Abstract
Niemann–Pick type C1 (NPC1) protein is needed for cellular utilization of low-density lipoprotein-derived cholesterol that has been delivered to lysosomes. The protein has 13 transmembrane domains, three large lumenal domains, and a cytoplasmic tail. NPC1’s lumenally oriented, N-terminal domain binds cholesterol and has been proposed to receive cholesterol from NPC2 protein as part of the process by which cholesterol is exported from lysosomes into the cytosol. Using surface plasmon resonance and affinity chromatography, we show here that the second lumenal domain of NPC1 binds directly to NPC2 protein. For these experiments, a soluble NPC1 lumenal domain 2 was engineered by replacing adjacent transmembrane domains with antiparallel coiled-coil sequences. Interaction of NPC2 with NPC1 lumenal domain 2 is only detected at acidic pH, conditions that are optimal for cholesterol binding to NPC2 and transfer to NPC1; the pH is also appropriate for the acidic environment where binding would take place. Binding to NPC1 domain 2 requires the presence of cholesterol on NPC2 protein, a finding that supports directional transfer of cholesterol from NPC2 onto NPC1’s N-terminal domain. Finally, human disease-causing mutations in NPC1 domain 2 decrease NPC2 binding, suggesting that NPC2 binding is necessary for NPC1 function in humans. These data support a model in which NPC1 domain 2 holds NPC2 in position to facilitate directional cholesterol transfer from NPC2 onto NPC1 protein for export from lysosomes.
Keywords: cholesterol trafficking , Niemann–Pick type C disease
A major source of cellular cholesterol is endocytosed as low-density lipoprotein, which is delivered to late endosomes and lysosomes where cholesterol is released (1). Within late endosomes and lysosomes, Niemann–Pick type C1 (NPC1) and NPC2 proteins are required for the subsequent delivery of cholesterol to other intracellular compartments (2). NPC1 is a large, 1,254 residue, integral membrane protein that is predicted to span the bilayer 13 times (3, 4); it contains a lumenally oriented, N-terminal cholesterol binding site that interacts with the 3β-hydroxyl end of the cholesterol molecule (5–7). NPC2 is a much smaller, soluble lysosomal protein of 132 amino acid residues (8) that binds cholesterol in an opposite orientation via cholesterol’s isooctyl side chain (9–11). The importance of NPC1 and NPC2 for cholesterol and glycosphingolipid homeostasis is demonstrated in Niemann–Pick type C disease: patients carrying homozygous mutations in either of these proteins suffer neurodegeneration and die in childhood due to cholesterol and glycosphingolipid accumulation in the brain, liver, and lungs (12, 13).
NPC2 is thought to play an important role in extracting cholesterol from intralysosomal membranes that are rich in cholesterol and bis(monoacylglycerol) phosphate. Transfer of that cholesterol to NPC1 is proposed to facilitate passage of this hydrophobic sterol across the glycocalyx that lines the inner lysosomal membrane.
Recent studies using purified NPC2 and a soluble construct comprised of the NPC1 N-terminal, cholesterol binding domain have shown that NPC2 can facilitate transfer of cholesterol between NPC1 and liposomes in vitro (14). Transfer is optimal at pH 5.5, the pH characteristic of the lumen of late endosomes and lysosomes (14). Specific features of the transfer reaction and the NPC1 N-terminal domain structure have led to the proposal that NPC1 exists in a closed conformation that can be opened by interaction with NPC2 to facilitate cholesterol receipt (6, 14). Cholesterol transfer requires a cluster of NPC2 surface residues (79, 81, and 83) that are located immediately adjacent to the sterol-binding pocket (15). Wang et al. (15) also identified NPC1 residues L175/L176 and E191/Y192 as being critical for receipt of cholesterol from NPC2. Moreover, when introduced into cells, the L175Q/L176Q mutant NPC1 protein cannot restore egress of cholesterol from lysosomes (15). This observation is consistent with a requirement for interaction between NPC1 and NPC2 proteins as part of the normal cholesterol export process. Despite strong genetic and biochemical indications for interactions between NPC1 and NPC2 proteins, direct binding has not been demonstrated to date; moreover, gel filtration and surface plasmon resonance failed to detect interactions between NPC1 N-terminal domain and NPC2 protein (15). We show here that NPC1’s second lumenal domain binds NPC2 in a cholesterol-dependent manner, a process that may facilitate directional transfer of cholesterol from NPC2 onto NPC1’s N-terminal domain.
