Abstract
Human serum albumin (HSA) is widely used in clinical and cell culture applications. Conventional production of HSA from human blood is limited by the availability of blood donation and the high risk of viral transmission from donors. Here, we report the production of Oryza sativa recombinant HSA (OsrHSA) from transgenic rice seeds. The level of OsrHSA reached 10.58% of the total soluble protein of the rice grain. Large-scale production of OsrHSA generated protein with a purity >99% and a productivity rate of 2.75 g/kg brown rice. Physical and biochemical characterization of OsrHSA revealed it to be equivalent to plasma-derived HSA (pHSA). The efficiency of OsrHSA in promoting cell growth and treating liver cirrhosis in rats was similar to that of pHSA. Furthermore, OsrHSA displays similar in vitro and in vivo immunogenicity as pHSA. Our results suggest that a rice seed bioreactor produces cost-effective recombinant HSA that is safe and can help to satisfy an increasing worldwide demand for human serum albumin.
Keywords: crystal structure, recombinant human serum albumin, recombinant protein expression, rice endosperm, recombinant protein processing
Human serum albumin (HSA) is a soluble, globular, and unglycosylated monomeric protein; it functions primarily as a carrier protein for steroids, fatty acids, and thyroid hormones, and plays an important role in stabilizing extracellular fluid volume (1). HSA is widely used clinically to treat serious burn injuries, hemorrhagic shock, hypoproteinemia, fetal erythroblastosis, and ascites caused by cirrhosis of the liver (2, 3). HSA is also used as an excipient for vaccines or therapeutic protein drugs and as a cell culture medium supplement in the production of vaccines and pharmaceuticals (4). Many other novel uses for HSA in biological applications have recently been explored, such as carrier of oxygen (5), nanodelivery of drugs (6), and fusion of peptides (7).
The market demand for HSA is estimated at more than 500 tons per year worldwide. Currently, commercial production of HSA is primarily based on collected human plasma, which is limited in supply but of high clinical demand, not the least in China. It was reported that the shortage of human plasma led to a rapid increase in price of HSA, which in turn resulted in fake albumin appearing on the market (8). Furthermore, there is an increasing public health concern with plasma-derived HSA (pHSA) with its potential risk for transmission of blood-derived infectious pathogens such as hepatitis and HIV (9, 10). In fact, illegal plasma collection has caused HIV to spread rapidly, creating what are known as AIDS villages in Henan Province in China (11). To eliminate the potential risk of viral contamination, regulatory agencies have encouraged pharmaceutical companies to use non–animal-derived sources for pharmaceutical production (12). Thus, the development of a low-cost method for the production of recombinant HSA (rHSA) is essential as a safer and potentially unlimited alternative to pHSA.
Over the past decades, various expression systems have been used to produce rHSA, including Escherichia coli (13), Saccharomyces cerevisiae (14), Kluyveromyces lactis (15), Pichia pastoris (16), transgenic animals (17), and transgenic plants (18–21). Attempts to produce rHSA in tobacco leaves and potato tubers achieved expression levels of 0.02% of total soluble protein (TSP) (18), and expression was increased to 0.2% of TSP by targeting the rHSA to the apoplast of potato tubers (19). Recently, an expression level of 11.1% of TSP was obtained by expressing rHSA in tobacco leaf chloroplasts (20). More recently, an rHSA expression level of 11.5% of total proteins was achieved in a rice cell culture by a sugar starvation-induced promoter (21). Although rHSA has been successfully expressed in these systems, none of them has proven to be cost-effective at large scale.
Plant seeds, especially cereal crop seeds, are promising vehicles for producing recombinant proteins, because they can achieve high accumulation of recombinant protein, display high levels of protein stability, stored for long periods of time, and are well controlled on a production scale (22, 23). Human lysozyme and lactoferrin produced for oral administration have been successfully expressed in rice seeds (24, 25). Here, we report rice seeds as a bioreactor for large-scale production of Oryza sativa recombinant HSA (OsrHSA). OsrHSA can be highly and stably expressed in rice seeds and can be processed cost-effectively. OsrHSA was found to be equivalent to pHSA in terms of biochemical properties, physical structure, functions, and immunogenicity.
Results
OsrHSA Accumulates Highly and Specifically in the Transgenic Rice Endosperm.
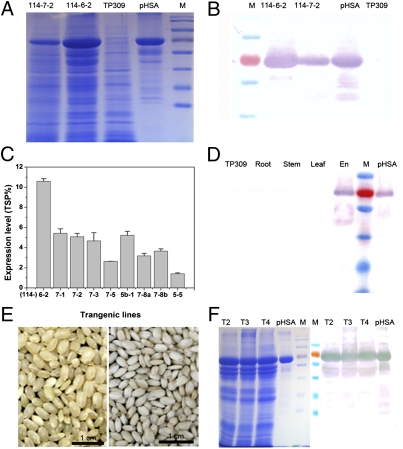
To obtain high expression levels of rHSA and to ensure cost-effective production, a strong endosperm-specific promoter, Gt13a and its signal peptide (26) were used to target rHSA into the protein storage vacuoles. Rice-preferred gene codons were used for transcription of the HSA gene (Fig. S1A). A total of 25 independent transgenic plants were obtained using a two-strain Agrobacterium-mediated transformation. Of these 25 plants, nine independent fertile transformants were obtained. The expression level of OsrHSA ranged from 1.40% to 10.58% of (Fig. 1 A–C and Fig. S1 B–D). A tissue-specific assay indicated that OsrHSA was expressed only in the endosperm and was not present in other tissues (Fig. 1D). The seeds of the transgenic line 114-6-2 (Fig. 1E) displayed an opaque phenotype compared with wild-type seeds. We monitored the genetic stability of transgenic line 114-6-2 and found that the expression of OsrHSA was highly stable from the T2–T4 generation (Fig. 1F); thus, this transgenic line was used for scale-up and further studies.
