Abstract
In laboratory studies, acquired resistance to long-term antihormonal therapy in breast cancer evolves through two phases over 5 y. Phase I develops within 1 y, and tumor growth occurs with either 17β-estradiol (E2) or tamoxifen. Phase II resistance develops after 5 y of therapy, and tamoxifen still stimulates growth; however, E2 paradoxically induces apoptosis. This finding is the basis for the clinical use of estrogen to treat advanced antihormone-resistant breast cancer. We interrogated E2-induced apoptosis by analysis of gene expression across time (2–96 h) in MCF-7 cell variants that were estrogen-dependent (WS8) or resistant to estrogen deprivation and refractory (2A) or sensitive (5C) to E2-induced apoptosis. We developed a method termed differential area under the curve analysis that identified genes uniquely regulated by E2 in 5C cells compared with both WS8 and 2A cells and hence, were associated with E2-induced apoptosis. Estrogen signaling, endoplasmic reticulum stress (ERS), and inflammatory response genes were overrepresented among the 5C-specific genes. The identified ERS genes indicated that E2 inhibited protein folding, translation, and fatty acid synthesis. Meanwhile, the ERS-associated apoptotic genes Bcl-2 interacting mediator of cell death (BIM; BCL2L11) and caspase-4 (CASP4), among others, were induced. Evaluation of a caspase peptide inhibitor panel showed that the CASP4 inhibitor z-LEVD-fmk was the most active at blocking E2-induced apoptosis. Furthermore, z-LEVD-fmk completely prevented poly (ADP-ribose) polymerase (PARP) cleavage, E2-inhibited growth, and apoptotic morphology. The up-regulated proinflammatory genes included IL, IFN, and arachidonic acid-related genes. Functional testing showed that arachidonic acid and E2 interacted to superadditively induce apoptosis. Therefore, these data indicate that E2 induced apoptosis through ERS and inflammatory responses in advanced antihormone-resistant breast cancer.
Keywords: aromatase inhibitor, antihormonal resistance, estrogen receptor, gene expression microarrays, selective estrogen receptor modulator
Elucidation of the basic structure function relationships of synthetic estrogens based on either stilbene (1) or triphenylethylene (2) was a landmark achievement that continues to have major therapeutic implications to this day. The first successful chemical therapy for the treatment of any cancer was the use of high-dose synthetic estrogen for the treatment of metastatic breast cancer (3). Response rates for patients who were more than a decade beyond menopause were about 30%. Importantly, treatment near menopause was ineffective, and therefore, tumor responsiveness was related to the duration of estrogen deprivation. In 1970, Alexander Haddow commented that “the extraordinary extent of tumor regression observed in perhaps 1% of postmenopausal cases [with oestrogen] has always been regarded as of major theoretical importance, and it is a matter for some disappointment that so much of the underlying mechanisms continues to elude us” (4). High-dose estrogen therapy using diethylstilbestrol (DES) remained the standard of care for the treatment of metastatic breast cancer in postmenopausal women for 30 y (1950s to late 1970s in the United States). However, triphenylethylene-based estrogens evolved into nonsteroidal antiestrogens (5), where the initial interest focused on their potential as postcoital antifertility agents. This application failed, and the compounds were subsequently reinvented as antiestrogens targeted to estrogen receptor (ER) for the treatment of all stages of breast cancer (6, 7). Subsequently, the nonsteroidal antiestrogens would again evolve and be reinvented as selective ER modulators (SERMs) (8). This new drug class exploited the observations that they blocked breast cancer development and growth as antiestrogens but lowered circulating cholesterol and maintained bone density as estrogens. This finding led to the idea that the treatment and prevention of osteoporosis would simultaneously prevent breast cancer (5, 9). Raloxifene is the first SERM of the class used to prevent both osteoporosis and breast cancer (10, 11).
The strategy of targeting ER and using long-term adjuvant tamoxifen therapy for breast cancer treatment (7) has increased 15-y survival rates (12, 13) and contributed significantly to the national reduction breast cancer mortality (14). From 1975 to 1990, breast cancer mortality rates held roughly steady, but from 1990 to 2000, they declined by 19.6%. It is estimated that about two-thirds of this reduction is because of therapy and one-third is because of mammography screening. Specifically, in ER-positive tumors, 5 y of tamoxifen therapy was estimated to have reduced the hazard of breast cancer mortality by 37% (14). Tamoxifen remains the antihormone treatment of choice for the adjuvant treatment of breast cancer in premenopausal patients, despite the development of the aromatase inhibitors (AIs) for the adjuvant treatment of postmenopausal patients with ER-positive breast cancer. The AIs provide a modest, but significant, improvement in disease-free survival for patients and a significant decrease in the incidence of both endometrial cancer and thromboembolism associated with tamoxifen therapy in postmenopausal women (15). Nevertheless, tamoxifen remains an important and cheap lifesaving drug, available in countries without a sophisticated healthcare infrastructure.
Despite the ability of long-term adjuvant antihormone therapy to enhance breast cancer patient survivorship, the consequence of any sustained therapy to control tumor growth is the development of resistance. Studies in vivo with MCF-7 cells inoculated into athymic mice showed that, although tamoxifen initially blocked tumor growth, eventually tumors would grow, despite continued tamoxifen treatment (16). Similar studies showed that tamoxifen, in fact, stimulated growth of resistant MCF-7 tumors (17). A new form of acquired drug resistance was described for breast cancer that grew in response to ER activation through either tamoxifen or the natural ligand 17β-estradiol (E2). This finding explained the observed resistance to tamoxifen in ER-positive metastatic breast cancer patients after ∼1–2 y of therapy but was inconsistent with the clinical observation that patients with stages I and II breast cancer could be routinely treated with 5 or more y of adjuvant tamoxifen therapy without developing tumor recurrence. A possible explanation would emerge from studies of acquired resistance to antihormone therapy that, at the same time, would expose a vulnerability of breast cancer cells and explain the mechanism of high-dose estrogen therapy for the treatment of breast cancer.
The continuous passage of MCF-7 tumors for more than 5 y in tamoxifen-treated athymic mice results in a reconfiguration of survival signaling pathways. Although tumors remain tamoxifen stimulated for growth, physiologic E2 now causes rapid tumor regression rather than growth (18, 19). Indeed, some tumors that regress and then regrow during continuous E2 treatment are exclusively E2-dependent, because tamoxifen or E2 withdrawal will impair tumor growth (19). The evolution of acquired resistance to SERMs (20) naturally raised the concern of the development of resistance to the new standard of care for adjuvant treatment of ER-positive breast cancer in postmenopausal patients, the AIs.
