Abstract
Negative-strand (NS) RNA viruses comprise many pathogens that cause serious diseases in humans and animals. Despite their clinical importance, little is known about the host factors required for their infection. Using vesicular stomatitis virus (VSV), a prototypic NS RNA virus in the family Rhabdoviridae, we conducted a human genome-wide siRNA screen and identified 72 host genes required for viral infection. Many of these identified genes were also required for infection by two other NS RNA viruses, the lymphocytic choriomeningitis virus of the Arenaviridae family and human parainfluenza virus type 3 of the Paramyxoviridae family. Genes affecting different stages of VSV infection, such as entry/uncoating, gene expression, and assembly/release, were identified. Depletion of the proteins of the coatomer complex I or its upstream effectors ARF1 or GBF1 led to detection of reduced levels of VSV RNA. Coatomer complex I was also required for infection of lymphocytic choriomeningitis virus and human parainfluenza virus type 3. These results highlight the evolutionarily conserved requirements for gene expression of diverse families of NS RNA viruses and demonstrate the involvement of host cell secretory pathway in the process.
Keywords: RNA interference, transcription and replication, host cell factors for virus infection
Negative-strand (NS) RNA viruses include a large group of human and animal pathogens that cause diseases ranging from mild flu-like symptoms to fatal hemorrhagic fever. Vesicular stomatitis virus (VSV), the prototype of the family Rhabdoviridae, is an enveloped virus with a nonsegmented NS RNA genome. VSV infects most vertebrate and many invertebrate cells and has a short infection cycle. These characteristics have earned appreciation for VSV as an excellent model for understanding virus entry, genome uncoating, replication, assembly, and budding processes, as well as for studying innate and adaptive immune defense mechanisms. VSV is also used as a viral vaccine vector, as an oncolytic agent, and for gene therapy (1).
VSV encodes five proteins: the nucleocapsid protein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the large polymerase protein (L) (2). The viral RNA exists in the virion core as N protein-bound nucleocapsid (NC) to which the viral polymerase is associated. During infection, VSV binds to susceptible cells, although the receptor(s) mediating virus entry remains unidentified (3). It enters cells by clathrin-mediated endocytosis, requiring endocytic adaptor protein AP-2, actin, and dynamin (4–6). Once in the cytoplasm, low pH-dependent fusion of viral envelope with the endosomal membrane leads to the release of NC in the cytoplasm for transcription and replication to occur. Progeny NCs are transported toward the cell periphery in a microtubule-dependent manner (7). Assembly of the viral components occurs at the plasma membrane, and nascent virions are released from the cells.
Viruses use key cellular pathways for their infection and replication (8, 9). Although much is known about the viral proteins in the biology of the virus, little is known about the host factors in VSV and other NS RNA virus infections. Identifying the cellular factors and studying the mechanisms of their involvement in these viral infections is important not only for understanding the biology of these pathogens, but also for development of antiviral therapeutics.
The advent of siRNA technology and the availability of genome-wide siRNA libraries have been useful in identifying host factors required for influenza virus, an NS RNA virus, and several positive-strand RNA viruses, as well as HIV (10–19). The lack of similar studies with other NS RNA viruses has limited the understanding of the role of host cell factors in replication of these viruses. Using VSV, we conducted a genome-wide siRNA screen to identify mammalian genes required for viral infection. Our studies revealed requirements for several cellular pathways and proteins in VSV infection. Many of the factors identified in the screen for VSV are also required for infection by two other NS RNA viruses: the human parainfluenza virus type 3 (HPIV3), a nonsegmented NS RNA virus in the family Paramyxoviridae, and the lymphocytic choriomeningitis virus (LCMV), a segmented genome NS RNA virus in the family Arenaviridae. Interestingly, for these three viruses representing diverse families of NS RNA viruses, viral gene expression required the function of the coatomer complex I (COPI), a coat protein complex involved in retrograde vesicular transport from the Golgi to the endoplasmic reticulum (ER) (20). Overall, the studies reveal a critical need for the cellular secretory pathway in gene expression of disparate families of NS RNA viruses.
Results
Genome-Wide RNAi Screen for Host Factors in VSV Infection.
