Abstract
Background
Patients with severe congenital neutropenia (SCN) often develop periodontitis despite standard medical and dental care. In light of previous findings that mutations in the neutrophil elastase gene, ELANE, are associated with more severe neutropenic phenotypes, we hypothesized an association between the genotype of SCN and development of periodontitis.
Methods
Fourteen Swedish patients with SCN or cyclic neutropenia harboring different genetic backgrounds were recruited for periodontal examination. Peripheral blood, gingival crevicular fluid (GCF), and subgingival bacterial samples were collected. The levels of cytokines and antibacterial peptides were determined in GCF and plasma by multiplex immunoassay and immunoblotting, respectively. Subgingival bacterial samples were analyzed using 16S rDNA pyrosequencing.
Results
ELANE mutations correlated with more severe periodontal status than the HAX1 or unknown mutations in patients with SCN. The subjects with mutant ELANE had higher levels of IL-1β in GCF. Using principal coordinate analysis of the subgingival microbiota, patients with ELANE mutations and reference subjects with periodontitis tended to cluster differently from patients with HAX1 or unknown mutations and non-periodontitis reference subjects.
Conclusion
This study demonstrates an association between ELANE mutations in SCN and the development of periodontitis with skewed subgingival microbiota, indicating a potential role of ELANE mutations in the pathogenesis of periodontitis.
Keywords: Kostmann neutropenia, neutrophil elastase 2, periodontal diseases, subgingival microflora
Introduction
Severe congenital neutropenia (SCN, also known as Kostmann disease) includes a heterogeneous group of disorders characterized by chronic low absolute neutrophil counts (ANC) (below 0.5 × 109/l) in the peripheral blood, early onset of bacterial infections, and mostly a maturation arrest of the myelopoiesis in the bone marrow at the level of promyelocyte/myelocyte stage [1–3]. Recent studies have revealed that a number of inherited gene mutations may cause SCN [4]. Heterozygous mutations in the ELANE gene (formerly named ELA2), encoding the neutrophil primary granule protease, neutrophil elastase, were demonstrated in approximately 50–60% of patients with SCN [5, 6], whereas homozygous mutations in the HAX1 gene, encoding the mitochondrial antiapoptotic protein HS1-associating protein X-1 (HAX-1), were identified in about 15% of patients [3]. In addition, around one third of the patients with SCN is still uncharacterized by any gene mutation. Cyclic neutropenia (CyN) is another hereditary form of severe chronic neutropenia in which the neutrophil count oscillates and patients present less severe clinical symptoms compared to SCN. In the majority of cases with CyN, ELANE mutations were determined to be responsible for the disease [7].
Patients with SCN or CyN currently receive recombinant human granulocyte colony-stimulating factor (G-CSF) therapy and more than 90% of patients respond to this treatment with increased peripheral neutrophil level, diminished vulnerability to bacterial infections and much improved quality of life [8]. However, there are still patients who exhibit unsatisfactory periodontal health despite having G-CSF-normalized neutrophil levels and receiving regular professional dental care [9–11].
The pathogenesis of gingivitis and periodontitis is multifactorial and includes complex interactions between oral microbes and host defense [12]. Neutrophils are key immune cells for oral health and neutrophil deficiency or dysfunction often results in periodontal disease [13]. Besides low levels of ANC patients with SCN also exhibit deficiencies in neutrophil granule-associated proteins, including the antimicrobial peptides pro-LL-37 (or hCAP-18) with its active peptide LL-37, and human neutrophil peptides 1–3 (HNP1–3) [14]. The lack of LL-37 and/or HNP suggests that these neutrophils are functionally deficient with respect to their antimicrobial capacity. Such deficiency in periodontal neutrophils may influence the subgingival microbiota composition in the periodontal pocket, and as a consequence, contribute to the pathogenesis of periodontal breakdown.
Although it has long been recognized that patients with SCN or CyN often suffer from early onset of severe periodontitis [15–19], the correlation between genotype and phenotype in terms of gene mutations in SCN and periodontal health is still unclear. Previous studies have demonstrated that ELANE mutations correlate with more severe disease manifestation in patients with SCN [20], and that patients with ELANE mutations require higher doses of G-CSF compared to patients with HAX1 mutations [3]. In light of these findings, we hereby address the hypothesis that ELANE gene mutations are associated with the occurrence of periodontitis in subjects with SCN. The underlying parameters that are believed to contribute to periodontitis were studied, including subgingival microbiota composition, proinflammatory cytokines, as well as innate immune components HNP1–3 and pro-LL-37/LL-37.
Materials and Methods
Participants
From 2006 to 2008, patients with SCN (n = 13) or CyN (n = 1) were recruited from Karolinska University Hospital, Sweden and numbered periodontitis–neutropenia (PN) 1-to-14 according to recruitment date. The subjects ranged in age from 6 to 50 years with various forms of SCN or CyN. Ethical permission was granted by the local ethical committee at Karolinska University Hospital (2006/176-31/4). All subjects or their parents provided informed consent before participating in this study.
Clinical Examination
The clinical examination involved recording visible plaque index (%), bleeding on probing (BOP, %), probing depth (mm), and radiographs which were taken in order to determine the occurrence of alveolar bone loss. The distance between enamel cement junction and marginal bone (mm) was measured on the radiographs and alveolar bone loss was diagnosed when the distance exceeded 3.0 mm. Based on the clinical examination, the patients were categorized as either being healthy, suffering from gingivitis or periodontitis, or edentulous, respectively. Gingivitis was diagnosed when BOP exceeded 25%, while periodontitis was diagnosed when the patient exhibited both alveolar bone loss for more than three teeth and periodontal pockets exceeding 4 mm for the same teeth.
Plasma, GCF, and Subgingival Bacteria Sampling
Peripheral blood was collected and coagulation was inhibited using EDTA. Following centrifugation, plasma was gathered from the top layer and subsequently stored at −80°C in aliquots.