Results
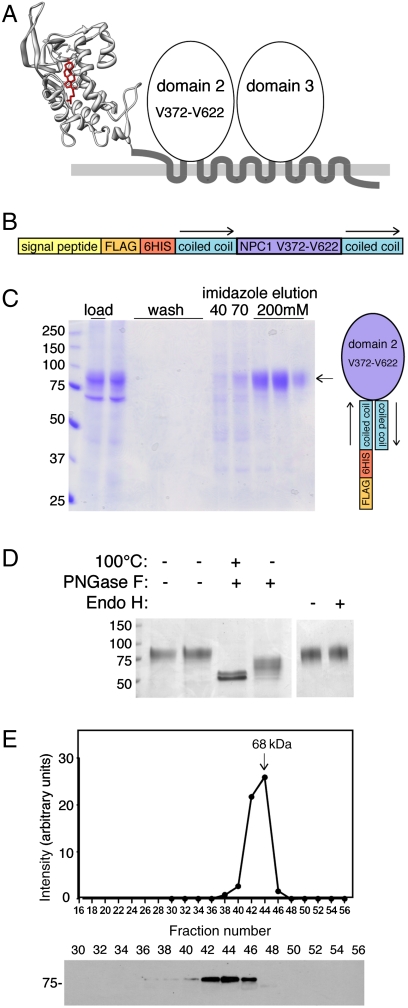
Fig. 1A shows a model of the proposed topology of NPC1 protein. The structure of the first, N-terminal, lumenal domain has been determined (6), but nothing is known about the structure and function of the remainder of this important glycoprotein. To study the biochemical properties of NPC1 lumenal domain 2, we replaced transmembrane domains 2 and 3 with sequences that form a stable, antiparallel coiled-coil structure (16). With the goal of generating a properly folded NPC1 domain 2, we presumed that the C terminus of transmembrane domain 2 would be adjacent to the N terminus of transmembrane domain 3 in the native protein; an antiparallel coiled coil would retain these junctions in an adjacent configuration. The protein was constructed such that it carried an N-terminal signal peptide to permit entry into the secretory pathway of mammalian cells, as well as FLAG and 6-His tags for detection and purification (Fig. 1B).
Fig. 1.
Engineering a soluble NPC1 lumenal domain 2. (A) Schematic diagram of the topology and domain organization of NPC1 protein. The structure of the N-terminal domain (6) is shown. (B) Diagram of the engineered NPC1 domain 2 construct. (C) Coomassie blue stained SDS-PAGE of NPC1 domain 2 purification. Load, starting material secreted from 293F cells, and imidazole elutions are shown. At right is a depiction of the coiled-coil orientation. (D) Domain 2 carries complex oligosaccharide modification. Coomassie blue stained gel of purified protein after treatment with protein N-glycanase F (PNGase F) or endoglycosidase H (endo H). Mobility of molecular weight markers is indicated at left in kilodaltons. (E) Gel filtration of purified NPC1 domain 2 analyzed by immunoblot (below) using anti-FLAG antibodies.