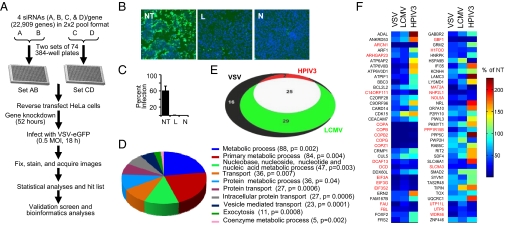
To identify host proteins required for VSV infection, a genome-wide siRNA screen was conducted. Four independent siRNAs, grouped into a 2 × 2 pool format (Fig. 1A), targeting each of 22,909 mammalian genes, were used. VSV-eGFP, a recombinant virus encoding enhanced green fluorescent protein (eGFP) (7), was used to infect HeLa cells. Expression of eGFP in the cells would indicate virus infection and gene expression. AllStars nontargeting (NT) siRNA and siRNAs targeting the VSV L and N mRNAs were used as controls. At 52 h post-siRNA transfection (hpt), cells were infected with VSV-eGFP (hereafter VSV), and at 18 h postinfection (hpi), they were fixed, stained for nuclei, and processed for automated image analysis. Cell number and percent infection were obtained for each well. Infection of NT siRNA-transfected cells was optimized to yield, on average, 60% infection rate. Under these conditions, the rate of infection of cells transfected with L and N siRNAs was reproducibly <1% (Fig. 1 B and C). Wells with low cell number due to combined effects of siRNA toxicity and VSV cytopathic effects (SI Experimental Procedures) were excluded from further analyses. Sum rank analysis (21) was used, and 233 host genes (P < 0.01) were identified as required for VSV infection.
Fig. 1.
The RNAi screen. (A) Schematic of the RNAi screen. (B) Representative images of cells treated with AllStars NT, L, and N siRNAs and infected with VSV-eGFP. (C) Bar graph showing the percent infection (mean ± SD) of cells for control siRNAs in the genomic screen. (D) Overrepresented Panther categories of biological processes of cellular genes identified in the screen. The number of genes in each category and statistical significance (P) are shown. (E) The extent of overlap among the validated genes for VSV, LCMV, and HPIV3 is shown in the Venn diagram. (F) Percent infections for the validated genes normalized to that of NT siRNA are presented as a heat map. Data represents average of four experiments for VSV and two experiments for LCMV and HPIV3. Genes required for all three viruses are shown in red.
We found several expected genes, including those of the canonical ribosomal proteins, key parts of the cellular translational machinery. We deleted the ribosomal protein genes and narrowed down the list to 173 genes (Dataset S1). Because our screen design used a 2 × 2 siRNA pool format, the 173 genes exhibited the same phenotype with at least two different siRNAs (at least one from each pool), a recommended criterion for true hit identification (22). Thus, the 173 genes identified may represent factors required for VSV infection. The biological process categories of cellular genes overrepresented in the list based on the Panther classification system (23) include genes involved in metabolic processes, nucleic acid metabolism, intracellular protein transport, vesicle-mediated transport, and exocytosis, among others (Fig. 1D). Pathway analysis revealed that protein functions involved in at least eight major cellular pathways were significantly enriched and showed possible interconnections between these pathways for VSV infection (Fig. S1A). Comparison of the mammalian genes identified from genome-wide siRNA screens for other RNA viruses (10–13, 15, 16, 24, 25) with those from the VSV screen revealed that genes in several major biological functions categories are shared by RNA viruses (Fig. S1B).
We then conducted a validation screen for the 173 identified genes using siRNAs from another source, Dharmacon ON-TARGETplus pool of four siRNAs. The validation screen was performed in four replicates, and analysis of the results (SI Experimental Procedures) led to identification of 72 out of the 173 genes as required for VSV infection (Fig. 1F and Dataset S2).
HPIV3 and LCMV Infection Share Many Factors Required for VSV Infection.
To identify genes and pathways used by diverse families of NS RNA viruses, the involvement of the 72 genes identified for VSV was examined in HPIV3 and LCMV infection. These viruses belong to distinct families of NS RNA viruses and have common as well as unique strategies for entry, uncoating, replication, and virus assembly. GFPs expressing LCMV and HPIV3 (26, 27) were used, and the screen was conducted as described for VSV in duplicate. By using this strategy, 54 and 27 genes were identified as required for LCMV and HPIV3 infection, respectively, whereas 25 genes were required for infection by all three viruses (Fig. 1E). The genes identified for infection by the three viruses along with their known and putative functions, can be found in Datasets S2 and S3, respectively. The normalized percent infection for the 72 genes identified as required for VSV are shown in the heat map (Fig. 1F) and are compared with those found for LCMV and HPIV3.