For each subject, GCF was collected from the mesial surface of an incisor or for PN2 from a deciduous molar by inserting a paper strip (PerioPaper, Oralflow Inc.) into the gingival sulcus for 15 s. The strip was then analyzed using a Periotron Model 8000 (Oralflow Inc.), and the volume was calculated by interpolation from a standard curve. The two edentulous patients (PN4 and PN9) did not provide GCF samples. Individual strips were then placed into a sterile tube containing 120 μl PBS buffer (pH = 6.8), 0.01 M EDTA, 0.3% bovine globulin, 0.005% Triton X-100, and 0.05% sodium azide. The samples were then stored at −80°C.
Subgingival bacteria samples were collected using a paper strip from the distal surface of an incisor or from a deciduous molar for PN2. Since there was lack of data and references in the literature regarding the subgingival microbiota assessed using 454 pyrosequencing, we collected subgingival bacterial samples from nine systemically healthy individuals aged from 5 to 19 years, with three samples from sites of periodontitis and six from healthy sites or those of gingivitis, in order to provide references for samples from neutropenic cases in the 454 analysis. After collection, all samples were stored at −80°C until analysis.
Luminex Cytokine Immunoassay
Plasma and GCF samples were analyzed for IL-1β, IL-4, IL-6, IL-17, IFN-γ, and TNF-α concentrations using fluorescent bead-based Luminex cytokine immunoassays, which were performed using the Bio-Plex system (Bio-Rad laboratories). Samples were thawed on ice and homogenized in a vortex mixer for 1 min before analysis. The cytokine concentrations were determined using a human cytokine LINCOplex kit (Millipore) according to the manufacturer’s instructions and were expressed as ng/ml in GCF and pg/ml in plasma.
Gel Electrophoresis and Immunoblotting
Plasma and GCF samples were analyzed for pro-LL-37 and mature LL-37 peptide content using Western blotting. GCF samples were further tested for HNP1–3 using the same method. The GCF samples were treated with 60% acetonitrile containing 1% trifluoroacetic acid for 2 h on a shaker at 4°C to extract small peptides from the periopaper. Following centrifugation, the extraction supernatant was then transferred into a sterile tube, kept at −80°C, and lyophilized until dry. The GCF extract and plasma were dissolved in NuPAGE SDS sample buffer (Invitrogen) and electrophoresed in 1.0 mm 4–12% NuPAGE Bis–Tris gels (Invitrogen) under reducing conditions. Immunoblotting was performed as previously described [21] using the following antibodies: rabbit anti-LL-37 (Innovagen, Sweden), mouse anti-alpha defensin 1+2+3 antibody (Abcam), goat anti-rabbit, and goat anti-mouse immunoglobulins (Dako, Denmark). Detection was carried out using chemiluminescence (SuperSignal West Pico, Pierce).
454 Pyrosequencing
The microbiota of subgingival bacterial samples from the patients and reference individuals was analyzed using a 454 FLX pyrosequencing facility according to previously described methods with minor modifications [22, 23]. Briefly, DNA extraction was performed using DNeasy Blood and Tissue kit (Qiagen) with proteinase K treatment at 56°C for 16 h. For each extracted DNA sample, three 50 μl PCR mixes were prepared containing 1× PCR buffer, 200 μM dNTP PurePeak DNA Polymerization Mix (Pierce Nucleic Acid Technologies), 0.5 μM of each primer, 0.5 U Phusion F-530L enzyme (Finnzyme), and 2 μl template-DNA. The primer pairs, amplifying the hypervariable 16s rRNA gene V3-V4 regions, were: 341f (5′ CCTACGGGNGGCWGCAG) with adaptor B and 805r (5′ GACTACHVGGGTATCTAATCC) with adaptor A and a sample-specific sequence barcode. The PCR conditions were 95°C for 5 min, 26 cycles of 95°C for 40 s, 58°C for 40 s, and 72°C for 1 min, followed by 72°C for 7 min. A PCR reaction without template was also used as a control for each primer pair. After analyses in agarose gel (1% w/v in TBE buffer), the samples with the same barcode were pooled and PCR reactions were purified using an Agencourt AMPure system (Beckman Coulter Genomics). The DNA concentrations were measured using Qubit (Invitrogen), and the quality control was performed with a Bioanalyzer 2100 using the DNA 1000 chip (Agilent Technologies). The samples were diluted to 3 ng/μl, and 5 μl of each sample was pooled. Region V4 was sequenced using 454 pyrosequencing with a standard amplicon kit and run in the 454-FLX (Roche, Switzerland) [24].
Sequences were excluded if there was no perfect match with the primer or barcode, ambiguous nucleotides, or the sequence was shorter than 200 nucleotides excluding the primer/barcode. Non-redundant reads with the primer/barcode removed were aligned and sorted into operational taxonomic units (OTU) using complete linkage clustering and a 3% distance threshold, which was performed using the Pyrosequencing Pipeline at Ribosomal Database Project (RDP) [25]. 16S rRNA gene sequences from RDP 10.22 were converted into a local BLAST database. The OTUs were BLAST searched against the database with a 95% identity threshold over at least 180 nucleotides. Different OTU hits were sorted to the taxonomic level for further analysis.
The different sequence identification levels were analyzed and visualized with regards to relative abundance as a heat map using MultiEperiment Viewer v4.6 software [26]. Principal coordinate analysis (PCoA) was performed and visualized in Fast Unifrac (http://bmf.colorado.edu/fastunifrac/) [27] using normalized weighted abundance. The Shannon diversity index was calculated using the R package vegan (http://CRAN.R-project.org/package=vegan) for each sample, and the significance was tested using the Wilcoxon rank sum test.
Results
Clinical Findings
The medical history of the patients (n = 14) is presented in Table I. All patients except PN14 were diagnosed before 1 1/2 years of age. Of all the patients with SCN, six exhibited ELANE mutations, four HAX1 mutations, and three unknown mutation(s). Two patients (PN8 and PN9) had received HSCT before they were recruited into the study, and three patients (PN7, PN8, and PN9) had not received G-CSF treatment by the time the clinical examinations were performed.