Fig. 1.
Generation of transgenic rice expressing recombinant HSA. (A and B) Expression of OsrHSA in transgenic lines 114-6-2 and 114-7-2 as characterized using SDS/PAGE (A) and Western blotting (B). 114-6-2 and 114-7-2 are two transgenic lines expressing OsrHSA; TP309 is the nontransgenic rice; pHSA is plasma-derived HSA, M represents molecular marker. (C) The expression levels of OsrHSA in different transgenic lines as quantified using ELISA. Expression levels were calculated according to the percentage of total soluble protein (TSP %). (D) Endosperm-specific expression of OsrHSA in transgenic rice as tested using Western blot. (E) Phenotype of OsrHSA-expressing seeds (114-6-2; Right) and wild-type seeds (Left). (F) Stable expression of OsrHSA in transgenic line 114-6-2. T2, T3, and T4 are the second, third, and fourth generation of 6-2, respectively. En, endosperm; M, molecular marker; pHSA, plasma-derived HSA; TP309, nontransgenic plants.
OsrHSA Is Structurally and Biochemically Equivalent to pHSA.
The expressed OsrHSA has the same molecular mass, amino acid sequence, N- and C terminus, and melting point as pHSA (Fig. S2 A–C and F). The circular dichroism (CD) spectrum of OsrHSA in the near- and far-UV region matched that of pHSA (Fig. S3 A and B), indicating that both the secondary and tertiary structure of OsrHSA are identical to those of pHSA. Spectroscopic analysis further confirmed that OsrHSA has the same conformation as pHSA (Fig. S2 D and E).
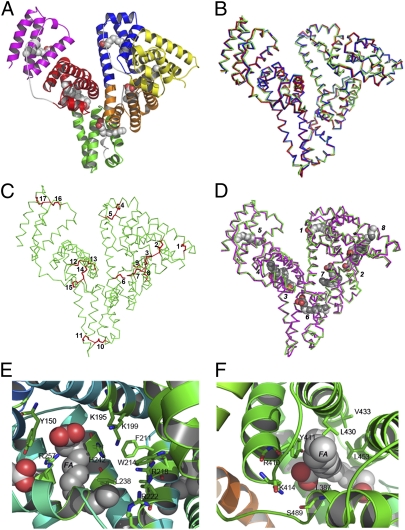
To characterize the structure of OsrHSA, an OsrHSA–myristic acid complex was crystallized, and the structure was solved by molecular replacement using the previously determined HSA structure at a refinement of 2.5 Å (PDB ID code 1BJ5), presenting a final R-free value of 0.303 (Table S1). A crystal of two independent molecules of OsrHSA was obtained in an asymmetric unit. The overall structure of OsrHSA is a heart shape formed by three helical domains, designated I, II, and III, and with myristic acids bound at the hydrophobic cavities (Fig. 2A and Fig. S3C). A superposition of all α-carbon atoms of the crystal structures of OsrHSA and pHSA resulted in a rmsd of 0.67 Å for PBD ID code 2I2Z and 0.69 Å for 1BJ5. No obvious differences in main-chain conformations were identified between pHSA and OsrHSA (Fig. 2B), which demonstrates that OsrHSA in rice endosperm cells is folded into the identical structure as pHSA in blood plasma. As expected, 17 disulfide bonds were identified as genuine disulfide bonds, and a free cysteine (Cys) residue was observed at position 34 (Fig. 2C and Fig. S3D).
Fig. 2.
Crystal structure of OsrHSA (PDB ID code 3SQJ). (A) Overall structure of OsrHSA. Bound mysritic acid molecules are depicted in a sphere representation. Subdomains IA, IB, IIA, IIB, IIIA, and IIIB are colored yellow, blue, orange, green, red, and magenta, respectively. (B) Superposition of molecule of OsrHSA (green) and HSA from PDB files with codes 2I2Z (blue) and 1BJ5 (red); rmsd (2I2Z) = 0.67 Å, rmsd (1BJ5) = 0.69 Å. (C) Disulfide profile of OsrHSA with disulfides shown as red sticks, each disulfide from domain I to III are labeled, cysteines involving in disulfides formation are (1) C53–C62; (2) C75–C91; (3) C90–C101; (4) C124–C169; (5) C168–C177; (6) C200–C246; (7) C245–C253; (8) C265–C279; (9) C278–C289; (10) C316–C361; (11) C360–C369; (12) C392–C438; (13) C437–C448; (14) C461–C477; (15) C476–C487; (16) C514–C559; and (17) C558–C567. (D) Comparison of location of bound myristic acids (light gray) in OsrHSA (green) and those (dark gray) in HSA from PBD with codes 1E7G (red), rmsd = 0.72 Å. (E) Drug-binding site I in OsrHSA. (F) Drug-binding site II in OsrHSA. Myristic acids bound near a drug site are presented as spheres.
HSA is known to act as a carrier protein, mainly through two docking sites for drugs and 7–9 sites for fatty acids (27). Eight myristic acids were observed bound to the recombinant HSA molecule, and all fatty acid binding sites in OsrHSA were identified at the same locations as in pHSA (Fig. 2D). Two drug-binding sites found in pHSA were also present in OsrHSA. Site I is a hydrophobic cavity located in subdomain II, delineated by the residues F211, W214, A215, and L238. Two clusters of polar residues responsible for ligand interaction were populated by amino acids Y150, H242, R257, and K195, K199, R218, R222, respectively (Fig. 2E). Site II was another hydrophobic pocket in subdomain IIIA composed of L387, Y411, L453, V433, and L430. One polar region, made up of R410, K414, and S489, was found in this binding site (Fig. 2F). The structural topology of these binding sites in OsrHSA suggests that it can perform the same biological functions as pHSA.