Identification of Genes Involved in Various Stages of VSV Infection.
The screen used a multicycle VSV infection assay that included all stages of the virus infection cycle, such as entry and uncoating, transcription and replication, and assembly and release. Our screen identified several subunits of vATPase as necessary for VSV infection (Fig. 1F and Fig. S2), confirming the known role of vATPase in endocytosis and virus uncoating (28).
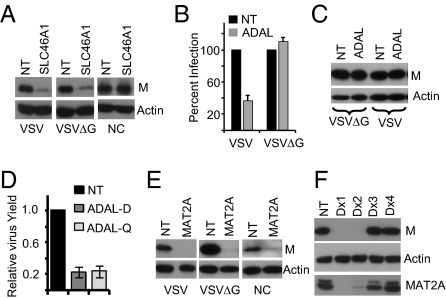
The screen identified several solute carriers localized to the plasma membrane, including the solute carrier family 46 member 1 [SLC46A1; proton-coupled folate transporter (PCFT)]. We examined the role of SLC46A1 in VSV infection. Results showed that in siRNA-treated cells infected with VSV or VSVΔG [a virus that lacks the G gene and cannot produce infectious virus, so the infection with this virus is limited to single cycle only (29)], VSV gene expression (levels of M protein) was reduced (Fig. 2A). We then transfected VSV NC to cells treated with siRNAs for SLC46A1 to bypass the endosome-mediated entry and uncoating and examined the effects of depletion of the protein on gene expression. VSV NCs are not infectious per se when added to cells but can initiate the viral genome transcription and replication when delivered into the cytoplasm by transfection. Results show that virus gene expression from transfected NCs remained unaffected (Fig. 2A). Multiple siRNAs from two different sources also led to reduced viral gene expression as well as depletion of the SLC46A1 protein (Fig. S3A), indicating that the inhibitory effect is specific. Together, these results suggest that SLC46A1 may be required for VSV entry and/or uncoating.
Fig. 2.
Identification of genes for VSV entry/uncoating, assembly/budding, and gene expression. (A) Cells were transfected with SLC46A1 siRNA for 72 h and infected with viruses (VSV, 0.005 MOI; VSVΔG, 0.2 MOI) or transfected with NC. Cell extracts prepared at 14 hpi (VSV), 5 hpi (VSVΔG), or 6 hpt (NC) were examined for M expression. (B) Experiment was performed as in A by using ADAL siRNAs, and percent infection was determined. Values are normalized to NT siRNA and represent mean ± SD from three experiments. (C) Cells transfected with ADAL siRNA were infected with VSV (0.1 MOI) or VSVΔG (0.2 MOI), and M protein expression was examined at 5 hpi. (D) Relative VSV yield (mean ± SD) at 14 hpi (0.001 MOI) after ADAL siRNAs from Dharmacon (ADAL-D) or Qiagen (ADAL-Q) treatment from three experiments. (E) Effect of siRNA for MAT2A on viral gene expression in cells infected with VSV or VSVΔG or transfected with NCs. Experimental conditions are as in A. (F) Cells were transfected with individual siRNA duplexes (Dharmacon) for MAT2A and infected with VSV (0.005 MOI) for 12 h. Cell lysates were examined for levels of M, actin, and MAT2A proteins.
To identify genes involved in virus assembly and release, we screened the 72 genes using both multicycle (with VSV) and single-cycle (with VSVΔG) infection assays. We found that in ADAL (Adenosine Deaminase Like) siRNA-treated cells infected with VSV, the percent infection was ~threefold less compared with the NT siRNA-treated cells, whereas the percent infection of cells with VSVΔG was similar in both ADAL and NT siRNA-treated cells (Fig. 2B). We found no differences in the level of viral gene expression in cells treated with either siRNAs and infected with either VSVΔG or VSV (Fig. 2C). However, infectious virus production was reduced in VSV-infected cells treated separately with two different sources of ADAL siRNA (Fig. 2D). These results suggest that ADAL may facilitate VSV assembly and/or release.