Table I.
Medical background of the patients with congenital neutropenia
| Subjects | Age | Sex | Diagnosis | Gene mutation | Age at diagnosis | ANCc (×109/L) | G-CSF/Durationd/Dose (μg/kg/day) | Other important clinical findings | |
|---|---|---|---|---|---|---|---|---|---|
| ELANE | HAX1 | ||||||||
| PN1 | 14 | M | SCN | D89H | wt | 4 months | 2.1 | Yes/Since diagnosis/7.8 | Bronchial asthma |
| PN2 | 6 | M | SCN | C122S | wt | 1 week | 0.9 | Yes/Since diagnosis/25 | G-CSF resistant; T-S prophylaxis |
| PN3 | 17 | M | SCN | C26S | wt | 1 week | 0.5 | Yes/Since 1991/5.0 | T-S prophylaxis |
| PN4 | 50 | F | SCN | C26S | wt | 3 weeks | 0.5 | Yes/Since 1991/2.5 | |
| PN5 | 19 | F | SCN | C26S | wt | 3 months | 0.2 | Yes/Since 1991/4.5 | Atopic dermatitis |
| PN6 | 17 | M | SCN | wt | W44X | 1 1/2 years | 1.5 | Yes/Since diagnosis/4.3 | Ureter reflux and kidney infection |
| PN7 | 14 | M | SCN | wt | Q190X and E31KfsX54b | 2 months | 1.0 | No | |
| PN8 | 23 | M | SCN (HSCTa) | wt | Q190X | 2 months | 1.7 | No | HSCT at 7 months of age; Epilepsy; ADHD |
| PN9 | 31 | F | SCN (HSCTa) | L92H | wt | 2 weeks | 3.1 | No | Developed G-CSFR mutations; HSCT 2004 |
| PN10 | 21 | F | CyN | wt | wt | 11 months | 9.2 | Sporadically | |
| PN11 | 19 | F | SCN | wt | Q190X | 13 months | 7.0 | Yes/Since 1991/3.0 | Developed G-CSFR mutations; Epilepsy |
| PN12 | 8 | M | SCN | wt | wt | 14 months | 1.6 | Yes/Since diagnosis/9.0 | Motor proficiency difficulties |
| PN13 | 17 | M | SCN | wt | wt | 1 year | 5.7 | Yes/Since diagnosis/4.2 | Special school; Splenomegaly |
| PN14 | 27 | F | SCN | wt | wt | 5 years | 1.2 | Yes/Since 1991/1.5 | |
PN periodontitis–neutropenia, wt wildtype, T-S trimethoprim–sulfamethoxazole, G-CSFR G-CSF receptor, ADHD attention deficit hyperactivity disorder
aHSCT before participating in this study. PN8 had mixed chimerism and had been treated with low dose (0.5 μg/kg/day) of G-CSF until adult age; PN9 had 100% donor and normal ANC
bCompound heterozygous HAX1 (cHAX1) mutations [61]
cReference range is 2.0–8.0
dG-CSF treatment started in Sweden in 1991
The periodontal conditions of the patients are described in Table II. Except for PN4 and PN9 who were edentulous and PN11 who underwent professional dental care 1 year previously, the other patients had had a last dental visit between 2-to-6 months prior to the clinical examination. Their toothbrushing habits were at least once per day. Of all the patients with SCN, four were classified as being periodontally healthy, two as having gingivitis, five as having periodontitis, and two subjects as edentulous due to periodontitis at an early age. Of the six patients with ELANE mutations, five were diagnosed with periodontitis or edentulous. Conversely, within SCN cases, the subjects with HAX1 or unknown mutations were mostly (six out of seven) classified as being healthy or having gingivitis. ELANE mutations are significantly correlated with the occurrence of periodontitis/edentulism compared to HAX1 or unknown mutations (P = 0.025; Table III). The single patient with CyN displayed a healthy periodontium.
Table II.
Periodontal status of the patients with congenital neutropenia
| Subjects | Gene mutation | Full mouth | Periodontal status | Tested site | |||||
|---|---|---|---|---|---|---|---|---|---|
| ELANE | HAX1 | VPI (%) | BOP (%) | PD > 4 mm | Tooth | PD (mm) | ECJ-MB (mm) | ||
| PN1 | D89H | wt | >50 | >50 | No | Gingivitis | 41, 31 | 3 | 2 |
| PN2 | C122S | wt | >50 | >50 | Yes | Periodontitis | 55, 85 | 4, 3 | 4, 3 |
| PN3 | C26S | wt | >50 | >50 | Yes | Periodontitis | 41, 31 | 6 | 9 |
| PN4 | C26S | wt | – | – | – | Edentulous | – | – | – |
| PN5 | C26S | wt | >50 | >50 | Yes | Periodontitis | 41, 31 | 6 | 5 |
| PN6 | wt | W44X | >50 | >50 | Yes | Periodontitis | 41, 31 | 4 | 3.5 |
| PN7 | wt | Q190X and E31KfsX54 | <15 | 0 | No | Healthy | 41, 31 | 2 | 2 |
| PN8 | wt | Q190X | <25 | <15 | No | Healthy | 41, 31 | 2 | 1.5 |
| PN9 | L92H | wt | – | – | – | Edentulous | – | – | – |
| PN10 | wt | wt | <15 | <15 | No | Healthy | 41, 31 | 2 | 2 |
| PN11 | wt | Q190X | 0 | 0 | No | Healthy | 41, 31 | 2 | 1.2 |
| PN12 | wt | wt | <15 | <15 | No | Healthy | 41, 31 | 2 | 2 |
| PN13 | wt | wt | >50 | >50 | Yes | Periodontitis | 41, 31 | 4 | 3.5 |
| PN14 | wt | wt | <25 | >25 | Yes | Gingivitis | 41, 31 | 3 | 2.5 |
PN periodontitis–neutropenia, wt wildtype, VPI visible plaque index, BOP bleeding on probing, PD probing depth, ECJ-MB enamel cement junction–marginal bone
Subgingival microbiota analysis from tooth 41, or from tooth 55 in PN2; cytokine and AMP analysis from tooth 31, or from tooth 85 in PN2
Table III.
