leu145, Lys148, Val149, and Ala152. These single-site alanine (or glycine for Ala152) mutants were found to exhibit very similar biophysical properties and yields compared with WT CD1d, suggesting that the mutations did not affect the conformation of CD1d. We generated CD1d-α-GalCer tetramers and measured the affinity of each mutant for thymus-derived Vβ8.2, Vβ7, and Vβ2 NKT cells (Fig. 4). For all of the mutants tested, none completely abrogated binding to the NKT cells. The pattern of reactivity against these mutants was approximately similar across all of the Vβ8.2, Vβ7, and Vβ2 NKT cells, indicating an equal energetic contribution of these CD1d residues in interacting with the NKT TCRs. For example, the Val149Ala mutant had the most marked effect on Vβ2, Vβ7, and Vβ8.2 NKT TCR interaction, a residue that interacts with the CDR1β, CDR3β, and CDR3α loop of the Vβ2 NKT TCR, whereas for the Vβ8.2 and Vβ7 NKT TCRs, Val149 exclusively contacted the CDR3α loop; therefore, the effect of this mutation is attributable to disruption of the interactions mediated through the invariant TCR α-chain (Table S3). Additionally, Lys86Ala had a most marked effect for the Vβ2 NKT cells. The impact of the Lys86Ala is attributable to disrupting the salt bridge to Glu57β in the Vβ2 NKT TCR, which was consistent with this TCR residue being energetically important (13, 14). Although this Glu57β-Lys86 salt bridge interaction is also present in the Vβ8.2 NKT TCR-CD1d-α-GalCer complex (Glu56β-Lys86) and a vdw interaction occurs between Ser56β-Lys86 in the Vβ7 complex, the Lys86Ala mutant did not appreciably impact on Vβ7 NKT TCR and only moderately impacted on Vβ8.2 NKT TCR-mediated recognition. This finding indicates that the Lys86-mediated contact in the Vβ8.2 or Vβ7 NKT TCR-CD1d-α-GalCer complexes is less energetically important. Collectively, our data suggest that the CDR2β loop of the Vβ2 NKT TCR is less optimally configured to interact with CD1d compared with the Vβ8.2 and Vβ7 NKT TCR.
Fig. 4.
Impact of CD1d mutants on NKT TCR binding. Mouse CD1d-α-GalCer tetramers (WT or mutants) were assessed for their ability to bind Vβ2+, Vβ7+, and Vβ8.1/8.2+-enriched thymic NKT cells. CD3+NK1.1+ Vβ+ cells were examined for mean fluorescence intensity (MFI) of mCD1d/α-GalCer tetramer. Data are shown as a percentage of WT and are from three independent experiments with each experiment represented by a symbol.
Antigen Recognition by Vβ2+ NKT Cells with Diverse CDR3β Loops.
Given the extensive role of the Vβ2 NKT TCR CDR3β loop in mediating interactions with CD1d, we next determined how well the isolated Vβ2 NKT TCR (with only one CDR3β sequence) used in our molecular studies was representative of the Vβ2+ NKT cell population and to what extent CDR3β diversity contributed to CD1d-Ag recognition. To address this question, we used a CD1d tetramer dilution assay to compare the staining intensity of freshly isolated NK1.1+CD3+ NKT cells (with variable CDR3β use) to compare staining of Vβ2+ cells with Vβ8+ and Vβ7+ cells for the range of glycolipid Ags tested in our SPR studies (Fig. S3). With this approach, the vast majority of cells are NKT cells (as seen using the α-GalCer–loaded CD1d tetramer), although a small percentage failed to stain with this tetramer, which would correspond with non-NKT cells that are known to fall within the NK1.1+CD3+ population (4). Whereas similar staining of Vβ2, Vβ8, and Vβ7 NKT cells was achieved for α-GalCer (C26), α-GalCer (C20:2), and α-GlcCer (C20:2), we observed reduced staining for 3′4″-deoxy α-GalCer and 4′,4″-deoxy α-GalCer and very low staining for OCH for Vβ2+ NKT cells, which was depicted by lower median fluorescence intensity in Fig. S3A and also, tetramer dilution analysis in Fig. S3B. Interestingly, for each of 3′,4″-deoxy α-GalCer, 4′,4″-deoxy α-GalCer, and OCH, the staining intensity of Vβ2+ NKT cells ranged from very low to high (Fig. S3A), which is consistent with a more prominent role for the hypervariable CDR3β loop in Vβ2 NKT TCR-mediated Ag recognition. Also, Vβ7+ NKT cells stained less brightly than Vβ8+ NKT cells when CD1d tetramer was loaded with OCH, consistent with our earlier observation that Vβ7+ NKT cells were underrepresented when OCH was used to drive NKT cell proliferation in vitro (18). Collectively, our tetramer dilution studies, in agreement with our SPR data, support the notion that Vβ2 NKT TCRs are less tolerant to modifications in the glycolipid Ag than Vβ8+ and Vβ7+ NKT cells.