Parallel studies to replicate the clinical expression of acquired resistance to estrogen deprivation (i.e., resistance to an AI) (21, 22) were initiated in vitro 20 y ago using ER-positive MCF-7 breast cancer cells. When cells were grown under long-term estrogen-deprived conditions (>1 y), cells lost their dependency on estrogen for proliferation but maintained expression of ER. Subsequent studies of E2 action on the growth of long-term estrogen-deprived MCF-7 cells in vitro at high (23) and low concentrations in vitro and in vivo (24, 25) indicated that the concept of “an estrogen purge” (19) to destroy antihormone-resistant cells could be applied to the treatment of breast cancer. This concept has now been translated to clinical trials.
A pivotal study of high-dose DES therapy (15 mg daily) in 32 patients with metastatic breast cancer who had been treated exhaustively with antihormonal therapies produced a 30% objective response rate (26). There were 4 of 32 complete responses, and one patient maintained a complete response for an additional 7 y, even after stopping estrogen (27). A recent study in patients whose breast cancer had responded but then failed AI treatment (28) showed that low-dose E2 treatment (6 mg daily) would produce the same clinical benefit as high-dose E2 (30 mg daily) but with fewer toxic side effects. Thus, laboratory observations with low doses of estrogen treatment translate to clinical practice, and a mechanism is now emerging to explain the original observations by Haddow (3, 4). The goal of future translational research is to discover molecular mechanisms to amplify the estrogen-induced apoptotic trigger.
The question arises as to the precise sequence of events that lead to E2-induced apoptosis. By describing and defining these molecular events, refractory cells may be manipulated to respond to estrogen-induced apoptosis. To begin to address the question, we have developed a series of MCF-7 variants that are either estrogen-dependent for growth (MCF-7:WS8 cells) (29–31) or resistant to estrogen deprivation (ED) and refractory (MCF-7:2A) (25, 30, 31) or sensitive (MCF-7:5C) (24, 29, 32) to E2-induced apoptosis. We previously reported changes in gene expression among these cell lines by Affymetrix-based microarray analysis under estrogen-free conditions (33). Thus, these identified differentially expressed genes were associated with progression to an ED-resistant phenotype. We have also recently reported a proteomic analysis of 5C compared with WS8 cells after 2 h of E2 exposure to identify proteins that may initiate apoptosis (34). Here, we seek to identify genes differentially regulated by E2 over a 2–96 h time course, which overlaps with actively occurring apoptosis. Therefore, we interrogated these models for changes in E2-regulated global gene expression as a function of time using Agilent 4 × 44 K oligonucleotide microarrays. We developed a method termed differential area under the curve (dAUC) analysis to identify genes that exhibited significantly altered regulation by E2 across time specifically in the apoptosis-sensitive 5C cells compared with both the estrogen-dependent WS8 and apoptosis-refractory 2A cells. Examination of the identified 5C-specific genes and functional testing indicated that E2-elicited endoplasmic reticulum stress (ERS) and inflammatory stress responses that led to apoptosis.
Results and Discussion
Cell Line Characterization.
Before gene expression microarray studies were carried out, the estrogen-dependent WS8 (29–31), ED-resistant but apoptosis-refractory 2A (25, 30, 31), and apoptosis-sensitive 5C cells (24, 29, 32) were characterized to confirm previously reported growth responses, biomarker status, and estrogen response element (ERE) -regulated transcriptional activity (SI Results and Discussion, SI Methods, and Fig. S1). The apoptotic responses of 5C cells to E2 were also characterized according to loss of plasma membrane integrity (SI Results and Discussion, SI Methods, and Fig. S2). The 5C cells exhibited an EC50 for apoptosis of 3 × 10−11 M E2 after 96 h of exposure (Fig. S2B). Additionally, 10−9 M E2, the concentration used for the microarray studies, caused apoptosis ranging from ∼30% to 42% of the 5C cells, depending on the experiment (Fig. S2 B and C). The pure antiestrogen fulvestrant completely blocked apoptosis induced by E2 and DES, showing that apoptosis was ER-dependent (Fig. S2C).
Global Gene Expression Across Time.
To identify genes and pathways/processes associated with E2-induced apoptosis, differential regulation of global gene expression in response to E2 was interrogated in ED-resistant/apoptotic-sensitive 5C cells vs. estrogen-dependent WS8 and ED-resistant/apoptotic-refractory 2A cells. Each cell line was treated with 10−9 M E2 or vehicle control over a 96-h time course consisting of seven time points (2, 6, 12, 24, 48, 72, and 96 h) using six biological replicates per condition. cRNA probes from individual E2-treated samples were competitively hybridized against time-matched pooled control probes using two-color Agilent 4 × 44 K human oligonucleotide microarrays. The resulting gene expression values were log2 ratios of mRNA levels in E2/control-treated cells that, when plotted across time, form a curve. A measure of change in E2-mediated regulation of expression over a defined time interval was then calculated as the difference in AUCs or dAUCs for a given gene between two cell lines. Genes that showed a 50% change in AUCs between two cell lines (corresponding to a dAUC = 0.58 on a log2 scale) at a P value < 0.00005 (Methods has details on P value determination) were defined as significantly different. The dAUC method was applied to identify differentially regulated genes at 2–96, 2–24, and 24–96 h to identify overall, relatively early, and late-responding genes, respectively.
To identify genes specifically associated with E2-induced apoptosis, genes were selected with regulation that differed significantly with E2 in the 5C cells vs. both the WS8 and 2A cells. A total of 1,142 genes were identified as significantly differentially regulated by E2 specifically in the 5C cells (Dataset S1). These genes were examined for overrepresentation of those genes mapping to a particular curated pathway/network (Fig. S3A). As expected, estrogen signaling and apoptosis genes were significantly enriched. Within the apoptosis category, ERS was the most enriched apoptosis subcategory (Fig. S3B). Inflammatory response genes were also enriched. The overlapping distribution of genes mapping to estrogen signaling (Dataset S2), apoptosis (Dataset S3), and inflammatory responses (Dataset S4) is shown in the Venn diagram in Fig. S3C.
Estrogen Signaling Genes.