Upon transfection into cultured HEK-293F cells, the protein was efficiently processed for secretion and could be collected from the conditioned medium (Fig. 1C, left lane). Application of this material onto an Ni-nitrilotriacetate resin permitted elution of a highly purified protein upon addition of 200 mM imidazole (Fig. 1C, right lanes). The secreted protein contained asparagine-linked oligosaccharide chains, as evidenced by sensitivity to cleavage by protein N-glycanase; as is common for this enzyme, cleavage was most efficient after protein denaturation by incubation at 100 °C (Fig. 1D). A doublet seen after cleavage suggests that NPC1 domain 2 may also receive some O-linked glycosylation. That the protein passed through the Golgi complex is confirmed by the presence of endoglycosidase H-resistant oligosaccharide chains (Fig. 1D). These data show that NPC1 domain 2 can be prepared in a glycosylated, secreted form from cultured cells.
The quality control machinery of the secretory pathway recognizes misfolded proteins and targets them for degradation (17). To be sure that domain 2 was properly folded, we tested its oligomeric state by gel filtration chromatography. The protein eluted as would be expected for a monomeric globular protein (Fig. 1E), consistent with proper folding in the secretory pathway.
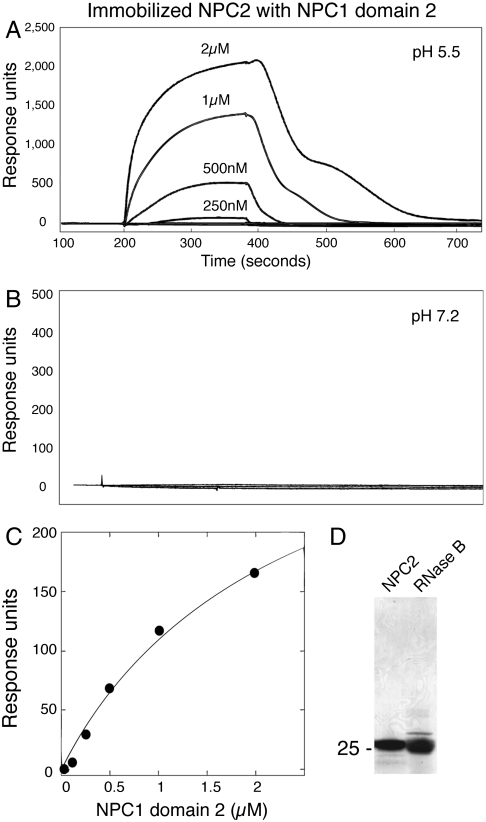
Surface plasmon resonance was used to test whether purified NPC1 domain 2 can bind NPC2. For these experiments, NPC2, purified from raw cow milk (Fig. 2D), was attached covalently to a chip and candidate partner proteins flowed over the surface. In control reactions, no interaction was detected when NPC2 protein itself, the similarly sized RNase B glycoprotein (Fig. 2D), or BSA were flowed over immobilized NPC2. In contrast, binding was readily detected when NPC1 domain 2 was added (Fig. 2A). Binding was concentration dependent, of approximately 2-μM affinity, and seen at pH 5.5 but not at pH 7.2 (Fig. 2 B and C). The pH at which NPC1 N-terminal domain optimally accepts cholesterol from NPC2 protein (14) and at which NPC2 optimally binds cholesterol (18) is pH 5.5. Because one of the binding partners (NPC2) was randomly oriented and constrained by attachment to the surface, these data likely reflect an underestimate of binding affinity.
Fig. 2.
Binding of NPC2 to NPC1 lumenal domain 2 is pH dependent. (A) Surface plasmon resonance of NPC1 domain 2 binding to immobilized bovine NPC2 protein at pH 5.5 (50 mM MES buffer, 150 mM NaCl). No cholesterol sulfate was present. The biphasic dissociation profile likely reflects two processes: loss of cholesterol from chip-bound NPC2 and NPC1-domain 2 protein dissociation (see Fig. 4). (B) Same as A but at pH 7.2 (10 mM Hepes, 150 mM NaCl). (C) Concentration dependence of NPC1 domain 2 binding to NPC2 from surface plasmon resonance data (10 mM Hepes at pH 5.5, 150 mM NaCl). (D) Coomassie blue stained SDS-PAGE of purified proteins used for binding experiments: bovine NPC2 or ribonuclease B (RNase B). Mobility of molecular weight marker is indicated at left in kilodaltons.