To further examine the role of CDR3β diversity in Vβ2+ NKT TCR binding to CD1d-Ag, we next established the importance of the residues within this loop. To assess this importance, we used retroviruses to generate a CDR3β library encoding Vβ2 chains in which four positions at the tip of the CDR3β loop were randomized. The library was estimated to encode ∼15,000 different sequences, and retroviruses were used to transduce a Vα14-expressing TCRβ-negative hybridoma, which was previously described (19, 26). Transduced cells were sorted for TCRβ expression and stained with the CD1d-α-GalCer tetramer. Hybridomas expressing a Vβ6 chain or Vβ8.2 chain with the DO.11.10 CDR3β sequence were used as negative and positive controls of the staining, respectively. Approximately 20% of the CDR3β sequences in the context of Vβ2-expressing TCRs interacted with tetramer (Fig. S4A), indicating that only a fraction of the CDR3β sequences are compatible with CD1d-α-GalCer recognition, which is in marked contrast to the lack of dependency of the CDR3β loop in Vβ8.2 and Vβ7 NKT TCR-mediated interactions with CD1d-α-GalCer (13). To determine the nature of the CDR3β responsible for this reactivity, CD1d-α-GalCer tetramer positive and negative cells from the Vβ2+ TCR library were sorted (Fig. S4B). mRNA was extracted from each population, and after cDNA synthesis, the Vβ2 TCRs were amplified by PCR using appropriate primers, cloned into the retroviral vector, and sequenced. In addition, each Vβ2 TCR was expressed separately with the invariant Vα14 chain into the 5KC hybridoma and stained with CD1d-α-GalCer tetramer. Multiple diverse CDR3β sequences in the Vβ2 chain, with no particular motif being favored, could bind the CD1d-α-GalCer tetramer [mean fluorescence intensity (MFI) > 600] when paired with the canonical Vα14 NKT TCR chain (Table S4). The lack of the requirement of a CDR3β motif within the Vβ2 NKT TCR engendering CD1d-α-GalCer reactivity is in contrast to the CDR3β motif required for Vβ8.2 NKT TCR-mediated autoreactivity (19). Taken together, these results verify the findings from our structural data, showing that, in stark contrast to Vβ8.2 and Vβ7 NKT TCRs, for Vβ2 NKT TCRs there is a greater dependency of the CDR3β loop in mediating interactions with CD1d-α-GalCer.
Discussion
Structures of NKT TCRs have been determined in complex with CD1d bound to various Ags (14, 15, 17–22). Despite the variability among the glycolipid Ags and a diverse but limited NKT TCR Vβ repertoire, a conserved docking mode has been observed. A fundamental question arises from these observations: what drives this innate style NKT TCR-CD1d docking mode? Previous structural and mutational studies have suggested that the conserved docking topology could arise from either the Jα18-encoded region and/or the germline-encoded CDR2β loop of the NKT TCR (13, 14, 26, 27). Within the human Vβ11 NKT TCR and the mouse Vβ8.2 and Vβ7 NKT TCRs, tyrosine residues encoded within their respective CDR2β loops make a series of energetically important and conserved contacts with CD1d. Thus, it was uncertain whether the Vβ2 NKT TCR would adopt the consensus NKT TCR-CD1d-Ag docking topology. Our structural data on the Vβ2 NKT TCR-CD1d-α-GalCer complex indicate that the tyrosine residues within the CDR2β-encoded loop do not play an exclusive role in determining the conserved docking mode. Consequently, our data suggest that the Vα chain and specifically, the Jα18-encoded loop, which contacts CD1d and the Ag, drive the pattern recognition receptor properties of the NKT TCR (26). However, recent studies have also identified a population of α-GalCer–reactive semi-invariant NKT cells in which the TCR α-chain is comprised of the Vα10-Jα50 genes and the TCR β-chain is largely restricted to the Vβ8 and Vβ7 genes (23). Despite the markedly different sequences of the Jα18- and Jα50-encoded gene segments, the Vα10-Jα50 NKT TCR docked onto CD1d-Ag in a very similar manner to the docking of the Vα14-Jα18 NKT TCR-CD1d-Ag complexes. Thus, although varied gene use of αβTCRs results in a wide variety of TCR-pMHC docking modes (6, 37), differing gene use by NKT TCRs converges to arrive at the same solution to bind the monomorphic CD1d. Why this occurrence happens is unclear, but it illustrates a fundamental difference between peptide MHC- and lipid CD1d-mediated immunity. In this context, it will be interesting to establish how other TCRs, such as the TCRs expressed by Type II NKT cells, which differ from Type I NKT cells in Vα and Vβ use, dock onto CD1d (16).