Estrogen signaling genes selectively regulated by E2 in 5C cells relative to both WS8 and 2A cells are listed in Dataset S2, and examples discussed are shown in Fig. 1. Multiple genes were differentially regulated by E2 in 5C cells compared with WS8 and 2A cells, which would diminish ERα activity and hence, the apoptotic stimulus. For example, genes that negatively modulate ERα activity (i.e., AR, CYP1B1, FHL2, HSD17B11, INHBA, NR2F1/COUP-TF1, SNAI1/Snail 1, and THRA/TRα) were selectively up-regulated, whereas those genes that promote ERα activity were selectively down-regulated (AREG, CAV1, and PIK3CB) by E2 in 5C cells. The up-regulated estrogen metabolizing enzymes CYP1B1 and HSD17B11 would decrease intracellular E2 pools. SETD7/SET7/SET9 methylates ER to stabilize the protein (Dataset S5, ref. 1); hence, its down-regulation by E2 in 5Cs would accelerate ERα protein degradation. ERα activity would be suppressed by up-regulation of transcription factors that repress ERα RNA expression (i.e., FHL2 and Snail-1) (Dataset S5, refs. 2 and 3) or compete with ERα for binding-extended ERE half-sites, which overlap with many natural EREs (i.e., COUP-TF1 and TRα) (Dataset S5, refs. 4 and 5). AR failed to down-regulate in response to E2 in 5C cells, allowing greater AR activity. AR and ERα interact in complexes, and androgens inhibit E2-stimulated growth of MCF-7 cells (Dataset S5, ref. 6); thus, AR can oppose ERα. ERα activity can also be suppressed by activin-A, a TGFβ superfamily ligand, in a SMAD3-dependent manner in MCF-7 cells (Dataset S5, ref. 7). Both INHBA and SMAD3 were selectively induced in 5C cells, and INHBA homodimerizes to form activin-A, which signals to SMAD3; SMAD3 interacts with ERα at promoters to repress transcription. AREG and PIK3CB failed to increase in response to E2 in 5C cells. This failure to increase may have prevented increased ERα activity, because AREG activates EGFR, which leads to ERα-Ser118 phosphorylation, and PIK3CB is the catalytic subunit of PI3K, which through Akt, targets ERα-Ser167 phosphorylation (Dataset S5, ref. 8). CAV1 expression also failed to increase, which again prevents increased ERα activity, because CAV1 interacts with and promotes activity of membrane-localized ERα (Dataset S5, ref. 9). However, not all of ERα's activities were suppressed. In particular, ERα interacts with and directs transcription through AP-1 transcription complexes in addition to EREs (Dataset S5, ref. 10). AP-1 complexes consist of FOS, JUN, and JUND subunits, which were all selectively induced by E2 in 5C cells [FOS and JUN were verified by quantitative PCR (qPCR) in Fig. S4]. Importantly, AP-1 complexes play important roles in apoptosis and inflammatory responses (discussed later), and thus, ERα interaction with AP-1 provides a mechanism for E2 to target such genes.
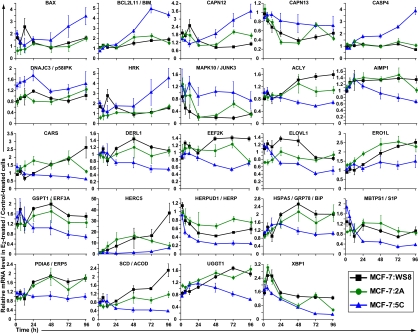
Fig. 1.
Examples of estrogen signaling genes. Full annotation, dAUC values, and P values of all estrogen signaling genes are given in Dataset S2.
Apoptosis Genes.
The identified apoptosis genes are listed in Dataset S3, and discussed examples are shown in Fig. 2. Enrichment analysis indicated ERS-mediated apoptosis as the top-scoring individual pathways within the apoptosis category (Fig. S3). The endoplasmic reticulum is a key site for protein folding. When cellular stresses perturb energy levels, the redox state, or Ca2+ concentrations, unfolded proteins accumulate and protein aggregation occurs; this condition is referred to as ERS (Dataset S5, refs. 11 and 12). To relieve ERS, an unfolded protein response (UPR) is triggered by the chaperone HSPA5/GRP78/BiP. In addition to binding unfolded proteins, GRP78 binds and prevents oligomerization of the endoplasmic reticulum transmembrane receptors EIF2AK3/PERK, IRE1α/ERN1, and ATF6. When unfolded proteins accumulate, GRP78 is released from binding the transmembrane receptors, allowing them to oligomerize and autophosphorylate to initiate a UPR signal. The UPR signals to attenuate protein translation, induce expression of additional chaperones, and export misfolded proteins to the cytosol for degradation. If the UPR fails to relieve the stress, the function of the UPR switches from promoting cell survival to promoting cell death. Thus, excessive or prolonged ERS typically induces apoptosis.
Fig. 2.
Examples of apoptosis genes. Full annotation, dAUC values, and P values of all apoptosis genes are given in Dataset S3.
Growth stimulation of hormonally responsive cells by E2 leads to increases in requirements for folding nascent polypeptides and clearance of malfolded proteins. However, in 5C cells compared with WS8 and 2A cells, E2-regulated expression changes indicated a deficiency in these functions. In 5C cells, E2 failed to sufficiently up-regulate endoplasmic reticulum-localized protein folding genes, including GRP78 (verified by qPCR in Fig. S4), ERO1L, PDIA6, and UGGT1. Cytoplasmic protein folding genes, including HSP90AB1/HSP90B, PPIAL4A, and PPIF (also FKBP10), also failed to up-regulate. Additionally, in 5C cells, E2 preferentially down-regulated HERPUD1/HERP1 and DERL1, factors that promote degradation of endoplasmic reticulum-resident proteins. A deficiency in up-regulating UPR genes in 5C cells may have resulted in part by the pronounced E2-mediated repression of MBTPS1/S1P, which cleaves ATF6, activating its translocation to the nucleus to induce transcription of UPR genes, including XBP1. Thus, decreased S1P may have led to decreased ATF6 and XBP1 activity, thereby preventing induction of multiple UPR genes.
E2-mediated gene expression alterations in 5C cells indicated widespread inhibition of protein translation compared with E2-treated WS8 and 2A cells. Within 2 h, E2 had up-regulated DNAJC3/p58IPK, which binds to and inactivates EIF2AK3/PERK, leading to reduced global translational initiation (Dataset S5, ref. 12). The aminoacyl tRNA synthetase interacting protein AIMP1 and tRNA synthetases, including CARS (also LARS, SARS, and YARS) failed to increase in response to E2 in 5Cs. Other translational factors that failed to induce in 5C cells include EEF2K and GSPT1/ERF3A (also EEF1A1, ETF1, and PABPC4).