HPLC-MS characterization showed that the lipids bound to OsrHSA were mainly lysophosphatidylcholine (lyso-PC), phosphatidylethanolamine (lyso-PE), and phosphatidylcholine (PC). The lipids bound to OsrHSA thus were highly similar to the lipids found associated with pHSA. In total, 15 fatty acids were identified in complex with OsrHSA. Four lyso-PE and nine lyso-PC molecules bound to OsrHSA were the same as the lipids found associated with pHSA (Table S2). Two lipids found on OsrHSA, (14:0)-lyso-PC and (14:0)-lyso-PE, were not detected at pHSA; however, both (14:0)-lyso-PC and (14:0)-lyso-PE are common phospholipids in human blood, which mainly bind to low-density lipid protein rather than HSA in vivo (28, 29). Those have been applied to prepare liposomes for drug delivery or stabilization (30, 31).
OsrHSA Functions Equivalently to pHSA in Terms of Binding Capacity and Promotion of Cell Growth.
One of the most important biological activities of HSA is its binding capacity for various ligands; thus, two site-specific drug markers, warfarin (site I) and naproxen (site II), were used to evaluate the binding capacity of OsrHSA. The binding assay showed that the affinities of warfarin and naproxen to OsrHSA were 1.64 ± 0.10 (×104 M−1) and 1.58 ± 0.30 (×106 M−1), respectively. These values for OsrHSA are similar to the binding affinities observed for warfarin and naproxen to pHSA (1.62 ± 0.08 × 104 M−1 and 1.21 ± 0.01 × 106 M−1), indicating that no significant difference in drug-binding affinity exists between OsrHSA and pHSA (Fig. S4 A–D).
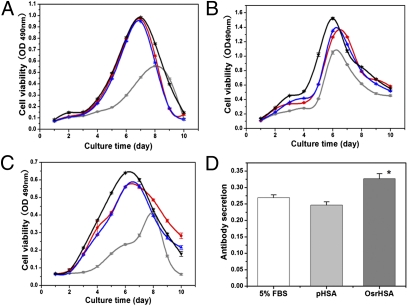
To test whether OsrHSA was functionally equivalent to pHSA in terms of promotion of cell growth, studies were performed with three common cell lines: Chinese hamster ovary (CHO), Vero, and SP2/0. Treatment with either pHSA or OsrHSA (1 g/L) in DMEM containing 5% FBS dramatically increased the cell density and viability compared with control treatment (5% FBS only) in all cell lines (Fig. 3 A–C and Fig. S4 E and F). The levels of growth enhancement observed with OsrHSA and pHSA were nearly the same. The promotion of cell growth was comparable to that of CHO cells on 10% FBS. Specifically, OsrHSA showed a 20% increase in maximum viability and density compared with pHSA in Vero cells (Fig. 3B). Notably, the titer of antibody (IgG1+ κ) measured in the culture medium of SP2/0 cells supplemented with OsrHSA was significantly higher than that observed for the same cells supplemented with pHSA (P = 0.015; Fig. 3D). Based on these results, we conclude that OsrHSA performed as well as pHSA in the promotion of cell growth, and thus can effectively replace pHSA in cell culture media.
Fig. 3.
OsrHSA functions equivalently to pHSA in vitro. (A–C) The effects of OsrHSA (blue) and pHSA (red) on cell viability in CHO (A), Vero (B), and SP2/0 (C) cell lines. Five percent FBS was used as a negative control (gray) and 10% FBS as a positive control (black). One gram per liter of OsrHSA or pHSA was added to DMEM supplemented with 5% FBS for all cell cultures. (D) Antibody productivity of SP2/0 cells cultured with 5% FBS (white), OsrHSA (dark gray), or pHSA (gray; *P < 0.05 OsrHSA vs. pHSA).
OsrHSA Functions Equivalently to pHSA in the Reduction of Rat Liver Ascites.
To evaluate the efficacy of OsrHSA in treatment of liver ascites, a rat liver ascite model was used. Compared with a normal group, rats with liver cirrhosis showed a dramatic decrease in body weight (232 ± 25 g, n = 53, P < 0.01) and urine output (0.6 ± 0.5 mL⋅mg−1⋅h−1, n = 53, P < 0.01) accompanied with a significant increase in abdominal circumference (abdominal circumference/body weight, 69 ± 5 cm/kg, n = 53, P < 0.01). The livers of these rats were seriously damaged, as demonstrated by intensive proliferation of connective tissue.
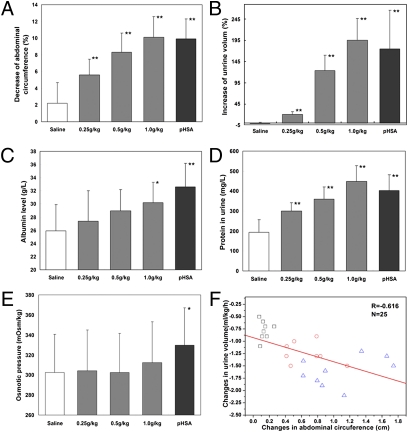
Various dosages of OsrHSA and a single dose of pHSA (1.0 g/kg) were delivered via i.v. infusion for efficacy tests. Abdominal circumference decreased dramatically after OsrHSA treatment in a dose-dependent manner, with a decrease of 5.6 ± 1.9%, 8.3 ± 2.3%, and 10.1 ± 2.5% for the 0.25, 0.5, and 1.0 g/kg dosages, respectively (Fig. 4A and Table S3). Similar effects on abdominal circumference were observed in rats treated with the same doses (1.0 g/kg) of pHSA and OsrHSA. Urine outputs were also significantly and dose-dependently increased in rats treated with OsrHSA, with increases of 20.6 ± 6%, 123.9 ± 35.9%, and 195.5 ± 50.9% for 0.25, 0.50, and 1.0 g/kg dosages, respectively. Rats administered the same dose (1.0 g/kg) of pHSA or OsrHSA showed similar increases in urine output (P < 0.01; Fig. 4B and Table S3).