Within this common docking framework, fine specificity differences between the Vβ8.2, Vβ7, and Vβ2 NKT TCRs are apparent. For example, although the Vβ8.2 NKT TCR showed a greater dependency on the 4′-OH position of α-GalCer compared with the 3′-OH moiety (18), the affinity of the Vβ2 NKT TCR interaction was dramatically reduced by either modification. Moreover, the OCH analog, with a truncated sphingosine chain, impacted markedly on Vβ2 and Vβ7 NKT TCR recognition, whereas the Vβ8.2 NKT TCR was less affected. These observations are consistent with the NKT TCR being able to sense modifications within the F′ pocket through an induced fit mechanism (17, 18, 38) but also highlight that the Vβ2 and Vβ7 NKT TCRs are less tolerant to such perturbations, possibly as a result of the latter two NKT TCRs possessing nonoptimal CDR2β sequences for CD1d engagement. Nevertheless, the affinity of the Vβ8.2, Vβ7, and Vβ2 NKT TCRs for CD1d-α-GalCer were quite comparable, with an affinity hierarchy of Vβ8.2 ∼ Vβ2 > Vβ7, indicating that the Vβ2 NKT TCRs need not necessarily be of lower affinity compared with the Vβ7 and Vβ8.2 NKT TCRs. The high affinity of the Vβ2 NKT TCR was most likely attributable to the compensatory role of the CDR3β loop, which made a clear contribution to the NKT TCR-CD1d interface. In line with this work, it has emerged that the CDR3β loop can also significantly enhance the binding affinity of Vβ8 and human Vβ11 NKT TCRs, thus contributing to NKT TCR autoreactivity against self-lipid antigens presented by CD1d (12, 19–22). Thus, in this regard, the CDR3β and CDR2β loops collaborate to enable functional recognition of CD1d-Ag (13). Furthermore, many randomized CDR3β residues failed to support staining by CD1d-α-GalCer, whereas freshly isolated NKT cells with diverse CDR3β were all stained by this reagent; this finding suggests that permissive CDR3β loops are selected in the thymus during the process of NKT cell development. Although no particular motif was favored for CDR3β loop recognition, analysis of the mouse NKT ternary complexes solved to date that involve a role for the CDR3β loop (17, 19–23) suggests a focal point within CD1d, comprising residues Lys148, Val149, and Ala152, underpins recognition by this loop, regardless of CDR3β amino acid sequence.
Accordingly, our studies have indicated that NKT TCRs dock onto CD1d in a conserved manner, regardless of TCR β-chain use. Regarding Vβ2 NKT cells, these interactions are heavily influenced by CDR3β diversity and interactions between this loop and CD1d. Taken together, although the invariant TCR α-chain exerts a major influence in facilitating CD1d-Ag recognition, TCR β-chain diversity fine tunes and modulates the NKT cell response.
Materials and Methods
Cloning and Expression of Genes Encoding the Mouse Vβ2 NKT TCRs.
RNA was extracted from NKT-expressing mouse thymocytes (purified by flow cytometric sorting of thymocytes stained with CD1d-α-GalCer tetramers) and reverse-transcribed. cDNAs encoding the mouse Vα14 and Vβ2 NKT TCR chains were amplified by PCR and cloned into P-GEM Easy (Promega). We were unable to refold the intact ectodomains of murine NKT TCRs and instead used the human constant domains of the NKT TCR to aid in refolding, which was described previously (14). The C-terminal sequences were PEDTFFPSPENDGGGCK for the α-chain and AEAWGRADQDRGGGCD for the β-chain, similar to the sequences previously described (39).
Vα14 and Vβ2 NKT TCR chains were expressed in BL21 Escherichia coli, and inclusion body protein was prepared, refolded, and purified essentially as previously described. The functional integrity of the NKT TCRs was confirmed by gel filtration and gel shift experiments.
Cloning and Expression of mCD1d, Mutagenesis, and Loading of CD1d-Ag.
Cloning and expression of mCD1d, mutagenesis, and loading of CD1d-Ag are described in SI Materials and Methods.