Further, we found that methionine adenosyltransferase 2A (MAT2A) siRNA treatment reduced VSV gene expression significantly (Fig. 2E). MAT2A is involved in l-methionine metabolism and catalyzes formation of S-adenosylmethionine (30). MAT2A siRNA treatment also reduced gene expression in cells infected with VSVΔG or in cells transfected with NCs (Fig. 2E). Multiple siRNAs for MAT2A led to depletion of the MAT2A protein and corresponding reduction in VSV gene expression (Fig. 2F). Viral mRNA and anti-genome levels were reduced in cells treated with MAT2A siRNA (Fig. S3B). These results indicate that MAT2A may have a role in viral gene expression.
COPI Is Necessary for VSV Infection at the Level of Viral Gene Expression.
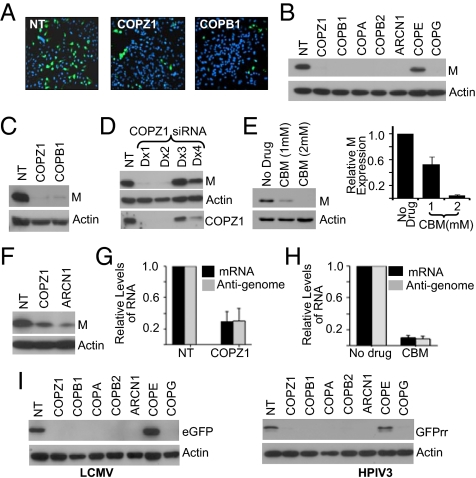
Network analysis revealed vesicle trafficking as one of the top scoring pathways required for infection. COPI is involved in retrograde vesicular transport of luminal and membrane proteins from the Golgi to the ER and intra-Golgi transport (20). We focused on COPI because multiple proteins in this pathway were identified and siRNAs for COPI subunits (except COPE) exhibited strong inhibition of VSV infection (Fig. 1F). Fig. 3A shows inhibition in VSV infection in cells treated with COPZ1 or COPB1 siRNAs.
Fig. 3.
COPI is required for VSV, LCMV, and HPIV3 infection. (A) HeLa cells transfected with siRNAs for COPI subunits were infected with VSV (MOI = 0.01), and at 14 hpi, they were fixed, stained with DAPI, and imaged at 10× magnification. (B) Cells transfected with siRNAs for COPI subunits for 48 h were infected with VSV (0.5 MOI) for 4 h. VSV gene expression was assessed by immunoblotting for M. (C) Cells transfected with siRNAs for the COPI subunits as above were infected with VSVΔG virus. At 5 hpi, viral gene expression was assessed by immunoblotting for M. (D) Cells transfected with individual siRNAs were infected with VSV for 4 h. VSV gene expression (M level) and COPZ1 levels were assessed by immunoblotting. (E) Cells pretreated with CBM for 1 h were infected with VSV as in B, and viral gene expression was assessed by immunoblotting for M. (Right) Histogram shows relative levels of viral gene expression from three experiments. (F) VSV NCs were transfected into cells treated with siRNAs for COPZ1 and ARCN1. At 6 h after NC transfection, viral gene expression was examined by immunoblotting for M. (G) VSV mRNA and anti-genomic RNA levels in COPZ1-depleted cells, determined by qRT-PCR using primers and probe concentrations shown in Tables S1 and S2 (SI Experimental Procedures). Experimental conditions were as in B. Values show mean ± SE of measurement (SEM) of duplicate reactions from two experiments after normalizing to NT control. (H) Cells infected with VSV as in B were treated with 2 mM CBM at 1 hpi. Viral mRNA and anti-genome levels were determined as in G. (I) Cells transfected with siRNAs for COPI as in B were infected with 1 MOI of LCMV for 7 h or HPIV for 14 h. Viral gene expression was assayed by immunoblotting for eGFP or Renilla reniformis-GFP (GFPrr).
Because the early secretory pathway is necessary for VSV G protein processing (31), it is possible that the COPI depletion might have affected G protein processing and, consequently, virion assembly and release. To determine whether COPI plays any role in other steps of the VSV infection cycle, we examined the effect of COPI subunit siRNAs on VSV gene expression at 4 hpi, a time at which viral gene expression is readily detectable. At this time, VSV gene expression was inhibited by all COPI siRNAs (except COPE) (Fig. 3B). Furthermore, VSVΔG virus infection of cells depleted of COPZ1 and COPB1 showed a significant reduction of viral gene expression (Fig. 3C), indicating that COPI plays a role in facilitating viral entry, uncoating, and/or gene expression. The degree of inhibition of VSV gene expression correlated with the level of depletion of COPZ1 (Fig. 3D) and COPB1 (Fig. S4A). Multiple siRNA for each of the COPI subunits (except COPE) reduced VSV gene expression (Fig. S4B). We then used 1,3-cyclohexanebismethylamine (CBM), an inhibitor of COPI function (32, 33), to examine involvement of COPI in VSV gene expression. Treatment of cells with CBM 1 h before infection reduced VSV M protein levels in a dose-dependent manner (Fig. 3E) without adversely affecting the cell viability (Fig. S5). These studies suggest that COPI is necessary for VSV infection.