Periodontal status of patients with SCN harboring different genetic mutations
| Variables | ELANE mutations (n = 6) | HAX1 or unknown mutations (n = 7) | P value |
|---|---|---|---|
| Receiving G-CSF | 5 | 5 | |
| Periodontal status | |||
| Healthy | 0 | 4 | |
| Gingivitis | 1 | 1 | |
| Periodontitis/edentulousa | 5 | 2 | 0.025b |
aDue to periodontitis
bSpearman correlation test
Antimicrobial Peptide and Cytokine Levels in GCF and Plasma
Subjects who had received HSCT and the edentulous patients (n = 3) were excluded from the GCF analysis. HSCT-transplanted patients (n = 2) were excluded from the plasma analysis.
Neither ANC (P = 0.116), plasma pro-LL-37 (P = 0.106), GCF α-defensin (P = 0.703), or GCF pro-LL-37/LL-37 (P = 0.450) levels were significantly different in patients with ELANE mutations compared to HAX1 or unknown mutations. However, an interesting observation was that patients with unknown gene etiology exhibited low levels of both HNP1–3 and LL-37, maybe reflecting a similar gene defect. The LL-37 levels in all but one patient with SCN were 10-to-1,000-fold lower than in those two patients (PN8 and PN10) that exhibited normal ANC levels without G-CSF treatment at the time of sampling (Tables I and IV).
Table IV.
Antimicrobial peptides in GCF and plasma from patientsa
| Diagnosis | Gene mutation | Subjects | Periodontal status | GCF HNP1–3 (μg/ml) | GCF pro-LL-37 and LL-37c | Plasma pro-LL-37 c, d |
|---|---|---|---|---|---|---|
| SCN | ELANE | PN1 | Gingivitis | 192 | 0.25 | 0.01 |
| PN2 | Periodontitis | 428 | 0.82 | <0.01 | ||
| PN3 | Periodontitis | <4 | 10.56 | <0.01 | ||
| PN5 | Periodontitis | <4 | 0.05 | <0.01 | ||
| HAX1 | PN6 | Periodontitis | 40 | 6.21 | 0.03 | |
| PN7 | Healthy | <4 | 6.93 | <0.01 | ||
| PN11 | Healthy | 247 | 78.18 | 0.17 | ||
| unknown | PN12 | Healthy | 5 | 0.08 | 0.06 | |
| PN13 | Periodontitis | 4 | 0.38 | 0.10 | ||
| PN14 | Gingivitis | 4 | 0.13 | 0.09 | ||
| SCN (HSCTb) | HAX1 | PN8 | Healthy | 840 | 324.0 | 0.31 |
| CyN | unknown | PN10 | Healthy | 227 | 132.8 | 0.54 |
PN periodontitis–neutropenia
aTwo edentulous patients were excluded
bHSCT before participating in this study
cRelative to reference plasma
dRange in healthy individuals is 0.35–1.5
In addition, the level of IL-1β (P = 0.038) in GCF was significantly higher in the ELANE mutation group compared to HAX1 or unknown mutation group (Table V). Although the levels of IL-17, IL-6, and TNF-α were not statistically different in the GCF, it may be noted that mean and median values in ELANE mutation group are higher than in HAX1 or unknown mutation group. The GCF cytokine levels of IFN-γ and IL-4 were both below the detection limit in all samples. Moreover, there was no significant difference in terms of the cytokine levels in plasma between the two subgroups of patients.
Table V.
Cytokines in GCF from patients with SCN harboring different genetic mutations
| Cytokines (ng/ml) | ELANE mutations (n = 5) | HAX1 or unknown mutations (n = 6) | P valuea | ||
|---|---|---|---|---|---|
| Median | Mean ± SD | Median | Mean ± SD | ||
| IL-1β | 9.3 | 13.1 ± 9.3 | 2.9 | 5.1 ± 6.8 | 0.038 |
| IL-17 | 0.5 | 0.5 ± 0.4 | 0.1 | 0.2 ± 0.1 | 0.131 |
| IL-6 | 3.0 | 3.8 ± 3.8 | 0.8 | 0.8 ± 0.5 | 0.186 |
| TNF-α | 0.5 | 0.5 ± 0.4 | 0.01 | 0.03 ± 0.05 | 0.089 |
aMann–Whitney test
Subgingival Microflora
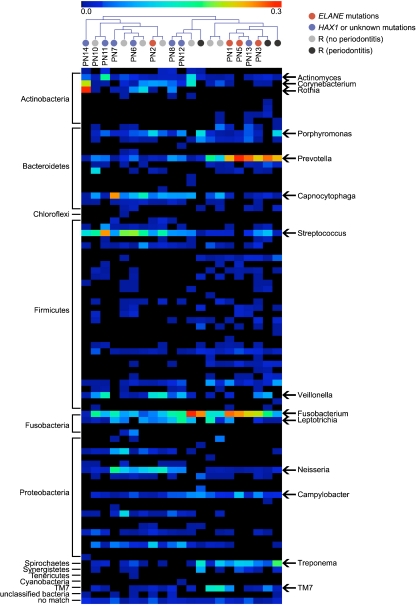
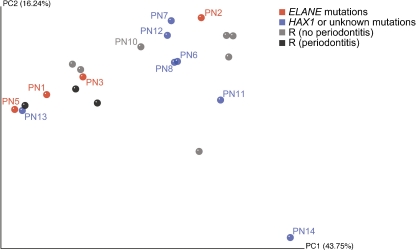
We generated 8,810 high-quality 16S rRNA gene sequences from 12 subgingival bacterial samples (PN4 and PN10 were excluded due to edentulism). The sequences represented seven major phyla: Actinobacteria (19.5%), Bacteroidetes (19.5%), Firmicutes (22.2%), Fusobacteria (16.6%), Proteobacteria (15.9%), Spirochaetes (2.0%), and candidate domain TM7 (1.3%). The predominant genera were Fusobacterium, Streptococcus, Rothia, Prevotella, Corynebacterium, Neisseria, Veillonella, Capnocytophaga, and Leptotrichia, which in total accounted for 66% of all sequences. The Shannon diversity index was similar in patients with ELANE mutations and HAX1 or unknown mutations according to Wilcoxon rank sum test (P = 0.788). To illustrate subgingival microbiota composition of individuals, a heat map was generated to reveal the abundance of different taxa in samples from both patients with neutropenia and references (Fig. 1). A sample tree was also created using hierarchical clustering using Pearson correlation of absolute distance and complete linkage clustering (Fig. 1). In both hierarchical clustering and UniFrac PCoA (Fig. 2), three out of four samples from patients with ELANE mutations were clustered together with all reference samples of periodontitis.