Under severe ERS, the UPR can shut down lipogenesis as cells commit to death (Dataset S5, ref. 12). This was likely the case in E2-treated 5C cells since they showed a lack of induction of critical genes involved in fatty acid synthesis, including ACLY, SCD/ACOD, and ELOVL1. ACLY is the primary enzyme responsible for synthesis of acetyl-CoA, the basic building block of fatty acids. SCD introduces a C-C double bond in fatty acyl-CoA substrates, including stearoyl-CoA and palmitoyl-CoA, a key step in producing monounsaturated fatty acids. ELOVL1 condenses both saturated and monounsaturated fatty acids. Notably, SCD and ELOVL1 are localized to the endoplasmic reticulum membrane.
In response to severe ERS, specific BCL2 and Bcl-2 homology domain 3 (BH3) -only family members are targeted to initiate apoptosis (Dataset S5, ref. 11). Prototypical BCL2 inhibits cell death by binding and inactivating proapoptotic members such as BAX. BH3 only-containing proteins like BCL2L11/BIM indirectly activate BAX by binding BCL2 (through the BH3 motif), thereby releasing BAX from the complex. BAX then permeabolizes the mitochondrial outer membrane, allowing cytochrome C release to the cytoplasm. Under ERS, BAX also interacts with and activates IRE1α. IRE1α then signals to JNK to simultaneously activate BIM and inhibit BCL2 (Dataset S5, ref. 11). A variety of ERS inducers stimulate BIM expression, and BIM is essential in ERS-induced apoptosis in a wide range of cell types (Dataset S5, ref. 13). This apoptotic pathway was likely activated by E2 in 5C cells. E2 failed to repress MAPK10 (JNK3) in 5C cells, indicating higher JNK3 activity. Meanwhile, E2 selectively up-regulated BAX, BIM (verified by qPCR in Fig. S4), and another BH3-only proapoptotic factor, HRK (also BBC3/PUMA but PUMA did not make the significance cutoff) (Fig. S5). Importantly, E2 repressed BCL2 in 5C cells but induced it in WS8 cells. However, E2 also repressed BCL2 in 2A cells, and therefore, it was not a 5C-specific gene (Fig. S5). We previously verified the importance of BAX and BIM by showing that they were selectively induced by E2 at the protein level in 5C vs. WS8 cells and that their depletion by RNAi blocked E2-induced apoptosis (31). Therefore, ERS may have triggered mitochondrial-mediated apoptotic cell death in E2-treated 5C cells.
After prolonged ERS, specific caspases are activated to enact cell death. Examination of the caspases revealed that only CASP4 met the stringent statistical significance criteria in the microarray data. CASP1, CASP5, and CASP8 also showed up-regulation in 5C cells but did not meet our significance threshold (Fig. S5). CASP4 along with CASP1 and CASP5 are inflammatory caspases, because they are involved in cytokine maturation (Dataset S5, ref. 14). CASP4 specifically localizes to the endoplasmic reticulum and undergoes cleavage in response to ERS-inducing agents/proteins but not other apoptotic agents, and its blockade using z-LEVD-fmk or depletion by RNAi can prevent endoplasmic stress-induced apoptosis in multiple model systems (Dataset S5, refs. 15–20). Importantly, CASP4 autoactivates by dimerizing and undergoing interdomain cleavage (Dataset S5, ref. 21), and thus, simply overexpressing CASP4 is sufficient to induce cleavage of downstream caspases (Dataset S5, ref. 22) and cause apoptosis (Dataset S5, ref. 23). Under ERS, CASP4 can also be activated by calpain (Dataset S5, refs. 24 and 25), and CAPN12 and CAPN13 were selectively up-regulated in 5C cells.
Inflammatory Response Genes.
The inflammatory response genes are listed in Dataset S4, and discussed examples are shown in Fig. 3. In 5C cells, E2 elicited up-regulation of many proinflammatory cytokine/cytokine receptors, including IL-4R (verified by qPCR in Fig. S4), IL-6R, IL-6ST/gp130, IL-17RD/Sef, and VEGFA. IL-4R was induced with early kinetics, indicating that it may be a primary response. IL-6R was up-regulated shortly after IL-4R, whereas IL-6ST/gp130, also an IL-4R subunit, was already up-regulated by 2 h. Hence, IL-6 signaling was likely activated in 5Cs. IL-17RD/Sef not only mediates IL-17 signaling, but its overexpression also leads to JNK activation and apoptosis (Dataset S5, ref. 26), which links inflammatory responses and ERS. VEGFA also leads to activation of JNK in tamoxifen-resistant MCF-7 cells (Dataset S5, ref. 27). An IFN response was likely activated, because the IFN IFNL1 and the IFN-responsive genes IFI6 and IFI16 (Dataset S3) were up-regulated. CASP4 can also be induced by IFN (Dataset S5, ref. 28).
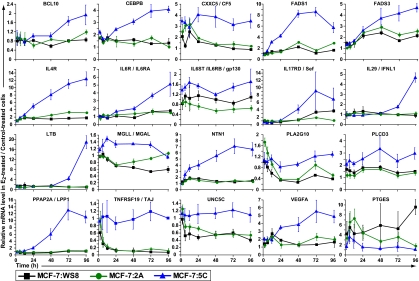
Fig. 3.
Examples of inflammatory response genes. Full annotation, dAUC values, and P values of all inflammatory response genes are given in Dataset S4.
A number of other proinflammatory genes, such as CEBPB, NTN1 (verified by qPCR in Fig. S4), and UNC5C, were selectively up-regulated in E2-treated 5C cells with relatively early kinetics, indicating possible mechanistic roles. CEBPB is important in induction of IL-6, is activated by ERS (Dataset S5, ref. 29), is required for nuclear import of the key ERS protein CHOP/GADD153 (Dataset S5, ref. 30), and enhances NF-κB signaling (Dataset S5, refs. 31 and 32). NTN1 is a secreted inflammatory marker, but it protects tissues from inflammatory injury by suppressing cytokine production, repulsing leukocyte infiltration, and acting as an antiinflammatory and antiapoptotic ligand of its receptors DCC and the UNC-5 family members (Dataset S5, refs. 33 and 34). In the context of E2-induced apoptosis, NTN1 may have been up-regulated to limit or resolve the inflammatory response. Interestingly, E2 rapidly down-regulated UNC5C in WS8 and 2A cells within 6 h but failed to do so in 5C cells, resulting in higher UNC5C expression. UNC5C may have a proinflammatory role, because synovial cells from patients with rheumatoid arthritis and osteoarthritis dramatically overexpress UNC5C (769-fold) compared with those cells of healthy donors (Dataset S5, ref. 35).