Since COPI is involved in endosomal transport (34), it is possible that the inhibition of VSV gene expression could be due to disruption of endosomal transport required for VSV entry and uncoating. However, inhibition of viral gene expression was observed in NC-transfected cells depleted of COPI subunits (Fig. 3F), suggesting that COPI is required for viral gene expression, independent of entry and uncoating steps. Additionally, we found reduced levels of mRNA (transcription product) and anti-genome (replication product) in COPZ1-depleted cells (Fig. 3G). Furthermore, when cells were treated with CBM 1 h after VSV infection, a time-frame in which the majority of the virus would have uncoated their NCs (35), reduced levels of mRNA and anti-genome were also observed (Fig. 3H), suggesting that COPI is required for VSV gene expression.
We then examined whether COPI is also required for gene expression of LCMV and HPIV3. Cells treated with siRNA for COPI subunits were infected with GFP-encoding LCMV or HPIV3. The level of GFP protein was examined in cell extracts collected at the earliest time point when GFP expression was seen in infected cells (7 hpi for LCMV or 14 hpi for HPIV3) (26). Depletion of the COPI subunits (except COPE) significantly inhibited LCMV and HPIV3 gene expression (Fig. 3I), suggesting that these viruses also require COPI functions.
Viral Gene Expression Requires ARF1 and GBF1, the Upstream Effectors of COPI.
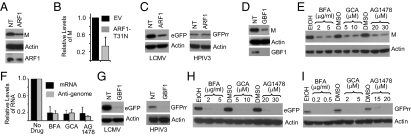
Activation of ADP ribosylation factor 1 (ARF1) is required for the recruitment of COPI complex onto the Golgi membranes (36). In ARF1 siRNA-treated cells infected with VSV, the viral gene expression was reduced by twofold with concomitant similar reduction in the levels of ARF1 protein (Fig. 4A and Fig. S6A), suggesting that moderate inhibition of viral gene expression may be due to insufficient depletion of ARF1. In cells transfected with a plasmid encoding a dominant-negative mutant, ARF1-T31N (37), viral gene expression was inhibited (Fig. 4B). ARF1 depletion also inhibited LCMV gene expression but surprisingly had no effect on HPIV3 gene expression (Fig. 4C). It is possible that moderate level of ARF1 depletion might not have shown measurable inhibitory effect on HPIV3 gene expression.
Fig. 4.
Viral gene expression requires ARF1 and GBF1. (A) Cells transfected with ARF1 siRNAs for 72 h were infected with VSV (0.05 MOI). M protein and ARF1 expression were examined at 6 hpi by immunoblotting. (B) Cells were transfected with empty vector (EV) or ARF1-T31N encoding vector. At 24 hpt, cells were infected with VSV (0.5 MOI) for 4 h. M protein levels from three experiments are shown as mean ± SD. (C) Cells transfected with ARF1 siRNAs were infected with LCMV or HPIV3 (as in Fig. 3I). Viral gene expression was examined by immunoblotting for eGFP or GFPrr. (D) Cells transfected with GBF1 siRNAs for 72 h were infected with VSV (0.5 MOI) for 4 h. M and GBF1 were examined by immunoblotting. (E) Cells infected with VSV (1 MOI) were treated with the drugs at 1 hpi. Cell lysates at 4 hpi were assessed for M. (F) Cells infected with VSV were treated with BFA (2 μg/mL), GCA (10 μM), and AG1478 (30 μM) at 1 hpi for 2 h. Fold change in mRNA and anti-genome levels were determined as in Fig. 3H. Mean ± SEM (n = 2) is shown. (G) GBF1 siRNA transfected cells were infected with LCMV or HPIV3, and viral gene expression was measured as described in Fig. 3I. (H and I) Cells infected with 1 MOI of LCMV (H) or HPIV3 (I) were treated at 1 hpi with the drugs. Viral gene expression was assessed by immunoblotting for eGFP or GFPrr at 7 (LCMV) or 14 (HPIV3) hpi.