Fig. 2.
Principal coordinate analysis (PCoA) of microbiota in individual subjects using 1% distance threshold with normalized weighted abundance. The scatterplots are for the first two principal components (PC1 and PC2); each point represents a sample. PN8 is stem cell transplanted. R represents reference samples and patient PN10 (CyN)
Fig. 1.
Individual subgingival microbiota composition. The heat map depicts the abundance of the different taxa in individual subgingival samples. The color scale of relative abundance ranges from 0% (black) to 30% (red). The phylum level is shown on the left-hand side. The dominant taxa (genus level, except TM7) are displayed according to the relative abundance per sample. The sample tree was generated using hierarchical clustering with Pearson correlation of absolute distance and complete linkage clustering. PN8 is stem cell transplanted. R represents reference samples and patient PN10 (CyN)
Discussion
Although the prognosis and quality of life of patients with congenital neutropenia were improved dramatically following the introduction of G-CSF therapy 20 years ago [28], some patients still suffer from frequent periodontal infections despite efficient and adequate oral hygiene. In the current study, we demonstrate for the first time a link between mutations affecting ELANE encoding neutrophil elastase and the occurrence of periodontitis.
The periodontal condition of the patients in the present study varied from healthy to severe periodontitis and two patients were edentulous due to periodontitis, indicating great variation of the chronic inflammatory response in the periodontium among patients with congenital neutropenia. Recent genetic studies have identified multiple gene mutations in SCN, with the most common mutations affecting the ELANE (ELA2) gene. To date, more than 45 distinct ELANE mutations have been described in SCN and CyN [29]. Our current findings demonstrate a correlation between ELANE mutations and periodontitis in patients with SCN, concurring with the view that ELANE mutations are correlated with more severe disease manifestations and having a relatively poorer response to G-CSF treatment [3, 20]. However, the number of patients in our study is limited since SCN is a rare disease. Further investigation involving a larger cohort will be needed to confirm our findings.
Many of the patients with ELANE mutations had low ANC and therefore may be expected to have more severe periodontal disease. However, it is both neutrophil functionality and neutrophil counts over time that can affect the outcome of periodontal disease. Moreover, factors such as oral hygiene and diet may influence the outcome of oral health, which we have attempted to account for by using questionnaires for patients and their dentists.
The antimicrobial peptides HNP1–3 and LL-37 are produced during neutrophil maturation in the bone marrow and are stored as pro-peptides in neutrophil granules. The pro-LL-37 (hCAP18) is also detectable in plasma from healthy individuals [30]. We previously demonstrated that plasma pro-LL-37 levels were low in patients with SCN in spite of G-CSF elevated neutrophil levels, thus reflecting impaired neutrophil development [21, 31]. In the current study, pro-LL-37 levels in plasma were low in all patients with SCN and did not differ between the ELANE mutation group compared to HAX1 or unknown mutation group.
In GCF, elevated levels of both HNP1–3 and LL-37 have been reported in subjects with chronic periodontitis, most likely due to enhanced neutrophil influx [32]. In our study, the GCF levels of HNP1–3 and LL-37 did not appear to be statistically different between patients harboring different mutations, possibly due to the wide range of values in both groups. The α-defensins HNP1–3 are stored in primary granules in neutrophils, and the GCF levels of HNP1–3 have been reported to vary widely in healthy subjects [33, 34]. The considerable difference of GCF HNP1–3 in the present study is compatible with our previous observation that HNP1–3 levels vary from deficiency to normal levels in neutrophils from patients with SCN [14]. To date, gingival LL-37 has been demonstrated to have two main sites of origin, neutrophils and epithelial cells [35]. In the absence of neutrophil-derived LL-37 in the GCF of patients with SCN, it was possible to determine the epithelial contribution to LL-37 levels, which was found to be noticeably low. LL-37 levels below bactericidal concentrations have been demonstrated to serve as a chemoattractant or modulator of host inflammatory responses in concert with other epithelial-derived cytokines [36–38]. Thus, in the absence of efficient neutrophil antibacterial clearance due to deficiency of neutrophil granule peptides, epithelial-derived peptides might even augment periodontal inflammation.
In the current study, we demonstrated that IL-1β levels were significantly higher in GCF samples from subjects with mutant ELANE. In addition, the mean and median levels of IL-17, IL-6, and TNF-α in patients with ELANE mutations were higher than in HAX1 or unknown mutations although these differences did not reach statistical significance, most likely due to the small size of the cohort and great variations within groups. It is known that elevated levels of IL-1β [39, 40], IL-6 [41], TNF-α [42], and IL-17 [43] in GCF are associated with severe periodontal disease, and that these cytokines may also be elevated in chronic periodontitis tissue [44–46]. Thus, the GCF from patients with ELANE mutations displays the presence of the strong proinflammatory cytokine IL-1β, which might be expected in the inflamed periodontium.