Arachidonic acid (AA; 20:4n-6) is a polyunsaturated fatty acid that plays a key role as an inflammatory mediator. Enzymes involved in AA biosynthesis were up-regulated by E2 in 5C cells, including FADS1 (verified by qPCR in Fig. S4), FADS3, PLA2G10, PLCD3, MGLL/MAGL, PPAP2A/LPP1 (verified by qPCR in Fig. S4), and SGMS1/SMS1. FADS3 and FADS1 catalyze the first and last steps in AA biosynthesis by introducing C-C double bonds in linoleic acid, producing γ-linolenic acid (18:3n-6), and dihomo-γ-linolenic acid (20:3n-6), producing AA. PLA2s hydrolyze phospholipids, releasing AA, whereas PLCD3 cleaves AA from diacylglycerol. MGLL converts monoacylglycerides such as 2-arachidonoylglycerol to free fatty acids including AA. PPAP2A/LPP1 converts phosphatidic acid to diacylglycerol, providing increased substrate levels for PLCD3 to release AA. As an inflammatory mediator, AA is used as a precursor by cyclooxygenase and lipoxygenase to generate inflammatory prostaglandins and leukotrienes, respectively. However, the cyclooxygenase pathway was unlikely to have been involved in E2-induced apoptosis, because induction of PTGES failed in 5C cells compared with WS8 and 2A cells. In hormone-dependent breast cancer cells, E2 is known to induce PTGES expression through an ERE, which may promote breast cancer proliferation, because the increased prostaglandin E2 may enhance aromatase expression and also promote local productions of estrogens (Dataset S5, ref. 36). Thus, a failure to induce PTGES may, ultimately, have served to prevent any potential increases in estrogen concentrations in 5C cells. Considering that ERS likely led to a block of fatty acid synthesis and conversion to monounsaturated fatty acids (i.e., no induction of ACLY and SCD), the selective increases in AA-related genes likely indicate the importance of AA in promoting an inflammatory response in E2-induced apoptosis.
Cross-Talk Between ERS and Inflammatory Stress.
As mentioned previously, ERS and inflammatory pathways intersect. The key ERS genes IRE1α, ATF6, and PERK can all activate NF-κB, which serves as a master regulator of inflammatory response gene transcription (Dataset S5, refs. 12 and 37). Many of the identified cytokine/cytokine receptors signal through NF-κB pathways. Other genes selectively induced by E2 in 5C cells, including BCL10 (Dataset S5, ref. 38), CXXC5 (Dataset S5, ref. 39), LTB (verified by qPCR in Fig. S4 and Dataset S5, ref. 40), and ITGB2 (Dataset S4; Dataset S5, ref. 41), activate NF-κB signaling as well. Additionally, SETD7/SET7/SET9, which negatively regulates NF-κB activity by methylating the RelA subunit to induce its degradation (Dataset S5, ref. 42), was down-regulated by E2 in 5Cs (Fig. 1). Furthermore, multiple 5C-specific genes are NF-κB–responsive, including BIM (Dataset S5, refs. 43 and 44), CASP4 (Dataset S5, ref. 45), CEBPB (Dataset S5, ref. 46), CP (Dataset S4; Dataset S5, ref. 47), NTN1 (Dataset S5, ref. 48), and VEGFA (Dataset S5, ref. 49). Moreover, ERα and NF-κB can interact to transcriptionally regulate promoters, providing a direct mechanism for E2 to target a diverse array of inflammatory and apoptotic genes. Therefore, NF-κB signaling was very likely involved in E2-induced apoptosis, and we are pursuing this hypothesis in future studies.
ERS also intersects with inflammatory responses through JNK. As mentioned, the ERS sensor IRE1α (Dataset S5, ref. 12) and the IL receptor 17RD/Sef can activate JNK (Dataset S5, ref. 26). The orphan TNF receptor TNFRSF19/TAJ, which failed to down-regulate in response to E2 in 5C cells, also activates JNK (Dataset S5, ref. 50). JNK then phosphorylates AP-1 complexes to induce expression of inflammatory response genes (Dataset S5, ref. 12). As mentioned earlier, the AP-1 subunits JUN, JUND, and FOS were selectively induced in E2-treated 5C cells.
Functional Involvement of AA and CASP4 in E2-Induced Apoptosis.
The involvement of ERS and inflammatory stress in E2-induced apoptosis was functionally examined. We first tested whether E2-induced apoptosis could be promoted by AA. AA was chosen, because (i) it is widely recognized as a proinflammatory agent; (ii) it induces apoptosis (Dataset S5, ref. 51), at least in part by depleting the endoplasmic reticulum of Ca2+ and inhibiting protein translation, thereby likely eliciting ERS (Dataset S5, ref. 52); (iii) it can activate NF-κB in mammary epithelial cells (Dataset S5, ref. 53); and (iv) several genes, which increase AA levels (e.g., FADS1 and PLA2G10), were up-regulated in response to E2 in 5C vs. WS8 and 2A cells. 5C cells were exposed to varying concentrations of both AA and E2 in a factorial design, and then, apoptosis was measured by flow cytometric analysis of YO-PRO-1 and 7-aminoactinomycin D staining (Fig. 4A). Because E2-induced apoptosis occurs maximally with 10−9 M E2 after 96 h of exposure, E2 was used at low concentrations of 2.5 and 5 × 10−11 M, and apoptosis was assayed at 72 h to allow observation of potential additional AA effects. The combination of AA plus E2 at all varied concentrations increased the percentage of apoptotic plus dead cells in a greater than additive manner relative to either agent alone. Fitting the data to a multiple regression model showed the rate of increase (slope) in apoptotic plus dead cells progressively and significantly increased comparing E2 alone with E2 + 10 μM AA or E2 + 20 μM AA. Therefore, AA and E2 interacted to superadditively induce apoptosis, indicating that their pathways functionally intersect.
