Abstract
The research community at large is expending considerable resources to sequence the coding region of the genomes of tumors and other human diseases using targeted exome capture (i.e., “whole exome sequencing”). The primary goal of targeted exome sequencing is to identify nonsynonymous mutations that potentially have functional consequences. Here, we demonstrate that whole-exome sequencing data can also be analyzed for comprehensively monitoring somatic copy number alterations (CNAs) by benchmarking the technique against conventional array CGH. A series of 17 matched tumor and normal tissues from patients with metastatic castrate-resistant prostate cancer was used for this assessment. We show that targeted exome sequencing reliably identifies CNAs that are common in advanced prostate cancer, such as androgen receptor (AR) gain and PTEN loss. Taken together, these data suggest that targeted exome sequencing data can be effectively leveraged for the detection of somatic CNAs in cancer.
Introduction
Recognition that copy number alterations (CNAs) in tumor genomes, which can result in the amplification of oncogenes or the deletion of tumor suppressors, contribute significantly to cancer etiology has led to the development of multiple techniques for their comprehensive identification. Initial global approaches for CNA detection relied primarily on array based technologies: whole-genome array comparative genomic hybridization (aCGH) tests the relative frequency of probe DNA segments between two genomes [1–4], whereas single-nucleotide polymorphism (SNP) arrays measure the probe intensities at known SNP loci to identify shifts in zygosity relative to another genome [5–9]. The recent advent of high-throughput sequencing has made the sequencing of whole human genomes feasible and has made possible the development of sequencing-based approaches to CNA identification [10–17].
The prohibitive cost and time constraints of whole-genome sequencing has necessitated further innovation, and hybridization-based approaches to high-throughput sequencing that focus on the human exome have been recently applied to detect novel somatic point mutations in tumor genomes [18–22]. Targeted exome sequencing allows one to achieve very high depths of coverage (100x coverage or greater) of regions of interest and thus provides advantages over whole-genome sequencing for mutation detection especially in the context of the highly deranged genomes of many tumors. Because targeted exome sequencing yields depth of coverage data, it is reasonable to ask whether exome sequencing data can also be used to detect CNAs, especially because a recent application to unmatched cancer cell lines indicated the potential of this approach [23]. In addition, the recent development of third-generation sequencing approaches has made sequencing of a tumor exome achievable within a week, making its application to detect somatic mutations in a clinical setting imminent. As a result of its wide applicability, there would be a clear and demonstrable advantage to applying exome sequencing to generate CNA data because it would obviate the need for performing aCGH or whole-genome sequencing to detect CNAs in patients awaiting treatment.
Here we propose a method for the detection of somatic CNAs from exome sequencing of a matched tumor/normal pair. By comparing depth of coverage across the exome between the tumor and normal samples, we detect regions with predicted copy gain or loss in the tumor sample. A comparison of these data to aCGH copy number data for the same samples demonstrates a high level of agreement between the two platforms. We apply our method to identify copy number aberrations from exome data generated for 17 prostate tumor-normal pairs, showing that our method identifies aberrations in multiple genes known to have copy number gains and losses in prostate cancer including AR, NCOA2, PTEN, RB1, and TP53 [24]. Together, these analyses show that exome sequencing data, in addition to being useful for detecting point mutations and indels, can be used in place of aCGH and whole-genome sequencing for the generation of CNA data.
Materials and Methods
Tissue Samples
Prostate tissues were from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program [25], both of which are part of the University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core. All samples were collected with informed consent of the patients and previous institutional review board approval.
High-Molecular Weight Genomic DNA Isolation
Frozen tissue samples were taken as chunks or sections from OCT-embedded, flash-frozen tissue blocks. Genomic DNA (gDNA) was isolated using the Qiagen DNeasy Blood and Tissue Kit (Valencia, CA) according to the manufacturer's instructions. Briefly, cell or tissue lysates were incubated at 65°C in the presence of proteinase K and SDS, purified on silica membrane-based mini columns, and eluted in buffer AE (10 mM Tris-HCl and 0.5 mM EDTA pH 9.0).