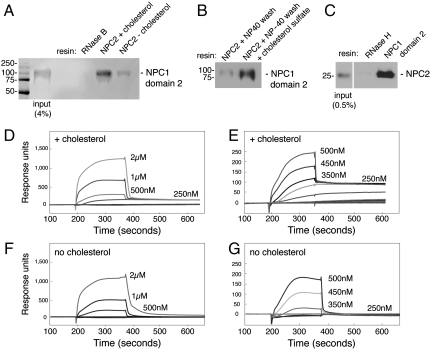
The significant binding detected by surface plasmon resonance should be detectable by more conventional methods. Indeed, purified NPC1 domain 2 showed binding to NHS-Sepharose-immobilized NPC2 protein (Fig 3A). In this scenario, immobilized NPC2 was preincubated with cholesterol sulfate. Then, bead-bound NPC2 was prewashed with either cholesterol sulfate or a low amount of Nonidet P-40 (NP-40) detergent to remove bound cholesterol (5). NPC1 domain 2 displayed binding to NPC2 bearing cholesterol and could be eluted from NPC2 by a simple wash with pH 8.0 buffer. No binding of NPC1 domain 2 was detected using a resin to which RNase B had been covalently attached, and the presence of cholesterol bound to NPC2 significantly enhanced binding (Fig. 3A). To ensure that NP-40 did not in any way alter NPC2 structure, the NPC2 resin washed with NP-40 could be reequilibrated by addition of cholesterol sulfate: Significant binding capacity was readily restored (Fig. 3B). Binding was also detected in the opposite orientation: NPC1 domain 2 immobilized using anti-FLAG antibody beads bound to recombinant, myc-tagged NPC2 protein obtained after expression in cultured cells, but not to myc-tagged RNase H protein (Fig. 3C). Thus, NPC1 domain 2 binds to NPC2 by affinity chromatography with either partner immobilized; binding is greatly stimulated by cholesterol sulfate bound to NPC2 protein.
Fig. 3.
NPC2 binding to NPC1 lumenal domain 2 requires cholesterol. (A) Immunoblot of NPC1 domain 2 binding to NHS-Sepharose to which either RNase B or NPC2 (2 mg/mL resin) has been covalently attached. NPC1 domain 2 (2 μM) was added for 30 min at 30 °C; for reactions with or without cholesterol (as indicated), samples received either cholesterol sulfate (60 μM) or 0.1% Nonidet P-40 for 10 min, followed by a wash with 10 volumes 50 mM MES (pH 5.5), 150 mM NaCl. Elution was with pH 8.0 buffer. (B) Binding of NPC1 domain 2 to immobilized NPC2 was carried out as in A except lane 2 shows a reaction to which cholesterol sulfate (60 μM) was readded for 20 min at room temperature after Nonidet P-40 treatment. (C) Immunoblot of NPC2 binding to FLAG–NPC1 domain 2 or FLAG–RNase H immobilized on anti-FLAG antibody resin. Myc-tagged NPC2 (2 μM), prebound with cholesterol sulfate (5 μM), was added for 30 min at 30 °C; columns were eluted with FLAG peptide (0.1 mg/mL). Eluted NPC2 protein was detected with anti-myc antibody. Mobility of molecular weight markers is indicated at left in kilodaltons. (D–G) Surface plasmon resonance of NPC1 domain 2 binding to immobilized bovine NPC2 protein at pH 5.5 (buffer B), either in the presence of 4 μM cholesterol sulfate during association and dissociation phases (D and E) or after incubation with 0.1% Nonidet P-40 followed by washing to remove detergent and without cholesterol sulfate addition (F and G). In addition, NPC2 was preincubated for 20 min at 25 °C with cholesterol sulfate (1.5-fold molar excess) prior to coupling to the chip. D–G compare binding detected at different concentrations of NPC1 domain 2.