Oral microbiota in the healthy population has been determined using high-throughput 16S rDNA pyrosequencing in several studies [47–49], providing a rather comprehensive view of the oral commensal microbial community. To our knowledge, this is the first time that the 16S rDNA pyrosequencing technique has been employed to map subgingival microbiota in subjects with a congenital immunodeficiency. The predominant taxa from periodontal sites of individuals with SCN were similar to the microbiota previously reported from healthy subjects [49]. Although two patients with ELANE mutations were not included in microbiota analyses due to edentulism, hierarchical clustering and UniFrac PCoA analysis revealed that three out of four samples from the ELANE mutation group were clustered with the periodontitis reference individuals. The skewed periodontal microbiota in both periodontitis references and the ELANE mutation group of SCN cases indicates that periodontal pathogens of the genera Fusobacterium, Prevotella, Treponema, and TM7 domain are more likely to grow in the gingival crevices in patients with ELANE mutations compared to HAX1 or unknown mutations [50]. As an outlier, PN2 did not cluster with other individuals with ELANE mutations that may partly be explained by the fact that periodontal pathogens of young children may differ from those of adolescent or adults [51, 52].
The neutropenia of patients with SCN arises as a consequence of bone marrow neutrophil precursor accelerated apoptosis [53]. The two most frequently reported gene mutations of SCN are HAX1 and ELANE [54]. HAX-1 is a mitochondrial protein involved in maintaining the mitochondrial membrane potential, signal transduction, and cell survival [55, 56]. The HAX1 mutations in SCN result in HAX-1-deficient neutrophils and neutrophil precursors, which have been demonstrated to show enhanced apoptosis [57]. The mechanism by which ELANE mutations result in apoptosis of neutrophil precursors is less obvious, but it has been suggested that the accumulation of misfolded elastase proteins activates the unfolded protein response, leading to apoptosis [58, 59]. Although rescued by G-CSF treatment, the neutrophils with ELANE mutations still carry mutated elastase proteins that may be aberrant in their localization and functions such as proteolytic processing of other proenzymes or cytokines, which in turn may affect the local periodontal immune response [60]. Thus, it is possible that in addition to the antimicrobial peptide deficiency that is shared by all patients with SCN, the ELANE mutations may confer an additional neutrophil dysfunctionality, leading to more severe periodontal disease.
Conclusion
We herein report that patients with SCN that harbor mutations in the gene ELANE coding for neutrophil elastase present with more severe periodontal disease as compared to patients with HAX1 mutations or with unknown mutations. The periodontal pockets of the former group display a skewed microflora and elevated levels of proinflammatory cytokine IL-1β. The correlation between ELANE mutations and periodontal disease indicates that the serine protease elastase may have an important role in the local defense in the gingival pocket.
Acknowledgments
The authors sincerely thank the patients and their families for their cooperation. The study was supported by grants from Swedish Children’s Cancer Foundation (G.C.), Söderbergs Foundation (L.E.), Swedish Research Council (K.P., M.A.), Åke Wiberg Foundation (K.P.), Swedish Society for Medicine (K.P.), and Swedish Patent Revenue Research Fund (T.M.).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Thomas Modéer and Katrin Pütsep have contributed equally to the study.
References
- 1.Kostmann R. Infantile genetic agranulocytosis (Agranulocytosis infantilis hereditaria): a new recessive lethal disease in man. Acta Paediatr Suppl. 1956;45:1–78. doi: 10.1111/j.1651-2227.1956.tb06875.x. [DOI] [PubMed] [Google Scholar]
- 2.Kostman R. Infantile genetic agranulocytosis. A review with presentation of ten new cases. Acta Paediatr Scand. 1975;64:362–8. doi: 10.1111/j.1651-2227.1975.tb03847.x. [DOI] [PubMed] [Google Scholar]
- 3.Zeidler C, Germeshausen M, Klein C, Welte K. Clinical implications of ELA2-, HAX1-, and G-CSF-receptor (CSF3R) mutations in severe congenital neutropenia. Br J Haematol. 2009;144:459–67. doi: 10.1111/j.1365-2141.2008.07425.x. [DOI] [PubMed] [Google Scholar]
- 4.Boztug K, Klein C. Novel genetic etiologies of severe congenital neutropenia. Curr Opin Immunol. 2009;21:472–80. doi: 10.1016/j.coi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Dale DC, Bolyard AA, Schwinzer BG, Pracht G, Bonilla MA, Boxer L, Freedman MH, Donadieu J, Kannourakis G, Alter BP, Cham BP, Winkelstein J, Kinsey SE, Zeidler C, Welte K. The Severe Chronic Neutropenia International Registry: 10-year follow-up report. Support Cancer Ther. 2006;3:220–31. doi: 10.3816/SCT.2006.n.020. [DOI] [PubMed] [Google Scholar]
- 6.Xia J, Bolyard AA, Rodger E, Stein S, Aprikyan AA, Dale DC, Link DC. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol. 2009;147:535–42. doi: 10.1111/j.1365-2141.2009.07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23:433–6. doi: 10.1038/70544. [DOI] [PubMed] [Google Scholar]
- 8.Dale DC, Bonilla MA, Davis MW, Nakanishi AM, Hammond WP, Kurtzberg J, Wang W, Jakubowski A, Winton E, Lalezari P. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (filgrastim) for treatment of severe chronic neutropenia. Blood. 1993;81:2496–502. [PMC free article] [PubMed] [Google Scholar]