The Golgi-associated brefeldin A resistant factor 1 (GBF1) is the guanine nucleotide exchange factor, which catalyzes GDP–GTP exchange to activate ARF1 for COPI recruitment onto the Golgi membranes (38). GBF1 siRNA reduced endogenous GBF1 protein levels by >90% and led to significant reduction in VSV M protein (Fig. 4D). Reduction in VSV mRNA and anti-genome levels was observed after GBF1 depletion (Fig. S6B), suggesting that VSV gene expression requires GBF1.
We then used inhibitors of GBF1 to probe its requirement in VSV gene expression. Brefeldin A (BFA) inhibits GBF1, BIG1 (brefeldin-inhibited guanine nucleotide exchange factor 1), and BIG2, whereas Golgicide A (GCA) and Tyrphostin AG1478 are specific inhibitors of GBF1 (39, 40). In the presence of these drugs, VSV gene expression was inhibited (Fig. 4 E and F). The drugs had no significant adverse effects on viability of uninfected cells (Fig. S5). We then examined VSV gene expression in Madin Darby canine kidney (MDCK) cells, which contain a natural mutation (M832L) in GBF1, rendering the cells resistant to BFA, GCA, and AG1478 (39–41). However, in these cells, BIG1 and BIG2 are sensitive to BFA (40). In MDCK cells infected with VSV, viral gene expression was not adversely affected, even at higher concentrations of the drugs (Fig. S6C). Studies with VSVΔG virus infection (Fig. S6D) and NC transfection (Fig. S6E) showed that GBF1 is required for VSV gene expression. In LCMV and HPIV3 infected cells, viral gene expression was reduced after depletion of GBF1 (Fig. 4G) and was sensitive to GBF1 inhibitors (Fig. 4 H and I), indicating that GBF1 is also required for these viruses.
Discussion
In the present study, using a genome-wide siRNA screen, we have identified host cell factors required for VSV infection. Because of our screen design (2 × 2 pool of siRNA format) and validation using a different source of siRNAs, the possibility of false positives in the list is likely to be low, but it may have compromised our ability to identify additional genes required for VSV infection. An integrated model (Fig. 5) revealing the host factors required for VSV infection demonstrates many previously undescribed functions and pathways used not only by VSV but also by other RNA viruses such as LCMV and HPIV3. This study provides a comparative analysis of cellular factors involved in replication of three disparate NS RNA viruses. Functions such as RNA processing, vesicular transport, transcription, and translation regulations, among others, are shared by all three viruses, which would suggest utilization of common cellular functions for their replication. It should be noted that the factors identified for LCMV and HPIV3 are only a subset of those identified for VSV and do not represent a complete set of factors required for either LCMV or HPIV3. However, it is evident that several of the cellular pathways and factors are shared by all three viruses, illustrating commonalities in the requirements for replication of these NS RNA viruses.
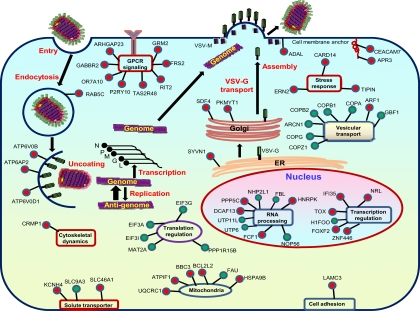
Fig. 5.
Integrated model of host factors required for VSV, LCMV, and HPIV3. Genes were placed at positions most likely relevant to VSV infection. Subcellular localization and biological functions of the proteins were determined by using Entrez Gene, Entrez PubMed, and UniProt. Genes shown as green circles are shared by all three viruses, whereas genes shown as red circles are either for VSV only or shared by VSV and LCMV or by VSV and HPIV3.
Although VSV is phylogenetically more closely related to HPIV3 than LCMV (42), only 27 of the 72 factors identified for VSV were required for HPIV3, whereas 54 of those were required for LCMV. We noted an enrichment of proteins involved in GPCR signaling, indicating that signaling through some of these GPCR components may be required for infection. The observations that SLC46A1 (PCFT) is specifically required in entry/uncoating, ADAL is required for assembly/budding, and MAT2A is required for gene expression of VSV are unique findings of this study. ADAL was recently discovered as a putative plasma membrane protein closely related to adenosine deaminase with unknown function. Further studies are required for understanding the molecular mechanisms of its involvement in VSV infection. Identification of MAT2A, in VSV RNA synthesis, suggests its possible involvement in VSV mRNA cap formation during transcription. Whether these identified proteins are directly involved in various steps in VSV infection or whether they mediate their activities through other cellular interacting partners (Fig. S7) is of interest for a mechanistic understanding of their role in VSV infection.