The importance of cholesterol for the NPC2–NPC1 domain 2 interaction was also seen by surface plasmon resonance (Fig. 3 D–G). In the absence of cholesterol, rapid and complete dissociation of NPC1 domain 2 was observed (Fig. 3 F and G). In contrast, when cholesterol sulfate was included during both association and dissociation phases of the experiment, biphasic dissociation was detected, with a substantial amount of residual binding even after 600 s (Fig. 3 D and E). These data are consistent with two classes of binding interactions: a weaker, cholesterol-independent interaction and a stronger, cholesterol-dependent interaction. Thus, by two independent methods, interaction of NPC1 domain 2 with NPC2 is greatly enhanced by cholesterol bound to NPC2 protein.
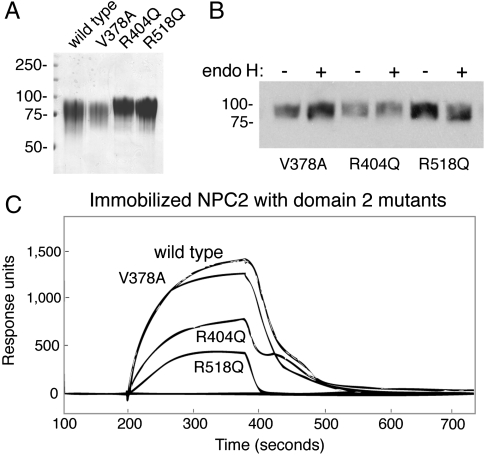
If interaction of NPC2 with NPC1 is important for function, disease-causing mutations in NPC1 protein may impede interaction. We generated NPC1 domain 2 versions of three disease-causing mutations: V378A, R404Q, and R518Q (19). R404Q (and R404W) have been reported in several patient studies and represent mutation of a residue conserved in Niemann–Pick C1-like 1 protein; R518Q (and R518W) seem to be more prevalent among Japanese NPC patients (20). The mutant glycoproteins were produced in the same quantity as the wild-type protein from conditioned media; all contained endoglycosidase H-resistant oligosaccharides (Fig. 4 A and B). Two of the mutant proteins showed significantly decreased binding to NPC2 using surface plasmon resonance (Fig. 4C). R518Q showed the greatest loss in binding; R404Q also showed half the binding of the wild-type protein. In contrast, V378A was essentially wild type. It is noteworthy that this experiment is monitoring primarily the weaker category of cholesterol-independent association between NPC2 and NPC1 domain 2—and even this weaker interaction is severely impacted by the R518Q mutation.
Fig. 4.
NPC1 domain 2 disease-causing mutations show decreased binding to NPC2 protein. (A) Coomassie blue stained gel of purified NPC1 domain 2 mutant proteins. (B) Endoglycosidase H (endo H) cleavage of purified proteins determined by immunoblot. Mobility of molecular weight markers is indicated at left in kilodaltons. (C) Surface plasmon resonance of NPC2 lumenal domain 2 mutants (1 μM) binding to immobilized bovine NPC2 protein at pH 5.5 as in Fig. 2 in the absence of additional cholesterol sulfate.