- 9.Carlsson G, Wahlin YB, Johansson A, Olsson A, Eriksson T, Claesson R, Hanstrom L, Henter JI. Periodontal disease in patients from the original Kostmann family with severe congenital neutropenia. J Periodontol. 2006;77:744–51. doi: 10.1902/jop.2006.050191. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson G, Fasth A. Infantile genetic agranulocytosis, morbus Kostmann: presentation of six cases from the original “Kostmann family” and a review. Acta Paediatr. 2001;90:757–64. doi: 10.1080/080352501750315663. [DOI] [PubMed] [Google Scholar]
- 11.van Winkelhoff AJ, Schouten-van Meeteren AY, Baart JA, Vandenbroucke-Grauls CM. Microbiology of destructive periodontal disease in adolescent patients with congenital neutropenia. A report of 3 cases. J Clin Periodontol. 2000;27:793–8. doi: 10.1034/j.1600-051x.2000.027011793.x. [DOI] [PubMed] [Google Scholar]
- 12.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–90. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 13.Deas DE, Mackey SA, McDonnell HT. Systemic disease and periodontitis: manifestations of neutrophil dysfunction. Periodontol 2000. 2000;32:82–104. doi: 10.1046/j.0906-6713.2003.03207.x. [DOI] [PubMed] [Google Scholar]
- 14.Andersson M, Karlsson J, Carlsson G, Pütsep K. Expression of granule-associated proteins in neutrophils from patients with severe congenital neutropenia. Blood. 2007;110:2772–3. doi: 10.1182/blood-2007-03-083063. [DOI] [PubMed] [Google Scholar]
- 15.Rylander H, Ericsson I. Manifestations and treatment of periodontal disease in a patient suffering from cyclic neutropenia. J Clin Periodontol. 1981;8:77–87. doi: 10.1111/j.1600-051X.1981.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 16.Scully C, MacFadyen E, Campbell A. Oral manifestations in cyclic neutropenia. Br J Oral Surg. 1982;20:96–101. doi: 10.1016/0007-117X(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 17.Prichard JF, Ferguson DM, Windmiller J, Hurt WC. Prepubertal periodontitis affecting the deciduous and permanent dentition in a patient with cyclic neutropenia. A case report and discussion. J Periodontol. 1984;55:114–22. doi: 10.1902/jop.1984.55.2.114. [DOI] [PubMed] [Google Scholar]
- 18.Mishkin DJ, Akers JO, Darby CP. Congenital neutropenia. Report of a case and a biorationale for dental management. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1976;42:738–45. doi: 10.1016/0030-4220(76)90096-7. [DOI] [PubMed] [Google Scholar]
- 19.Briars GL, Parry HF, Ansari BM. Dominantly inherited severe congenital neutropenia. J Infect. 1996;33:123–6. doi: 10.1016/S0163-4453(96)93081-9. [DOI] [PubMed] [Google Scholar]
- 20.Bellanne-Chantelot C, Clauin S, Leblanc T, Cassinat B, Rodrigues-Lima F, Beaufils S, Vaury C, Barkaoui M, Fenneteau O, Maier-Redelsperger M, Chomienne C, Donadieu J. Mutations in the ELA2 gene correlate with more severe expression of neutropenia: a study of 81 patients from the French Neutropenia Register. Blood. 2004;103:4119–25. doi: 10.1182/blood-2003-10-3518. [DOI] [PubMed] [Google Scholar]
- 21.Pütsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002;360:1144–9. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- 22.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 27.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonilla MA, Gillio AP, Ruggeiro M, Kernan NA, Brochstein JA, Abboud M, Fumagalli L, Vincent M, Gabrilove JL, Welte K, Souza LM, Oreilly RJ. Effects of recombinant human granulocyte colony-stimulating factor on neutropenia in patients with congenital agranulocytosis. N Engl J Med. 1989;320:1574–80. doi: 10.1056/NEJM198906153202402. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–24. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørensen O, Cowland JB, Askaa J, Borregaard N. An ELISA for hCAP-18, the cathelicidin present in human neutrophils and plasma. J Immunol Methods. 1997;206:53–9. doi: 10.1016/S0022-1759(97)00084-7. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson J, Carlsson G, Ramme KG, Hagglund H, Fadeel B, Nordenskjold M, Henter JI, Palmblad J, Pütsep K, Andersson M. Low plasma levels of the protein pro-LL-37 as an early indication of severe disease in patients with chronic neutropenia. Br J Haematol. 2007;137:166–9. doi: 10.1111/j.1365-2141.2007.06530.x. [DOI] [PubMed] [Google Scholar]
- 32.Puklo M, Guentsch A, Hiemstra PS, Eick S, Potempa J. Analysis of neutrophil-derived antimicrobial peptides in gingival crevicular fluid suggests importance of cathelicidin LL-37 in the innate immune response against periodontogenic bacteria. Oral Microbiol Immunol. 2008;23:328–35. doi: 10.1111/j.1399-302X.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKay MS, Olson E, Hesla MA, Panyutich A, Ganz T, Perkins S, Rossomando EF. Immunomagnetic recovery of human neutrophil defensins from the human gingival crevice. Oral Microbiol Immunol. 1999;14:190–3. doi: 10.1034/j.1399-302X.1999.140308.x. [DOI] [PubMed] [Google Scholar]
- 34.Lundy FT, Orr DF, Shaw C, Lamey PJ, Linden GJ. Detection of individual human neutrophil alpha-defensins (human neutrophil peptides 1, 2 and 3) in unfractionated gingival crevicular fluid—a MALDI-MS approach. Mol Immunol. 2005;42:575–9. doi: 10.1016/j.molimm.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Hosokawa I, Hosokawa Y, Komatsuzawa H, Goncalves RB, Karimbux N, Napimoga MH, Seki M, Ouhara K, Sugai M, Taubman MA, Kawai T. Innate immune peptide LL-37 displays distinct expression pattern from beta-defensins in inflamed gingival tissue. Clin Exp Immunol. 2006;146:218–25. doi: 10.1111/j.1365-2249.2006.03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang D, Chertov O, Oppenheim JJ. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell Mol Life Sci. 2001;58:978–89. doi: 10.1007/PL00000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Mookherjee N, Wee K, Bowdish DM, Pistolic J, Li Y, Rehaume L, Hancock RE. Host defense peptide LL-37, in synergy with inflammatory mediator IL-1beta, augments immune responses by multiple pathways. J Immunol. 2007;179:7684–91. doi: 10.4049/jimmunol.179.11.7684. [DOI] [PubMed] [Google Scholar]