Fig. 4.
Functional interrogation of E2-induced apoptosis. (A) AA and E2 interact to superadditively induce apoptosis. 5C cells were treated with combinations of AA and E2 as indicated for 72 h. (B) Screening of selective CASP inhibitors. The selectivity of the inhibitors for individual caspases is indicated according to the manufacturer. 5C cells were treated with 10−9 M E2 and 10 μM of each CASP inhibitor as indicated for 96 h. (A and B) Apoptosis according to altered plasma membrane permeability was determined by flow cytometric analysis of cells stained with the DNA-specific binding dyes YO-PRO-1 and 7-aminoactinomycin D. Double-negative staining cells were defined as viable, double-positive staining cells were defined as dead, and intermediately staining cells were defined as apoptotic. Data shown in B represent triplicates and associated SDs.
The importance of CASP4 was evaluated using a panel of irreversible caspase peptide inhibitors selectively targeting caspases-1 to -9 (except CASP3, which is not expressed in MCF-7 cells) (Dataset S5, ref. 54). 5C cells were treated with 10−9 M E2 plus each caspase inhibitor as indicated for 96 h to induce apoptosis, which was measured by altered plasma membrane permeability (Fig. 4B). The broad spectrum caspase inhibitor z-VAD-fmk was used as a positive control, because we previously reported that this inhibitor completely blocks E2-induced apoptosis (31), whereas the inactive inhibitor z-FA-fmk was used as a negative control. In an effort to prevent off-target caspase inhibition, the blockers were used at 10 μM, which was the concentration that reduced apoptosis by approximately one-half by the pan inhibitor z-VAD-fmk. The most active inhibitor was the CASP4 blocker z-LEVD-fmk, which was slightly more effective than the pan CASP inhibitor (Fig. 4B). The CASP8 inhibitor z-IETD-fmk was the next most active blocker but was significantly less potent than z-LEVD-fmk (P value = 0.0026). Therefore, in an unbiased comparison of caspases-1 to -9, CASP4 was validated as functionally critical in E2-induced apoptosis.
The functional activity of CASP4 was also studied. Real-time qPCR and immunoblotting confirmed induction of CASP4 expression at the mRNA and protein levels, respectively, occurred specifically in 5C cells in response to E2 (Fig. 5 A and B). Importantly, in 5C cells, z-LEVD-fmk at 20 μM completely blocked E2-induced PARP cleavage (Fig. 5B), reversed E2-inhibited growth (Fig. 5C), and prevented morphologic alterations associated with apoptosis in 5C cells (Fig. 5D). Because z-LEVD-fmk was used at 20 rather than 10 μM, we do not discount the possibility that some caspases in addition to CASP4 were also inhibited and that other caspases could still play an important role. Yet, our data establishes a critical role for CASP4 in E2-induced apoptosis.
Fig. 5.
Functional involvement of CASP4 in E2-induced apoptosis. E2-induced CASP4 at the (A) mRNA and (B) protein levels in 5C cells but not in WS8 or 2A cells. CASP4 mRNA and protein levels were measured by qPCR and immunoblotting, respectively. (B) In 5C cells, E2 led to cleavage of the apoptotic marker PARP, which was blocked by the CASP4 inhibitor z-LEVD-fmk. C, control; E, E2; I, inhibitor (z-LEVD-fmk); c-CASP4, cleaved CASP4; NS, nonspecific band. (C) E2-inhibited growth of 5C cells was completely reversed by z-LEVD-fmk. Proliferation was determined after 6 d of 10−9 M E2 exposure and measured by DNA mass per well. CASP4 Inh, CASP4 inhibitor z-LEVD-fmk. (D) Morphologic alterations after 96 h of 10−9 M E2 in 5C cells were completely reversed by z-LEVD-fmk (LEVD). (Scale bar: 100 microns.) (A–D) E2 was used at 10−9 M and z-LEVD-fmk at 2 × 10−5 M. Data in A and C represent the average and SDs of four and eight replicates, respectively.
Concluding Remarks.
We have interrogated E2-induced apoptosis by identifying differentially regulated genes across time associated with this process compared with E2-stimulated and -independent growth using a method we developed termed dAUC analysis. Overrepresentation analysis of the identified genes indicated that 5C cells respond to E2 by suppressing ERα signaling and producing endoplasmic reticulum and inflammatory stress. Estrogen signaling was suppressed by metabolically reducing intracellular E2 concentrations (increased CYP1B1 and HDS17B11) and up-regulating genes that antagonize ERα activity (SETD7, FHL2, Snail 1, COUP-TF1, TRα, AR, INHBA, and SMAD3) or repressing genes that promote ERα activity (AREG, PIK3CB, and CAV1). ERS was indicated by a deficiency in up-regulating genes involved in initiating a UPR (GRP78, XBP1, and S1P), protein folding (GRP78, PDIA6, and UGGT1), and degradation of malfolded proteins (HERP1 and DERL1), which would lead to accumulation of unfolded/misfolded proteins. Meanwhile, expression profiles indicated a widespread inhibition of protein translation (increased p58IPK and decreased aminoacyl tRNA synthetases, EEF2K, and ERF3A) and fatty acid synthesis (decreased ACLY and SCD), which combined with accumulation of unfolded proteins, would also promote stress and apoptosis. ERS was also indicated by induction of BIM, BAX, and the inflammatory caspase CASP4. We previously showed that depletion of BIM or BAX blocked E2-induced apoptosis (31), and here, we showed that blocking CASP4 with z-LEVD-fmk also blocked E2-induced apoptosis. Inflammatory stress was indicated by up-regulation of cytokines/cytokine receptors (IL-4R, IL-6R, IL-6ST/gp130, IL-17RD, and LTB), IFN/IFN responsive genes (IFNL1, IFI6, and IFI16), AA biosynthetic genes (FADS1 and PLA2G10), and other inflammatory markers (CEBPB, NTN1, and UNC5C). These findings indicate that inflammatory and ERS responses leading to apoptosis are highly interrelated and may cross-talk in part through NF-κB, JNK, and AP-1. Thus, additional stimulation of ERS and inflammatory responses by AA interacted with E2 to superadditively induce apoptosis.