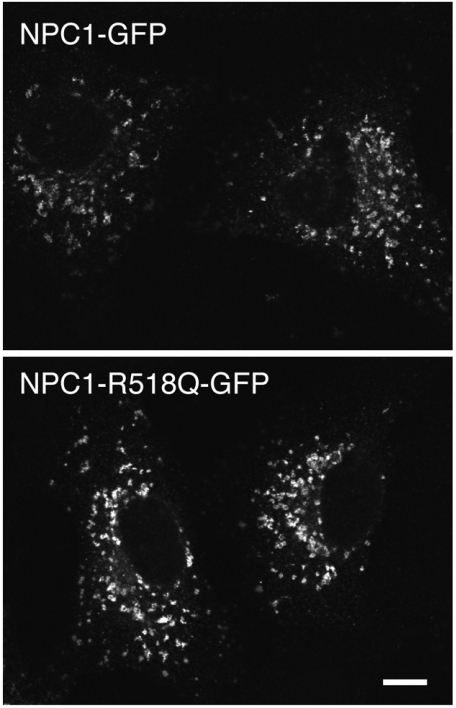
Many NPC1 mutant proteins misfold in the endoplasmic reticulum, and it is misfolding rather than true loss of function that is responsible for the disease phenotype (cf. ref. 21). Thus, before concluding that the R518Q mutation was a function-blocking mutation, it was important to verify that the NPC1 R518Q protein is capable of transport to lysosomes. For this purpose, we generated a full-length NPC1 protein harboring the R581Q protein and localized it in Vero cells 48 h after transfection. Fig. 5 shows immunofluorescence microscopy comparing the localizations of wild-type, full-length NPC1-GFP (Upper) with that of NPC1-R518Q-GFP (Lower). The overall morphologies were indistinguishable: R581Q showed the same punctate, lysosomal distribution as the wild-type protein, with no apparent accumulation in the endoplasmic reticulum. Thus, the cholesterol accumulation seen in patients carrying this mutation is most likely due to the mutant protein’s inability to bind NPC2 in lysosomes, as part of normal NPC1 protein function.
Fig. 5.
Immunofluorescence microscopy of Vero cells transfected with a plasmid encoding full length, murine NPC1-GFP (Upper) or full-length murine NPC1-R518Q-GFP under the control of the Cenp-A promoter to achieve low-level expression. Proteins were visualized 48 h after transfection with a Leica Sp2 confocal microscope and Leica confocal software using rabbit anti-GFP and Alexa 488-conjugated goat anti-rabbit antibodies (Invitrogen). (Scale bar: 10 μm.)
Discussion
We have shown here, using two independent methods, that NPC2 binds directly to the second lumenal domain of NPC1 protein and does so only at the compartment-appropriate pH and most strongly when NPC2 carries cholesterol. These findings provide a satisfying mechanism by which cholesterol can be transferred, directionally, from NPC2 onto the NPC1 N-terminal domain in the lumen of late endosomes and lysosomes. We propose that NPC2 bearing cholesterol binds the second lumenal domain of NPC1 protein to bring NPC2 into close proximity with NPC1’s N-terminal lumenal domain 1, which can accept cholesterol from NPC2. Loss of cholesterol from NPC2 will trigger its release from NPC1 domain 2; the NPC1-associated cholesterol can then be transferred into the membrane bilayer. Conformational changes in NPC2 upon release of cholesterol (11) will contribute to this cholesterol transfer cycle.
Disease-causing mutations R404 and R518Q in NPC1 interfere with the ability of NPC1 to bind NPC2. In addition, full-length NPC1 protein containing the R518Q mutation was shown to be competent for delivery to lysosomes, strongly suggesting that the defect seen in patients harboring this mutation is not due to protein mis-folding and, instead, is due to a functionally defective protein. Taken together, the interaction of NPC1 and NPC2 proteins appears to be central to the process by which NPC1 facilitates cholesterol egress from endolysosomal compartments. These biochemical findings also match well with the genetic relationships between NPC1 and NPC2 genes (22). If interaction with NPC2 is decreased in patients carrying mutations in NPC1 domain 2, enzyme replacement strategies to increase plasma NPC2 levels may have therapeutic benefit for these individuals.