Fig. 4.
Therapeutic efficacy of OsrHSA on ascites in rats with liver cirrhosis. (A) Decrease in abdominal circumference. (B) Increase in urinary volume. (C) Albumin content in the plasma of differently treated groups. (D) Urinary protein content seen with different treatments. (E) Changes in osmotic pressure of plasma after treatment with OsrHSA or pHSA. Bars in white, gray, and dark gray represent treatment with saline, OsrHSA, and pHSA, respectively (*P < 0.05, **P < 0.01 vs. saline group). (F) Correlation of abdominal circumference change and urine volume change. N, number of rats with each point representing a change from pretreatments. Black squares, red circles, and blue triangles represent data collected from treatment with OsrHSA at dosages of 0.25, 0.5, and 1.0 g/kg, respectively.
Treatment with OsrHSA or pHSA at a dose of 1.0 g/kg significantly increased the level of serum albumin compared with treatment with saline (P < 0.05; Fig. 4C and Table S3), but as known for pHSA, the increase was not significant at lower dosages of OsrHSA. The amount of protein in the urine dramatically increased with the administration of OsrHSA (Fig. 4D). Additionally, administration of OsrHSA increased the colloid osmotic pressure of blood with high OsrHSA dosage (1.0 g/kg), but the increase was not statistically significant as observed in treatment with pHSA (Fig. 4E). This may be caused by increased protein excretion resulting from increased urinary output. A negative correlation (R = −0.61; Fig. 4F) between changes in abdominal circumference and urine output was observed with increasing doses of OsrHSA. These results suggest that the efficacy of OsrHSA for the treatment of liver cirrhosis is equivalent to that of pHSA.
Production of OsrHSA from Transgenic Rice Seed Is Cost-Effective.
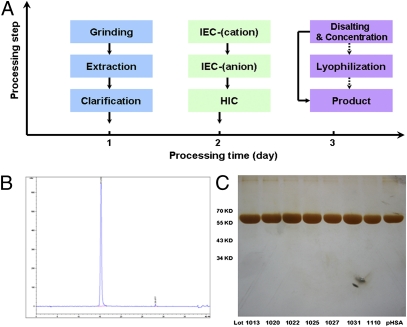
Production of highly purified OsrHSA is required for high-dosage applications in clinic treatments. Accordingly, a processing strategy for large-scale production of OsrHSA was developed. Processing comprised extraction and three chromatography steps with Capto-MMC, Q-Sepharose, and Phenyl-HP, followed by concentration/desalting and lyophilization (Fig. 5A). The complete purification process takes ∼48 h and can recover 45.57 ± 5.57% of the protein. The data from a scale-up purification showed that the recovery of OsrHSA was 55.75 ± 3.19%, equivalent to a yield of 2.75 g of OsrHSA per kilogram of brown rice (Table S4). The final product of OsrHSA from the large-scale purification showed a single peak in a reverse-phase HPLC (C4) analysis (Fig. 5B), and other proteins were barely visible in a silver-stained SDS/PAGE (Fig. 5C), together demonstrating that the purity of OsrHSA is comparable to a pHSA (>99%) control. The purity of OsrHSA from six different preparations was 99.45 ± 0.19% with a variability coefficient of 0.19%, indicating that the purification procedure is robust and highly reproducible. These results show that rice seed offers a cost-effective alternative for the production of rHSA.
Fig. 5.
Production of OsrHSA from transgenic rice seeds. (A) Large-scale process flow diagram of the purification of OsrHSA from transgenic rice seeds. (B) Purity analysis with reverse-phase HPLC (C4). (C) SDS/PAGE (silver staining) characterization of purified OsrHSA from large-scale production of lot nos. 1013 (99.09%), 1020 (99.42%), 1022 (99.60%), 1025 (99.45%), 1027 (99.59%), 1031 (99.55%), and 110 (99.09%). Purity was calculated by size-exclusion HPLC.
OsrHSA Displays the Same Immunogenicity as pHSA.
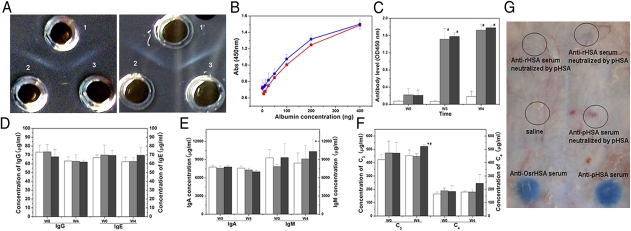
Although no allergic reaction was observed following i.v. administration of OsrHSA in rats treated for liver cirrhosis, potential safety concerns regarding clinical use of OsrHSA remain because of the need for large i.v. doses. Double-cross immunoprecipitation experiments between OsrHSA or pHSA and antiserum against pHSA or OsrHSA clearly showed that both OsrHSA and pHSA bound completely to antiserum against OsrHSA or pHSA (Fig. 6A). Additionally, in ELISA tests, polyclonal antibodies against pHSA reacted equally with OsrHSA (Fig. 6B). Taken together, these results indicate that the immunogenicity of OsrHSA is similar to that of pHSA in vitro.
Fig. 6.