Flow Cytometry.
Flow cytometry is described in SI Materials and Methods.
Surface Plasmon Resonance Measurements and Analysis.
The interaction between soluble NKT TCR and the CD1d-Ag complexes was analyzed by SPR with a Bio-Rad ProteOn XPR36 instrument essentially as described previously (14). Briefly, 50–300 response units of biotinylated CD1d-Ag were coupled to a streptavidin-coated GLC sensor chip (Bio-Rad), and soluble TCRs were serially diluted and simultaneously injected for 1–3 min at 30 μL/min over test and control (CD1d-endogenous) surfaces. The interactions were analyzed with ProteOn Manager version 2.1 (Bio-Rad) and Scrubber 2.0a software (Prot version; BioLogic Software). Steady state KD values were derived at equilibrium, and association rate (ka) and half-life (t1/2) were derived using a 1:1 Langmuir kinetic binding model.
CDR3β Libraries.
CDR3β libraries are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff at the MX2 beamline of the Australian synchrotron for assistance with data collection; Meena Thakur for the gift of the 4′,4″-deoxy α-GalCer analogue; and Christina Wang and Maria Sandoval for technical assistance. This work was supported in part by the Australian Research Council (ARC), the National Health and Medical Research Council of Australia (NHMRC), and the Cancer Council of Victoria; National Institutes of Health (NIH)/National Institute of General Medical Sciences Grant GM087136 (to A.R.H.); NIH Grant AI45889 (to S.A.P.); NIH Grant AI057485 (to L.G.); an ARC Federation Fellowship (to J.R.); and an NHMRC Principal Research Fellowship (to D.I.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3TO4).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109066108/-/DCSupplemental.
References
- 1.Godfrey DI, Kronenberg M. Going both ways: Immune regulation via CD1d-dependent NKT cells. J Clin Invest. 2004;114:1379–1388. doi: 10.1172/JCI23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 4.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: What's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 5.Brutkiewicz RR. CD1d ligands: The good, the bad, and the ugly. J Immunol. 2006;177:769–775. doi: 10.4049/jimmunol.177.2.769. [DOI] [PubMed] [Google Scholar]
- 6.Godfrey DI, Rossjohn J, McCluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 8.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 9.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci USA. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou D, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 12.Matulis G, et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallevaey T, et al. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 2009;31(1):60–71. doi: 10.1016/j.immuni.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31(1):47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448(7149):44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 16.Godfrey DI, et al. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin Immunol. 2010;22:61–67. doi: 10.1016/j.smim.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J Exp Med. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wun KS, et al. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 2011;34:327–339. doi: 10.1016/j.immuni.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallevaey T, et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity. 2011;34:315–326. doi: 10.1016/j.immuni.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aspeslagh S, et al. Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J. 2011;30:2294–2305. doi: 10.1038/emboj.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu ED, Girardi E, Wang J, Zajonc DM. Structural basis for the recognition of β-linked glycolipid antigens by invariant NKT cells. J Immunol. 2011;187:2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellicci DG, et al. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol. 2011;12:827–833. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uldrich AP, et al. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat Immunol. 2011;12:616–623. doi: 10.1038/ni.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjer-Nielsen L, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Florence WC, et al. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 2009;28:3579–3590. doi: 10.1038/emboj.2009.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 27.Wun KS, et al. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J Exp Med. 2008;205:939–949. doi: 10.1084/jem.20072141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 30.Dai S, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pyz E, et al. The complementarity determining region 2 of BV8S2 (V beta 8.2) contributes to antigen recognition by rat invariant NKT cell TCR. J Immunol. 2006;176:7447–7455. doi: 10.4049/jimmunol.176.12.7447. [DOI] [PubMed] [Google Scholar]
- 32.Raju R, et al. Synthesis and evaluation of 3″- and 4″-deoxy and -fluoro analogs of the immunostimulatory glycolipid, KRN7000. Bioorg Med Chem Lett. 2009;19:4122–4125. doi: 10.1016/j.bmcl.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jervis PJ, et al. Synthesis and biological activity of alpha-glucosyl C24:0 and C20:2 ceramides. Bioorg Med Chem Lett. 2010;20:3475–3478. doi: 10.1016/j.bmcl.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531–534. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 35.Kjer-Nielsen L, et al. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18(1):53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 36.Borg NA, et al. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat Immunol. 2005;6:171–180. doi: 10.1038/ni1155. [DOI] [PubMed] [Google Scholar]
- 37.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci USA. 2010;107:10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy C, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.