In examining structural models of NPC1 domain 1 in relation to NPC1’s first membrane-spanning domain, it appears that the entryway into the cholesterol binding site may lie close to, and face the membrane (Fig. 1A). The NPC1 domain 2, binding-essential residue R404 is only 25 residues away from the nearest transmembrane domain, consistent with the possibility that NPC1 domain 2 holds NPC2 close to the membrane where it can transfer cholesterol to NPC1 domain 1 (or even directly to the lipid bilayer). We cannot rule out the possibility that R404 is not directly in contact with NPC2 and instead is important for domain 2’s conformation. Structural studies of the soluble NPC1 domain 2 protein will provide critical information regarding the topology of this protein:protein interaction.
In summary, a model has been proposed for the transfer of cholesterol from NPC2 to NPC1 proteins (6, 14, 15). The biochemical findings reported here support the proposed directionality of this transfer process and provide human genetic evidence regarding the importance of the interaction between NPC1 and NPC2 proteins to accomplish cholesterol delivery from lysosomes into the cytoplasm.
Methods
DEAE CL-6B-, CM-, and SP-Sepharose, Ni-Sepharose, and NHS-Sepharose beads were from GE Healthcare and used according to the manufacturer; anti-FLAG M2 beads, sodium cholesteryl sulfate, bovine pancreatic ribonuclease B, and anti-FLAG M2 antibody were from Sigma. Ready Gel Tris·HCl 4–20% precast gradient gels were from Bio-Rad.
Plasmids and Transfections.
The pCMV-FLAG-His6-NPC1-domain 2 was cloned in the pFLAG-CMV-3 plasmid (Sigma). This construct was assembled to contain a preprotrypsin signal sequence, a FLAG tag, and a His tag; NPC1 residues V372–V622 of the 1,278 residue murine precursor (GenBank AAB63372.1) were then flanked by sequences that form a stable, antiparallel coiled coil. The precise sequence was as follows: MSALLILALVGAAVADYKDDDDKLAAANSSIDLMGSSHHHHHHSSGLVPRGSHMKRLEKELAQLEAELEELESKLWHLENENARLEKELAELEAELAESSS-(NPC1 V372–V622) SSEGDIMKRLKKKLAQLKAKLEENKSELWHLKNKLARLKKKLAELKAKLAE (16). The pCMV-FLAG-His6-NPC1m-domain2 R404Q, R518Q, V378A mutants were created by site-directed mutagenesis. The pcDNA3.1-NPC2m-myc-His6 was from M. Scott (Stanford University). Human 293-Freestyle cells (Invitrogen) were grown in GIBCO Freestyle 293 expression medium. A 30-mL culture (1 × 106 cells/mL) was transfected with 30 μg plasmid using 293 fectin (Invitrogen) according to the manufacturer. The medium was collected 72 h after transfection.
Purification of Bovine NPC2.
Bovine NPC2 was purified from raw cow milk (18). Three ion exchange chromatography steps were used: DEAE Sepharose, CM Sepharose column, and SP-Sepharose. The CM Sepharose column was eluted in steps of 10, 100, 150, 200 mM, and 1 M ammonium acetate pH 5.0. The protein eluted in the 100- and 150-mM fractions. A final step employed SP-Sepharose. A linear gradient of 0.0125–0.5 M ammonium acetate pH 4.5 (20 column volumes) was used. An immunoblot verified the presence of the protein and Coomassie-stained SDS-PAGE was used to verify protein purity.
FLAG-His6-NPC1-domain 2 and mouse NPC2-myc-His6 were purified from conditioned medium, collected 72 h after transfection. Cells were centrifuged at 2,383 × g for 5 min at 4 °C. The medium was filtered with a 0.2-μm syringe filter; 20 mM imidazole was added and purification was done in batch with a minimum of 4 h incubation with Ni-Sepharose beads. The proteins eluted at 200 mM imidazole in 50 mM MES pH 6.5, 150 mM NaCl; they were dialyzed into 50 mM MES pH 5.5, 150 mM NaCl to remove imidazole and were concentrated in an Amicon 30K for NPC1-domain 2 or an Amicon 10K for NPC2. Protein N-glycanase F (New England Biolabs) cleavage was carried out at 25 °C overnight with 10 μg of protein and 500 units of enzyme. Endoglycosidase H (Boehringer Mannheim) cleavage was carried out at 37 °C overnight with 5 μg of protein using 10 milliunits of endoglycosidase H.