Immunogenicity of OsrHSA. (A) Double-immunodiffusions of anti-OsrHSA serum (1) or anti-pHSA serum (1′) with pHSA (2) and OsrHSA (3). (B) Reactivity of OsrHSA (blue) and pHSA (red) against anti-pHSA polyclonal antibody. (C) IgG raised against HSA in serum of rats immunized with saline (white), OsrHSA (gray), or pHSA (dark gray) after 0 wk (W0), 3 wk (W3), and 4 wk (W4). (D) Total IgG and IgE in serum of rats immunized with saline (white), OsrHSA (gray), or pHSA (dark gray) after 0 wk (W0) and 4 wk (W4). (E) Total IgA and IgM in serum of rats immunized with saline (white), OsrHSA (gray), or pHSA (dark gray) after 0 wk (W0) and 4 wk (W4). (F) Level of C3 and C4 complement in serum of rats immunized with OsrHSA (gray) or pHSA (dark gray) after 0 wk (W0) and 4 wk (W4). #P < 0.05, HSA groups vs. saline group; *P′′ < 0.05, OsrHSA group vs. pHSA group. (G) Passive cutaneous anaphylaxis (PCA) assay in response to OsrHSA in guinea pig. Black circles indicate reaction with Evan's blue.
Analysis of the antibody titer in rabbit serum following immunization with OsrHSA revealed that specific IgGs against HSA increased quickly 21 d after the first immunization. The antibody titers demonstrated a significant increase in IgG against HSA in the first 3 wk after immunization with either OsrHSA (P, OsrHSA vs. saline) or pHSA (P′, pHSA vs. saline) compared with a saline control (P = 2.12 × 10−10, P′ = 1.17 × 10−6; Fig. 6C), whereas no significant difference was found between antibody titers for rabbits immunized with OsrHSA or pHSA (P′′ = 0.31, P′′, OsrHSA vs. pHSA). The total antiserum titer in pHSA- or OsrHSA-treated groups also displayed a similar antibody level as that in the saline group. Furthermore, the titers of individual immunoglobulins IgG (P = 0.937, P′ = 0.648), IgM (P = 0.465, P′ = 0.133), IgE (P = 0.937, P′ = 0.146), and IgA (P = 0.171, P′ = 0.012) showed no significant difference among the pHSA, OsrHSA, and saline groups (Fig. 6 D and E), and the titers of C4 complement were not significantly different among the OsrHSA, pHSA, and saline groups (Fig. 6F). However, the titers of C3 complement in the pHSA-immunized group were significantly higher than those in the OsrHSA group (P = 0.803, P′ = 0.008, P′′ = 9.20 × 10−5; Fig. 6F), which results from the presence of human blood impurities in the pHSA sample. To further assess the immunogenicity of OsrHSA, passive cutaneous anaphylaxis (PCA) was performed on guinea pigs. Anti-OsrHSA serum neutralized by pHSA or OsrHSA gave a negative response compared with the saline control (Fig. 6G), again indicating that OsrHSA displays the same immunogenicity as pHSA in vivo.
Discussion
HSA has been widely used in clinical applications and cell cultures for many decades. Currently, HSA is only obtained from human plasma. In this study, we successfully expressed rHSA in rice seeds and developed a cost-effective purification protocol for large-scale rHSA production. OsrHSA is biochemically, structurally, functionally, and immunologically equivalent to pHSA, demonstrating that rice endosperm is a safe and promising alternate system for the large-scale production of rHSA.
In general, downstream processing of recombinant pharmaceutical proteins are cost-consuming for many bioreactor systems, including those that use plants (23, 32). Our protocol for purification of OsrHSA is robust and economical, even at the kilogram scale. The recovery rate of OsrHSA was 2.75 g/kg brown rice, which is much higher than the estimated cost-effective threshold (0.1 g/kg) for production of rHSA in plants (19). Furthermore, recombinant protein production can be increased to kilogram scale within 1 y of obtaining a high-expressing transgenic line (Table S5). Here, transgenic rice seeds as bioreactor show great promise as a flexible system for the production and processing of rHSA with intact biochemical features on a large scale.
FBS is widely used in cell culture as a necessary nutritional supplement, but problems related to reliability, variability, and biological contaminants limit its use in industrial-scale cultures (33). In recent years, there has been an increased demand for animal-free components to replace serum- or animal-originated supplements to eliminate the risk of passage of prion-related diseases to humans (12). Supplementing 1 g/L of OsrHSA in serum-reduced media results in cell growth comparable to that seen using 10% FBS, indicating that OsrHSA can reduce the use of FBS in cell culture. Use of the rice seed bioreactor could provide an economical and safe approach for the production of non–animal-derived compounds, which will facilitate development of products from cell cultures.
HSA is clinically used in high dosages for i.v. administration. For instance, HSA of more than 10 g per dose is required for the treatment of hypoalbuminemia or traumatic shock (34). Continuous infusion of HSA improves the general condition of patients with cirrhosis (35). In our results, OsrHSA and pHSA displayed a similar efficacy when the same dosages (1.0 g/kg) were administrated to rats with ascites, suggesting the biological function of OsrHSA is identical to that of pHSA in vivo. Production of OsrHSA for treatment of patients suffering from liver cirrhosis with ascites could replace the use of pHSA.
Safety is a major public concern when a plant-made pharmaceutical is clinically applied. Immunogenicity of biopharmaceuticals is particularly important for clinical safety (36). Our data show that the total and specific antibodies induced by pHSA and OsrHSA were not significantly different. Even the change in the amount of complement factors, such as C4, did not differ between groups immunized with OsrHSA or pHSA. However, C3 complement titers from the OsrHSA group were significant lower than those of the pHSA group, suggesting that OsrHSA may be safer than HSA produced with other expression systems. Because of high antibody titers against P. pastoris components in human serum, rHSA produced in P. pastoris was required at a purity of more than 99.999% before it could be used clinically (37). Our results showed that OsrHSA with a purity of >99% had the same immunogenicity as pHSA with no observable side effects in the treatment of rats with liver cirrhosis. These data suggest that rice endosperm might provide a safer production platform for pharmaceutical proteins. The lower immnunogenicity of OsrHSA implies that humans can evolutionally adapt to antigens from rice seed by using rice as a staple food.