It should be noted that the differentially expressed genes identified here are associated with E2-induced apoptosis; hence, their causal role in apoptosis needs to be functionally validated. Additional characterization and functional validation of genes and pathways regulating E2-mediated apoptosis in 5C cells is currently being investigated using genome-wide, high-throughput RNAi profiling. Also, the results presented here are based on only MCF-7 derivative cell lines; hence, the findings may have limited applicability to the clinic. However, the MCF-7 cell line has accurately predicted clinical responses to antihormonal therapy in breast cancer (35). We and others have reported antihormonal-resistant MCF-7–based models besides 5C cells that exhibit E2-induced apoptosis in vitro and in vivo (18, 19, 23, 36). We are aware of only one other breast cancer model not derived from MCF-7 cells that exhibits this behavior (i.e., T47D cells stably expressing PKCα), but only when grown in vivo as xenograft tumors (37). Therefore, molecular markers of ERS and inflammatory stress need to be confirmed in low-dose E2 responding compared with nonresponding tumors in patients with estrogen-deprived metastatic breast cancer.
The identified 5C-specific genes may serve as biomarkers to predict response to estrogen therapy (e.g., the secreted factors IFNL1, LTB, and NTN1 could be readily measured in patients). The identified 5C-specific genes also provide the basis for potentially improving clinical response rates to estrogen by combining it with agents that promote ERS and/or tumor-specific inflammation. For example, neutralizing NTN1 antibodies, AA, or its precursor, conjugated LA (Dataset S5, ref. 51), may increase response rates without engaging systemic inflammatory responses. Furthermore, these findings lead to the hypothesis that antiinflammatory agents prescribed for ancillary clinical problems should not be used during antitumor estrogen therapy.
Methods
Generation and Validation of RNA Samples for Microarrays.
Each cell line was treated with or without 10−9 M E2 using six replicates per treatment for 2, 6, 12, 24, 48, 72, and 96 h. To validate that each isolated RNA sample was derived from cells appropriately treated with or without E2, expression of two classical E2-responsive genes, MYC and TFF1 (pS2), were measured using real-time qPCR. MYC exhibited early kinetics, and TFF1 exhibited later kinetics of E2 induction; together, induction of these markers spanned the entire time course. Successfully validated samples are shown in Figs. S6–S8.
dAUC Analysis.
Differentially labeled fluorescent cRNA probes for each individual E2-treated RNA sample (Cy3) and time point-matched, pooled, control-treated RNA samples (Cy5) were competitively hybridized to Agilent 4 × 44 K oligonucleotide microarrays using standard Agilent protocols. Gene expression values were extracted from arrays as relative log2 ratios of E2/control-treated cells. To determine whether a gene's regulation by E2 was significantly different between two cell lines, a method termed dAUC analysis was developed. In this method, the quantity of interest (dAUC) for a given probe is calculated as the signed area between the expression profiles for the two cell lines (using the average observed values at each time point). The null hypothesis is that dAUC is zero, and the distribution of dAUC values under the null hypothesis can be obtained by repeatedly permuting (n = 20,000) the cell line to which each log2 ratio value was assigned, while keeping the time points fixed. The two-sided P value of the observed dAUC can then be calculated as the proportion of permutations yielding a dAUC that exceeds the observed dAUC in absolute value. A probe was considered significantly different between two cell lines if the magnitude of the observed dAUC exceeded that obtained in all permutations (i.e., P < 0.00005). To exclude probes with statistically significant but numerically small differences, we imposed an additional condition that the probe's dAUC must have exhibited an average log2 fold change of 0.58 (1.5-fold on a linear scale) across a given time period. The dAUCs of each probe were calculated using all pairwise combinations of the three cell lines over the entire 2–96 h time course, and to delineate relatively early and late response genes, they were calculated over 2–24 and 24–96 h time periods.
Supplementary Material
Acknowledgments
We thank Olga Tchuvatkina for programming a sample tracking system. We thank Michael Bittner, Seungchan Kim, Samuel Litwin, Suraj Peri, Karthik Devarajan, and Yan Zhou for assistance with bioinformatic analyses. We thank John Taylor for technical assistance. This work was supported by Department of Defense Center of Excellence Grant W81XWH-06-1-0590 (to V.C.J.) and Weg and Genuardis funds (to V.C.J.) and National Institutes of Health Grants P30CA006927 (to Fox Chase Cancer Center) and P30CA051008 (to Lombardi Comprehensive Cancer Center, Georgetown University).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE29917).
See Profile on page 18876.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115188108/-/DCSupplemental.
References
- 1.Dodds EC, Goldberg L, Lawson W, Robinson R. Estrogenic activity of certain synthetic compounds. Nature. 1938;141:247–248. [Google Scholar]
- 2.Robson JM, Schönberg A. Oestrous reactions including mating produced by triphenylethylene. Nature. 1937;140:196. [Google Scholar]
- 3.Haddow A, Watkinson JM, Paterson E, Koller PC. Influence of synthetic oestrogens on advanced malignant disease. Br Med J. 1944;2:393–398. doi: 10.1136/bmj.2.4368.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haddow A. David A. Karnofsky memorial lecture. Thoughts on chemical therapy. Cancer. 1970;26:737–754. doi: 10.1002/1097-0142(197010)26:4<737::aid-cncr2820260402>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 5.Lerner LJ, Jordan VC. Development of antiestrogens and their use in breast cancer: Eighth Cain memorial award lecture. Cancer Res. 1990;50:4177–4189. [PubMed] [Google Scholar]
- 6.Jordan VC. Tamoxifen: A most unlikely pioneering medicine. Nat Rev Drug Discov. 2003;2:205–213. doi: 10.1038/nrd1031. [DOI] [PubMed] [Google Scholar]