The subunits of COPI complex emerged strongly as factors required for infection by all three viruses. The heptameric COPI complex associates with the Golgi membranes, resulting in the budding of COPI-containing vesicles, whose major function is to mediate transport of cellular proteins and cargo from the Golgi to the ER as well as the intra-Golgi transport (20). Depletion of COPI also perturbs the steady-state distribution of the Golgi enzymes (43) and thus inhibits processing of the VSV G protein and its transport to the plasma membrane (44, 45). Further, we have found that depletion of COPZ1 subunit had no adverse effect on VSV G protein-mediated entry of pseudotyped HIV (Fig. S8), consistent with recent studies that BFA has no significant effect on VSV G-mediated entry of retrovirus (46). The following observations suggest that COPI is involved in VSV gene expression: (i) VSV gene expression is inhibited in COPI-depleted cells that are independent of the viral G protein processing, (ii) pharmacologic inhibitor of COPI inhibits VSV gene expression as well as viral RNA levels, and (iii) VSV gene expression is reduced in NC transfected cells that have been depleted of COPI.
Multiple siRNA screens for influenza virus, an NS RNA virus that replicates in the nucleus, have identified COPI, but not COPII, as required for infection (11, 12, 24). Although COPI is required for influenza as well as VSV G-mediated virus entry (12), our results for VSV are in contrast to these studies. It is possible that the assay conditions, cell type used, and other unknown factors may have contributed to these disparate results. In this report and also in earlier reports (11, 15, 47), the siRNA for COPE did not inhibit VSV and other virus infections, indicating that this protein may not have been depleted sufficiently to observe the effect or that COPE may be dispensable for COPI function in viral infection (11, 15, 47). Transfection of siRNAs for COPII subunits resulted in statistically nonsignificant reduction of VSV infection (Fig. S9), indicating that COPII may not be required for VSV, consistent with the result that it was also not identified in the influenza virus screens (11, 12, 24). Our results point toward a specific role of COPI in VSV gene expression. The requirement for ARF1 and GBF1, the upstream activators of COPI assembly in modulating VSV RNA levels, suggests a role for the host cell secretory pathway in the process. Whether the COPI complex is directly involved in regulating the viral polymerase functions or whether it may signal through downstream effectors required for viral gene expression is of further interest.
The requirement of COPI for genome replication has been well documented for positive-strand RNA viruses, which replicate in association with cytoplasmic membranous structures (9, 15, 47). Viruses such as polio, hepatitis C, and coxsackie modulate the activities of ARF1, GBF1, and COPI for formation of intracellular organelles for replication (48, 49). Vaccinia virus, a DNA virus, requires COPI for its biogenesis, whereas HIV lacks a requirement for COPI (33). The studies presented here reveal a requirement for COPI in gene expression of VSV, and possibly other NS RNA viruses, suggesting a critical role of this secretory pathway in RNA virus infection. Most NS RNA viruses including VSV, LCMV, and HPIV3, replicate in the cytoplasm. The replication organelles, if any, for the cytoplasmically replicating NS RNA viruses are unknown. VSV replicates throughout the cytoplasm of infected cells (7, 50), although studies do not rule out the existence of specific replication organelles in the cytoplasm. Further biochemical and ultrastructural studies in cells infected with VSV and other NS RNA viruses will likely illuminate the nature of the replication organelles for these viruses.
In conclusion, this study has identified several previously undescribed candidates and pathways regulating infection of cells by VSV and two other NS RNA viruses. The requirement for host cell secretory pathway in infection by VSV, LCMV, and HPIV3 argues for a common mechanism by which this cellular pathway modulates NS RNA virus replication.
Experimental Procedures
The high-throughput primary siRNA screen was performed by using the Qiagen genome-wide siRNA library (Version 1.0). The validation screen was conducted by using Dharmacon ON-TARGETplus pool of four siRNAs. Details of the screen design, statistical analysis, hit identification, and other experimental procedures can be found in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank G. Belov, P. Collins, J. Donaldson, D. Lyles, D. McGavern, and E. Sztul for reagents and Z. H. Gill for excellent assistance in the laboratory. The work was supported in part by National Institutes of Health Grant R01AI34956.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113643108/-/DCSupplemental.