Due to the high dosage of HSA in clinical applications, large-scale production of OsrHSA requires field production of transgenic rice. Field trail of transgenic plant raises concerns of environmental safety, because rice is a staple food worldwide. Recently, rice was listed as a favorable host for molecular farming (38) for the following reasons: first, rice is a highly self-pollinated crop, and rice pollen is remarkably short-lived (<10 min) when it is out of the anther (39); with regard to biosafety assessment of transgenic rice it showed a very low frequency (0.04–0.80%) of pollen-mediated gene flow between genetically modified (GM) rice and adjacent non-GM plants (40, 41). This low frequency can be decreased to negligible levels by a short spatial isolation (42). To manage the environmental impacts of plant-made pharmaceuticals, the US Department of Agriculture (USDA) and European regulatory authorities have already issued guidance for field testing of GM organisms intended for industrial and pharmaceutical use (43, 44). Following the guidance, the USDA has approved 3358.3 ha and 13 plant hosts for field trial since 2004 (45). In the present work, an endosperm-specific promoter for HSA gene expression and a callus-specific promoter for a selective marker gene were used to generate OsrHSA transgenic rice, which could largely reduce the environmental risk from selective marker and target genes exposure. Furthermore, many strategies could be taken to prevent any possible escape of OsrHSA transgenic rice into the environment from the field trail, including an isolation zone (>100 m), a buffer zone around the field (>1.5 m), fencing, and established standard operation protocols for sowing, planting, harvesting, drying, transporting, processing and storage etc. These approaches will largely diminish the environmental impacts.
Material and Methods
Expression, Purification, and Biochemical Characterization of OsrHSA from Transgenic Rice.
Plasmids used in this study are described in the SI Materials and Methods or have been previously reported (26, 46). Transgenic lines expressing rHSA were obtained by Agrobacterium-mediated transformation as described previously (47). Purification and biochemical characterizations of OsrHSA are described in detail in SI Materials and Methods.
Crystallization and X-Ray Data Collection.
Crystals of HSA–myristic acid complexes were obtained as described in SI Materials and Methods. Diffraction data were collected from several crystals, and the best dataset was selected for structure determination and refinement. The structure was solved by molecular replacement with HSA coordinates from a previously solved structure (PDB ID code 1BJ5). The model was refined using Refmac 5 interspersed with manual rebuilding using Coot (48, 49). The refinement converged to an R-factor of 23.4% and R-free of 30.3%. The model was verified for proper geometry with 96% of residues in the most favorable region (Table S1). Figures depicting structure were prepared by PyMol.
Cell Culture.
To evaluate the cell growth promotion of OsrHSA, CHO-K, Vero, and hybridoma cell line SP2/0 were continuously cultured by media containing OsrHSA or pHSA, as presented in SI Materials and Methods. Cell viability and cell number were recorded according to the description in SI Materials and Methods.
Efficacy of OsrHSA on Liver Cirrhosis in Rats.
Specific pathogen-free male Wistar rats weighing 200 ± 20 g were used after acclimatization for 1 wk. Rats with liver cirrhosis were prepared according to previous reports (50, 51). Animals with a urinary protein excretion >51.3 mg/kg every 20 h, and those with a body weight decrease >10 g per day were excluded from this study. Data were collected and analyzed as presented in SI Materials and Methods.
Immunogenicity Evaluation of OsrHSA.
New Zealand rabbits aged 5–6 mo with 3.6 ± 0.3 kg body weight were randomly divided into three groups, with each group containing five rabbits. The rabbits were immunized as described in SI Materials and Methods. Serum samples were collected before immunization and 3 and 4 wk after the first immunization. The titer of anti-HSA IgG, total IgG, IgM, IgA, IgE, complement C3, and C4 levels were detected by ELISA. Double-immunodiffusions were performed as described in SI Materials and Methods. The P, P′, and P′′ values were calculated by t test using two tails. Female 10-wk-old guinea pigs were used for PCA assay. The PCA procedure is described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to the Biotechnology Research Institute of the National Research Council (Montreal, Canada) for the crystallization and X-ray diffraction studies of OsrHSA; Cowin Biotechnology Co. Ltd. (Beijing, China) for completing the cell culture assays, and the Genome Center Proteomics Core Facility at the University of California (Davis, CA) for the N-terminal sequence analysis. We also thank Medical College of Soochow University for osmotic pressure measurements, and Oil Crops Research Institute Chinese Academy Agricultural Science for fatty acids analysis. This work was supported by National High-Tech R&D Program 863 of China No. 2011AA100604, Major Projects of Genetically Modified Crop of China Grant 2011ZX08001-006, National Science and Technology Major Project for New Drug Development Grant 2009ZX09301-014, and Wuhan Institute of Biotechnology, Biolake, National Biological Industry Base in Wuhan.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystal structure of OsrHSA has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code: 3SQJ).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109736108/-/DCSupplemental.