- 7.Jordan VC. Tamoxifen: Catalyst for the change to targeted therapy. Eur J Cancer. 2008;44:30–38. doi: 10.1016/j.ejca.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan VC. Selective estrogen receptor modulation: A personal perspective. Cancer Res. 2001;61:5683–5687. [PubMed] [Google Scholar]
- 9.Jordan VC, Phelps E, Lindgren JU. Effects of anti-estrogens on bone in castrated and intact female rats. Breast Cancer Res Treat. 1987;10:31–35. doi: 10.1007/BF01806132. [DOI] [PubMed] [Google Scholar]
- 10.Cummings SR, et al. The effect of raloxifene on risk of breast cancer in postmenopausal women: Results from the MORE randomized trial. Multiple Outcomes of Raloxifene Evaluation. JAMA. 1999;281:2189–2197. doi: 10.1001/jama.281.23.2189. [DOI] [PubMed] [Google Scholar]
- 11.Vogel VG, et al. Effects of tamoxifen vs raloxifene on the risk of developing invasive breast cancer and other disease outcomes: The NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA. 2006;295:2727–2741. doi: 10.1001/jama.295.23.joc60074. [DOI] [PubMed] [Google Scholar]
- 12.Early Breast Cancer Trialists' Collaborative Group Tamoxifen for early breast cancer: An overview of the randomised trials. Early Breast Cancer Trialists’ Collaborative Group. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 13.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 14.Berry DA, et al. Modeling the impact of treatment and screening on U.S. breast cancer mortality: A Bayesian approach. J Natl Cancer Inst Monogr. 2006;36:30–36. doi: 10.1093/jncimonographs/lgj006. [DOI] [PubMed] [Google Scholar]
- 15.Dowsett M, et al. Meta-analysis of breast cancer outcomes in adjuvant trials of aromatase inhibitors versus tamoxifen. J Clin Oncol. 2010;28:509–518. doi: 10.1200/JCO.2009.23.1274. [DOI] [PubMed] [Google Scholar]
- 16.Osborne CK, Coronado EB, Robinson JP. Human breast cancer in the athymic nude mouse: Cytostatic effects of long-term antiestrogen therapy. Eur J Cancer Clin Oncol. 1987;23:1189–1196. doi: 10.1016/0277-5379(87)90154-4. [DOI] [PubMed] [Google Scholar]
- 17.Gottardis MM, Jordan VC. Development of tamoxifen-stimulated growth of MCF-7 tumors in athymic mice after long-term antiestrogen administration. Cancer Res. 1988;48:5183–5187. [PubMed] [Google Scholar]
- 18.Wolf DM, Jordan VC. A laboratory model to explain the survival advantage observed in patients taking adjuvant tamoxifen therapy. Recent Results Cancer Res. 1993;127:23–33. doi: 10.1007/978-3-642-84745-5_4. [DOI] [PubMed] [Google Scholar]
- 19.Yao K, et al. Antitumor action of physiological estradiol on tamoxifen-stimulated breast tumors grown in athymic mice. Clin Cancer Res. 2000;6:2028–2036. [PubMed] [Google Scholar]
- 20.Jordan VC. Selective estrogen receptor modulation: Concept and consequences in cancer. Cancer Cell. 2004;5:207–213. doi: 10.1016/s1535-6108(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 21.Katzenellenbogen BS, Kendra KL, Norman MJ, Berthois Y. Proliferation, hormonal responsiveness, and estrogen receptor content of MCF-7 human breast cancer cells grown in the short-term and long-term absence of estrogens. Cancer Res. 1987;47:4355–4360. [PubMed] [Google Scholar]
- 22.Welshons WV, Jordan VC. Adaptation of estrogen-dependent MCF-7 cells to low estrogen (phenol red-free) culture. Eur J Cancer Clin Oncol. 1987;23:1935–1939. doi: 10.1016/0277-5379(87)90062-9. [DOI] [PubMed] [Google Scholar]
- 23.Song RX, et al. Effect of long-term estrogen deprivation on apoptotic responses of breast cancer cells to 17beta-estradiol. J Natl Cancer Inst. 2001;93:1714–1723. doi: 10.1093/jnci/93.22.1714. [DOI] [PubMed] [Google Scholar]
- 24.Lewis JS, et al. Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst. 2005;97:1746–1759. doi: 10.1093/jnci/dji400. [DOI] [PubMed] [Google Scholar]
- 25.Lewis-Wambi JS, et al. Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis. Breast Cancer Res. 2008;10:R104. doi: 10.1186/bcr2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lønning PE, et al. High-dose estrogen treatment in postmenopausal breast cancer patients heavily exposed to endocrine therapy. Breast Cancer Res Treat. 2001;67:111–116. doi: 10.1023/a:1010619225209. [DOI] [PubMed] [Google Scholar]
- 27.Lønning PE. Additive endocrine therapy for advanced breast cancer—back to the future. Acta Oncol. 2009;48:1092–1101. doi: 10.3109/02841860903117816. [DOI] [PubMed] [Google Scholar]
- 28.Ellis MJ, et al. Lower-dose vs high-dose oral estradiol therapy of hormone receptor-positive, aromatase inhibitor-resistant advanced breast cancer: A phase 2 randomized study. JAMA. 2009;302:774–780. doi: 10.1001/jama.2009.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Mol Cell Endocrinol. 1992;90:77–86. doi: 10.1016/0303-7207(92)90104-e. [DOI] [PubMed] [Google Scholar]
- 30.Pink JJ, Jiang SY, Fritsch M, Jordan VC. An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer Res. 1995;55:2583–2590. [PubMed] [Google Scholar]
- 31.Pink JJ, Jordan VC. Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res. 1996;56:2321–2330. [PubMed] [Google Scholar]
- 32.Lewis JS, Osipo C, Meeke K, Jordan VC. Estrogen-induced apoptosis in a breast cancer model resistant to long-term estrogen withdrawal. J Steroid Biochem Mol Biol. 2005;94:131–141. doi: 10.1016/j.jsbmb.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 33.Lewis-Wambi JS, Cunliffe HE, Kim HR, Willis AL, Jordan VC. Overexpression of CEACAM6 promotes migration and invasion of oestrogen-deprived breast cancer cells. Eur J Cancer. 2008;44:1770–1779. doi: 10.1016/j.ejca.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu ZZ, et al. Proteomic analysis of pathways involved in estrogen-induced growth and apoptosis of breast cancer cells. PLoS One. 2011;6:e20410. doi: 10.1371/journal.pone.0020410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levenson AS, Jordan VC. MCF-7: The first hormone-responsive breast cancer cell line. Cancer Res. 1997;57:3071–3078. [PubMed] [Google Scholar]
- 36.Liu H, et al. Apoptotic action of 17beta-estradiol in raloxifene-resistant MCF-7 cells in vitro and in vivo. J Natl Cancer Inst. 2003;95:1586–1597. doi: 10.1093/jnci/djg080. [DOI] [PubMed] [Google Scholar]
- 37.Chisamore MJ, Ahmed Y, Bentrem DJ, Jordan VC, Tonetti DA. Novel antitumor effect of estradiol in athymic mice injected with a T47D breast cancer cell line overexpressing protein kinase Calpha. Clin Cancer Res. 2001;7:3156–3165. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.