References
- 1.Barber GN. VSV-tumor selective replication and protein translation. Oncogene. 2005;24:7710–7719. doi: 10.1038/sj.onc.1209042. [DOI] [PubMed] [Google Scholar]
- 2.Barr JN, Whelan SP, Wertz GW. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim Biophys Acta. 2002;1577:337–353. doi: 10.1016/s0167-4781(02)00462-1. [DOI] [PubMed] [Google Scholar]
- 3.Coil DA, Miller AD. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J Virol. 2004;78:10920–10926. doi: 10.1128/JVI.78.20.10920-10926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun X, Yau VK, Briggs BJ, Whittaker GR. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Das SC, Nayak D, Zhou Y, Pattnaik AK. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J Virol. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 11.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.König R, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tai AW, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao L, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherry S, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz J, Liu W. Insights into COPI coat assembly and function in living cells. Trends Cell Biol. 2006;16:e1–e4. doi: 10.1016/j.tcb.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Barrows NJ, Le Sommer C, Garcia-Blanco MA, Pearson JL. Factors affecting reproducibility between genome-scale siRNA-based screens. J Biomol Screen. 2010;15:735–747. doi: 10.1177/1087057110374994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 23.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Emonet SF, Garidou L, McGavern DB, de la Torre JC. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci USA. 2009;106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, et al. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J Virol. 2005;79:1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luyet PP, Falguières T, Pons V, Pattnaik AK, Gruenberg J. The ESCRT-I subunit TSG101 controls endosome-to-cytosol release of viral RNA. Traffic. 2008;9:2279–2290. doi: 10.1111/j.1600-0854.2008.00820.x. [DOI] [PubMed] [Google Scholar]
- 29.Dinh PX, Beura LK, Panda D, Das A, Pattnaik AK. Antagonistic effects of cellular poly(C) binding proteins on vesicular stomatitis virus gene expression. J Virol. 2011;85:9459–9471. doi: 10.1128/JVI.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Chantar ML, et al. L-methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: Role of S-adenosylmethionine. J Biol Chem. 2003;278:19885–19890. doi: 10.1074/jbc.M211554200. [DOI] [PubMed] [Google Scholar]
- 31.Orci L, et al. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 32.Hu T, Kao CY, Hudson RT, Chen A, Draper RK. Inhibition of secretion by 1,3-Cyclohexanebis(methylamine), a dibasic compound that interferes with coatomer function. Mol Biol Cell. 1999;10:921–933. doi: 10.1091/mbc.10.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, et al. A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc Natl Acad Sci USA. 2009;106:163–168. doi: 10.1073/pnas.0811631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitney JA, Gomez M, Sheff D, Kreis TE, Mellman I. Cytoplasmic coat proteins involved in endosome function. Cell. 1995;83:703–713. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 35.Das SC, Panda D, Nayak D, Pattnaik AK. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cells. J Virol. 2009;83:2611–2622. doi: 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu VW, Yang JS. Mechanisms of COPI vesicle formation. FEBS Lett. 2009;583:3758–3763. doi: 10.1016/j.febslet.2009.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters PJ, et al. Overexpression of wild-type and mutant ARF1 and ARF6: Distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell. 2003;14:2250–2261. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan H, et al. A novel small molecule regulator of guanine nucleotide exchange activity of the ADP-ribosylation factor and golgi membrane trafficking. J Biol Chem. 2008;283:31087–31096. doi: 10.1074/jbc.M806592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sáenz JB, et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol. 2009;5:157–165. doi: 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanke KH, et al. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol. 2009;83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Eighth report of the international committee on the taxonomy of viruses. London: Elsevier/Academic Press; 2005. In Virus Taxonomy. [Google Scholar]
- 43.Tu L, Tai WC, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- 44.Szul T, et al. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J Cell Sci. 2007;120:3929–3940. doi: 10.1242/jcs.010769. [DOI] [PubMed] [Google Scholar]
- 45.Manolea F, Claude A, Chun J, Rosas J, Melançon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol Biol Cell. 2008;19:523–535. doi: 10.1091/mbc.E07-04-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goueslain L, et al. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherry S, et al. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog. 2010;6:e1000958. doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.