- 38.Alalwani SM, Sierigk J, Herr C, Pinkenburg O, Gallo R, Vogelmeier C, Bals R. The antimicrobial peptide LL-37 modulates the inflammatory and host defense response of human neutrophils. Eur J Immunol. 2010;40:1118–26. doi: 10.1002/eji.200939275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai CC, Ho YP, Chen CC. Levels of interleukin-1 beta and interleukin-8 in gingival crevicular fluids in adult periodontitis. J Periodontol. 1995;66:852–9. doi: 10.1902/jop.1995.66.10.852. [DOI] [PubMed] [Google Scholar]
- 40.Engebretson SP, Grbic JT, Singer R, Lamster IB. GCF IL-1beta profiles in periodontal disease. J Clin Periodontol. 2002;29:48–53. doi: 10.1034/j.1600-051x.2002.290108.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee HJ, Kang IK, Chung CP, Choi SM. The subgingival microflora and gingival crevicular fluid cytokines in refractory periodontitis. J Clin Periodontol. 1995;22:885–90. doi: 10.1111/j.1600-051X.1995.tb01788.x. [DOI] [PubMed] [Google Scholar]
- 42.Yavuzyilmaz E, Yamalik N, Bulut S, Ozen S, Ersoy F, Saatci U. The gingival crevicular fluid interleukin-1 beta and tumour necrosis factor-alpha levels in patients with rapidly progressive periodontitis. Aust Dent J. 1995;40:46–9. doi: 10.1111/j.1834-7819.1995.tb05614.x. [DOI] [PubMed] [Google Scholar]
- 43.Vernal R, Dutzan N, Chaparro A, Puente J, Antonieta Valenzuela M, Gamonal J. Levels of interleukin-17 in gingival crevicular fluid and in supernatants of cellular cultures of gingival tissue from patients with chronic periodontitis. J Clin Periodontol. 2005;32:383–9. doi: 10.1111/j.1600-051X.2005.00684.x. [DOI] [PubMed] [Google Scholar]
- 44.Tervahartiala T, Koski H, Xu JW, Hayrinen-Immonen R, Hietanen J, Sorsa T, Konttinen YT. Tumor necrosis factor-alpha and its receptors, p55 and p75, in gingiva of adult periodontitis. J Dent Res. 2001;80:1535–9. doi: 10.1177/00220345010800061101. [DOI] [PubMed] [Google Scholar]
- 45.Ohyama H, Kato-Kogoe N, Kuhara A, Nishimura F, Nakasho K, Yamanegi K, Yamada N, Hata M, Yamane J, Terada N. The involvement of IL-23 and the Th17 pathway in periodontitis. J Dent Res. 2009;88:633–8. doi: 10.1177/0022034509339889. [DOI] [PubMed] [Google Scholar]
- 46.Cardoso CR, Garlet GP, Crippa GE, Rosa AL, Junior WM, Rossi MA, Silva JS. Evidence of the presence of T helper type 17 cells in chronic lesions of human periodontal disease. Oral Microbiol Immunol. 2009;24:1–6. doi: 10.1111/j.1399-302X.2008.00463.x. [DOI] [PubMed] [Google Scholar]
- 47.Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–20. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 48.Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, Francois P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79:266–71. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore WE, Holdeman LV, Smibert RM, Cato EP, Burmeister JA, Palcanis KG, Ranney RR. Bacteriology of experimental gingivitis in children. Infect Immun. 1984;46:1–6. doi: 10.1128/iai.46.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kononen E, Asikainen S, Saarela M, Karjalainen J, Jousimies-Somer H. The oral gram-negative anaerobic microflora in young children: longitudinal changes from edentulous to dentate mouth. Oral Microbiol Immunol. 1994;9:136–41. doi: 10.1111/j.1399-302X.1994.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 53.Cario G, Skokowa J, Wang Z, Bucan V, Zeidler C, Stanulla M, Schrappe M, Welte K. Heterogeneous expression pattern of pro- and anti-apoptotic factors in myeloid progenitor cells of patients with severe congenital neutropenia treated with granulocyte colony-stimulating factor. Br J Haematol. 2005;129:275–8. doi: 10.1111/j.1365-2141.2005.05428.x. [DOI] [PubMed] [Google Scholar]
- 54.Klein C, Welte K. Genetic insights into congenital neutropenia. Clin Rev Allergy Immunol. 2010;38:68–74. doi: 10.1007/s12016-009-8130-5. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki Y, Demoliere C, Kitamura D, Takeshita H, Deuschle U, Watanabe T. HAX-1, a novel intracellular protein, localized on mitochondria, directly associates with HS1, a substrate of Src family tyrosine kinases. J Immunol. 1997;158:2736–44. [PubMed] [Google Scholar]
- 56.Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature. 2008;452:98–102. doi: 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- 57.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, Rathinam C, Boztug K, Schwinzer B, Rezaei N, Bohn G, Melin M, Carlsson G, Fadeel B, Dahl N, Palmblad J, Henter JI, Zeidler C, Grimbacher B, Welte K. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 58.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 59.Kollner I, Sodeik B, Schreek S, Heyn H, von Neuhoff N, Germeshausen M, Zeidler C, Kruger M, Schlegelberger B, Welte K, Beger C. Mutations in neutrophil elastase causing congenital neutropenia lead to cytoplasmic protein accumulation and induction of the unfolded protein response. Blood. 2006;108:493–500. doi: 10.1182/blood-2005-11-4689. [DOI] [PubMed] [Google Scholar]
- 60.Meyer-Hoffert U, Wiedow O. Neutrophil serine proteases: mediators of innate immune responses. Curr Opin Hematol. 2011;18:19–24. doi: 10.1097/MOH.0b013e32834115d1. [DOI] [PubMed] [Google Scholar]
- 61.Carlsson G, Elinder G, Malmgren H, Trebinska A, Grzybowska E, Dahl N, Nordenskjold M, Fadeel B. Compound heterozygous HAX1 mutations in a Swedish patient with severe congenital neutropenia and no neurodevelopmental abnormalities. Pediatr Blood Cancer. 2009;53:1143–6. doi: 10.1002/pbc.22131. [DOI] [PubMed] [Google Scholar]