References
- 1.Peters T. All About Albumin: Biochemistry, Genetics, and Medical Applications. San Diego: Academic; 1995. [Google Scholar]
- 2.Alexander MR, Ambre JJ, Liskow BI, Trost DC. Therapeutic use of albumin. JAMA. 1979;241:2527–2529. [PubMed] [Google Scholar]
- 3.Hastings GE, Wolf PG. The therapeutic use of albumin. Arch Fam Med. 1992;1:281–287. doi: 10.1001/archfami.1.2.281. [DOI] [PubMed] [Google Scholar]
- 4.Marth E, Kleinhappl B. Albumin is a necessary stabilizer of TBE-vaccine to avoid fever in children after vaccination. Vaccine. 2001;20:532–537. doi: 10.1016/s0264-410x(01)00329-2. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchida E, et al. Artificial oxygen carriers, hemoglobin vesicles and albumin-hemes, based on bioconjugate chemistry. Bioconjug Chem. 2009;20:1419–1440. doi: 10.1021/bc800431d. [DOI] [PubMed] [Google Scholar]
- 6.Cai C, Zhou K, Wu Y, Wu L. Enhanced liver targeting of 5-fluorouracil using galactosylated human serum albumin as a carrier molecule. J Drug Target. 2006;14:55–61. doi: 10.1080/10611860600613324. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian GM, Fiscella M, Lamousé-Smith A, Zeuzem S, McHutchison JG. Albinterferon alpha-2b: A genetic fusion protein for the treatment of chronic hepatitis C. Nat Biotechnol. 2007;25:1411–1419. doi: 10.1038/nbt1364. [DOI] [PubMed] [Google Scholar]
- 8.Xinhua News Agency Drug watchdog tightens supervision of albumin medicine. Beijing Review. April 4, 2007. http://www.bjreview.com.cn/health/txt/2007-04/04/content_60856.htm.
- 9.Erstad BL. Viral infectivity of albumin and plasma protein fraction. Pharmacotherapy. 1996;16:996–1001. [PubMed] [Google Scholar]
- 10.Chamberland ME, Alter HJ, Busch MP, Nemo G, Ricketts M. Emerging infectious disease issues in blood safety. Emerg Infect Dis. 2001;7(3, Suppl):552–553. doi: 10.3201/eid0707.010731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Hu D, Xi Y, Zhang M, Duan G. Spread of HIV in one village in central China with a high prevalence rate of blood-borne AIDS. Int J Infect Dis. 2006;10:475–480. doi: 10.1016/j.ijid.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Merten OW. Virus contaminations of cell cultures—A biotechnological view. Cytotechnology. 2002;39:91–116. doi: 10.1023/A:1022969101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latta L, et al. Synthesis and purification of mature human serum albumin from E. coli. Biotechnology. 1987;5:1309–1314. [Google Scholar]
- 14.Sleep D, Belfield GP, Goodey AR. The secretion of human serum albumin from the yeast Saccharomyces cerevisiae using five different leader sequences. Biotechnology (N Y) 1990;8:42–46. doi: 10.1038/nbt0190-42. [DOI] [PubMed] [Google Scholar]
- 15.Fleer R, et al. Stable multicopy vectors for high-level secretion of recombinant human serum albumin by Kluyveromyces yeasts. Biotechnology (N Y) 1991;9:968–975. doi: 10.1038/nbt1091-968. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi K, et al. High-level expression of recombinant human serum albumin from the methylotrophic yeast Pichia pastoris with minimal protease production and activation. J Biosci Bioeng. 2000;89:55–61. doi: 10.1016/s1389-1723(00)88050-0. [DOI] [PubMed] [Google Scholar]
- 17.Barash I, et al. Synthesis and secretion of human serum albumin by mammary gland explants of virgin and lactating transgenic mice. Transgenic Res. 1993;2:266–276. doi: 10.1007/BF01968839. [DOI] [PubMed] [Google Scholar]
- 18.Sijmons PC, et al. Production of correctly processed human serum albumin in transgenic plants. Biotechnology (N Y) 1990;8:217–221. doi: 10.1038/nbt0390-217. [DOI] [PubMed] [Google Scholar]
- 19.Farran I, Sánchez-Serrano JJ, Medina JF, Prieto J, Mingo-Castel AM. Targeted expression of human serum albumin to potato tubers. Transgenic Res. 2002;11:337–346. doi: 10.1023/a:1016356510770. [DOI] [PubMed] [Google Scholar]
- 20.Fernández-San Millán A, Mingo-Castel A, Miller M, Daniell H. A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J. 2003;1:71–79. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang LF, Liu YK, Lu CA, Hsieh SL, Yu SM. Production of human serum albumin by sugar starvation induced promoter and rice cell culture. Transgenic Res. 2005;14:569–581. doi: 10.1007/s11248-004-6481-5. [DOI] [PubMed] [Google Scholar]
- 22.Lau OS, Sun SS. Plant seeds as bioreactors for recombinant protein production. Biotechnol Adv. 2009;27:1015–1022. doi: 10.1016/j.biotechadv.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Boothe J, et al. Seed-based expression systems for plant molecular farming. Plant Biotechnol J. 2010;8:588–606. doi: 10.1111/j.1467-7652.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- 24.Yang D, Guo F, Liu B, Huang N, Watkins SC. Expression and localization of human lysozyme in the endosperm of transgenic rice. Planta. 2003;216:597–603. doi: 10.1007/s00425-002-0919-x. [DOI] [PubMed] [Google Scholar]
- 25.Huang N, et al. Bioactive recombinant human lactoferrin, derived from rice, stimulates mammalian cell growth. In Vitro Cell Dev Biol Anim. 2008;44:464–471. doi: 10.1007/s11626-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 26.Ning T, et al. Oral administration of recombinant human granulocyte-macrophage colony stimulating factor expressed in rice endosperm can increase leukocytes in mice. Biotechnol Lett. 2008;30:1679–1686. doi: 10.1007/s10529-008-9717-2. [DOI] [PubMed] [Google Scholar]