Surface Plasmon Resonance.
Experiments were performed using a Biacore 3000 system at 25 °C with a flow rate of 20 μL min-1 in buffer A [10 mM Hepes (pH 7.4 or pH 5.5), 150 mM NaCl] or buffer B [50 mM MES (pH 5.5), 150 mM NaC]. Proteins were dialyzed into buffer A or B before injection. Bovine NPC2 was covalently coupled to CM5 sensor chips using amine coupling chemistry (according to the manufacturer) and different concentrations of NPC1-domain 2 (wild type or mutants) were injected over the NPC2 surface. Data were corrected for nonspecific interactions by subtracting the signal in a control flow cell that lacked immobilized ligand, and were analyzed using the BIAevaluation v.4.1 software (Biacore) or Scrubber 2 (Biologic Software).
Acknowledgments.
We are grateful to Dr. Pehr Harbury for advice, Drs. Martha Oakley for antiparallel coiled-coil sequences, Matt Scott for plasmids, Melissa Laughery for initiating this project, and Michael Eckart for help with surface plasmon resonance. This work was supported by grants from The Ara Parseghian Medical Research Foundation and the National Institutes of Health (DK37332).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–74. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Pentchev PG. Niemann-Pick C research from mouse to gene. Biochim Biophys Acta. 2004;1685:3–7. doi: 10.1016/j.bbalip.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Carstea ED, et al. Niemann-Pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 4.Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- 5.Infante RE, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 6.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu R, Lu P, Chu JW, Sharom FJ. Characterization of fluorescent sterol binding to purified human NPC1. J Biol Chem. 2009;284:1840–1852. doi: 10.1074/jbc.M803741200. [DOI] [PubMed] [Google Scholar]
- 8.Naureckiene S, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 9.Liou HL, et al. NPC2, the protein deficient in Niemann-Pick C2 disease, consists of multiple glycoforms that bind a variety of sterols. J Biol Chem. 2006;281:36710–36723. doi: 10.1074/jbc.M608743200. [DOI] [PubMed] [Google Scholar]
- 10.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 11.Xu S, Benoff B, Liou H-L, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J Biol Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pentchev PG, Vanier MT, Suzuki K, Patterson MC. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C-R, Beaudet A-L, Sly W-S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 2625–2639. [Google Scholar]
- 13.Vanier MT, Millat G. Niemann-Pick disease type C. Clin Genet. 2003;64:269–281. doi: 10.1034/j.1399-0004.2003.00147.x. [DOI] [PubMed] [Google Scholar]
- 14.Infante RE, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ML, et al. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12:166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClain DL, Woods HL, Oakley MG. Design and characterization of a heterodimeric coiled coil that forms exclusively with an anti-parallel relative helix orientation. J Am Chem Soc. 2001;123:3151–3152. doi: 10.1021/ja004099l. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, Sunryd JC, Hebert DN. Sorting things out through endoplasmic reticulum quality control. Mol Membr Biol. 2010;27:412–427. doi: 10.3109/09687688.2010.495354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedland N, Liou HL, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Natl Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Millat G, et al. Niemann-Pick C1 disease: Correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am J Hum Genet. 2001;68:1373–1385. doi: 10.1086/320606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park WD, et al. Identification of 58 novel mutations in Niemann-Pick disease type C: Correlation with biochemical phenotype and importance of PTC1-like domains in NPC2. Hum Mutat. 2003;22:313–325. doi: 10.1002/humu.10255. [DOI] [PubMed] [Google Scholar]
- 21.Gelsthorpe ME, et al. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem. 2008;283:8229–8236. doi: 10.1074/jbc.M708735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sleat DE, et al. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc Natl Acad Sci USA. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]