- 27.Ghuman J, et al. Structural basis of the drug-binding specificity of human serum albumin. J Mol Biol. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 28.Uran S, Larsen A, Jacobsen PB, Skotland T. Analysis of phospholipid species in human blood using normal-phase liquid chromatography coupled with electrospray ionization ion-trap tandem mass spectrometry. J Chromatogr B Biomed Sci Appl. 2001;758:265–275. doi: 10.1016/s0378-4347(01)00188-8. [DOI] [PubMed] [Google Scholar]
- 29.Kohno M, et al. Induction by lysophosphatidylcholine, a major phospholipid component of atherogenic lipoproteins, of human coronary artery smooth muscle cell migration. Circulation. 1998;98:353–359. doi: 10.1161/01.cir.98.4.353. [DOI] [PubMed] [Google Scholar]
- 30.Sapra P, Tyagi P, Allen TM. Ligand-targeted liposomes for cancer treatment. Curr Drug Deliv. 2005;2:369–381. doi: 10.2174/156720105774370159. [DOI] [PubMed] [Google Scholar]
- 31.Simon BJ, Svend H, Anders F, Bernd WM. US Patent Appl US2010/0190706A1. 2010 [Google Scholar]
- 32.Valdés R, et al. Hepatitis B surface antigen immunopurification using a plant-derived specific antibody produced in large scale. Biochem Biophys Res Commun. 2003;310:742–747. doi: 10.1016/j.bbrc.2003.08.149. [DOI] [PubMed] [Google Scholar]
- 33.Keenan J, Pearson D, Clynes M. The role of recombinant proteins in the development of serum-free media. Cytotechnology. 2006;50:49–56. doi: 10.1007/s10616-006-9002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters T., Jr . Serum albumin. In: Putnam FW, editor. The Plasma Protein. 2nd Ed. New York: Academic; 1975. pp. 133–181. [Google Scholar]
- 35.Romanelli RG, et al. Long-term albumin infusion improves survival in patients with cirrhosis and ascites: An unblinded randomized trial. World J Gastroenterol. 2006;12:1403–1407. doi: 10.3748/wjg.v12.i9.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schellekens H. Immunogenicity of therapeutic proteins: Clinical implications and future prospects. Clin Ther. 2002;24(11):1720–1740. doi: 10.1016/s0149-2918(02)80075-3. discussion 1719. [DOI] [PubMed] [Google Scholar]
- 37.Ohtani W, Ohda T, Sumi A, Kobayashi K, Ohmura T. Analysis of Pichia pastoris components in recombinant human serum albumin by immunological assays and by HPLC with pulsed amperometric detection. Anal Chem. 1998;70:425–429. doi: 10.1021/ac970596h. [DOI] [PubMed] [Google Scholar]
- 38.Loïc F, Véronique G. Recombinant Proteins from Plants: Methods and Protocols. Totowa, NJ: Humana; 2008. pp. 341–353. [Google Scholar]
- 39.Song Z, Lu B, Chen J. A study of pollen viability and longevity in Oryza rufipogon, O.sativa and their hybrid. Int Rice Res Notes. 2001;26:31–42. [Google Scholar]
- 40.Messeguer J, et al. Field assessments of gene flow from transgenic to cultivated rice (Oryza sativa L.) using a herbicide resistance gene as tracer marker. Theor Appl Genet. 2001;103:1151–1159. [Google Scholar]
- 41.Rong J, et al. Low frequency of transgene flow from Bt/CpTI rice to its non-transgenic counterparts planted at close spacing. New Phytol. 2005;168:559–566. doi: 10.1111/j.1469-8137.2005.01539.x. [DOI] [PubMed] [Google Scholar]
- 42.Rong J, et al. Dramatic reduction of crop-to-crop gene flow within a short distance from transgenic rice fields. New Phytol. 2007;173:346–353. doi: 10.1111/j.1469-8137.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- 43.UDSA–APHIS . Guidance for APHIS Permits for Field Testing or Movement of Organisms Intended for Pharmaceutical or Industrial Use. Washington, DC: US Department of Agriculture Animal and Plant Health Inspection Service Biotechnology Regulatory Services; 2008. [Google Scholar]
- 44.Authority EFS. Scientific opinion on fuidance for the risk assessment of genetically modified plants used for non-food or non-feed purposes. EFSA Journal. 2009;1164:1–42. [Google Scholar]
- 45.USDA–APHIS Release Permits for Pharmaceuticals, Industrials, Value-Added Proteins for Human Consumption, or for Phytoremediation Granted or Pending by APHIS as of July 18, 2011. 2011. http://www.aphis.usda.gov/brs/ph_permits.html.
- 46.Xie T, et al. A biologically active rhIGF-1 fusion accumulated in transgenic rice seeds can reduce blood glucose in diabetic mice via oral delivery. Peptides. 2008;29:1862–1870. doi: 10.1016/j.peptides.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 47.Yang D, Wu L, Hwang YS, Chen L, Huang N. Expression of the REB transcriptional activator in rice grains improves the yield of recombinant proteins whose genes are controlled by a Reb-responsive promoter. Proc Natl Acad Sci USA. 2001;98:11438–11443. doi: 10.1073/pnas.201411298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Horie S, Nagai H, Yuuki T, Hanada S, Nakamura N. Effectiveness of recombinant human serum albumin in the treatment of ascites in liver cirrhosis: Evidence from animal model. Gen Pharmacol. 1998;31:811–815. doi: 10.1016/s0306-3623(98)00064-0. [DOI] [PubMed] [Google Scholar]
- 51.Xu D, Wu Y, Liao ZX, Wang H. Protective effect of verapamil on multiple hepatotoxic factors-induced liver fibrosis in rats. Pharmacol Res. 2007;55:280–286. doi: 10.1016/j.phrs.2006.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.