Exome Capture Sequencing
Exome libraries of matched pairs of tumor/normal gDNAs were generated using the Agilent SureSelect Human All Exon Kit (Agilent, Santa Clara, CA; the 38-Mb kit, including 165,637 exon targets, was used on three tumor/normal matched pairs and the 50-Mb kit, including 213,050 exon targets, was used on the remaining 14; Table W2) and the Illumina Paired-End Genomic DNA Sample Prep Kit (Illumina, San Diego, CA) following the manufacturers' instructions. Three micrograms of each gDNA was sheared using a Covaris (Woburn, MA) S2 to a target peak size of 250 bp. Fragmented DNA was concentrated using AMPure XP beads (Beckman Coulter, Indianapolis, IN), and DNA ends were repaired using T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. 3′ A-tailing with exo-minus Klenow polymerase was followed by ligation of Illumina paired-end adapters to the gDNA fragments. The adapter-ligated libraries were electrophoresed on 3% Nusieve 3:1 (Lonza, Walkersville, MD) agarose gels, and fragments between 250 and 350 bp were recovered using QIAEX II gel extraction reagents (Qiagen). Recovered DNA was then amplified using Illumina PE1.0 and PE2.0 primers for nine cycles. The amplified libraries were purified using AMPure XP beads, and the DNA concentration was determined using a Nanodrop spectrophotometer (NanoDrop 8000; Thermo Scientific, Wilmington, DE). Five hundred nanograms of the libraries was hybridized to the Agilent biotinylated SureSelect Capture Library at 65°C for 65 hours. The targeted exon fragments were captured on DynalM-280 streptavidin beads (Invitrogen, Carlsbad, CA), washed, eluted, and enriched by amplification with the Illumina PE1.0/PE2.0 primers for eight additional cycles. After purification of the polymerase chain reaction products with AMPure XP beads, the quality and quantity of the resulting exome libraries were analyzed using an Agilent Bioanalyzer (Agilent). All captured DNA libraries were sequenced with the Illumina HiSeq 2000 (Illumina) in paired-end mode trimmed to yield 78-bp reads. The reads that passed the chastity filter of Illumina BaseCall software were used for subsequent analysis. Next, mate pairs were pooled and then mapped as single reads to the reference human genome (NCBI build 36.1, hg18), excluding unordered sequence and alternate haplotypes, using Bowtie [26], keeping unique best hits, and allowing up to two mismatched bases per read.
Array Comparative Genomic Hybridization
aCGH of six samples (matched tumor and normal) from three metastatic prostate cancer patients was performed using gDNA on Agilent's 244K aCGH microarrays (Human Genome CGH 244K Oligo Microarray) using Agilent's Standard Direct Method protocol and Wash Procedure B. Briefly, 1.5 to 3 µg of gDNA from prostate specimens (isolated as above) was restriction digested with AluI and RsaI, labeled with Cy-5 (test channel), purified using Microcon YM- 30 columns (Millipore, Hayward, CA), and hybridized with an equal amount of Cy-3 (reference channel)-labeled human male gDNA (Promega, Madison, WI) for 40 hours at 65°C. Posthybridization wash was performed with acetonitrile wash and Agilent Stabilization and Drying Solution wash. Scanning was performed on an Agilent scanner (Model G2505B; 5-µm scan with software v7.0), and data were extracted using Agilent Feature Extraction software v9.5 using protocol CGH-v4_95_Feb07.
For data analysis, quantifications for each probe were determined as rProcessedSignal/gProcessedSignal and analyzed on the log2 scale. To focus on somatic copy number changes, log2 ratios in the matched normal sample were subtracted from the log2 ratios in each tumor sample, and the resulting differences were used for analysis. Replicate probes on the array were summarized by computing the median value across replicates for each sample and using this median for analysis. The resulting log2 ratios were median centered for each tumor/normal matched pair.
Segmentation Analysis
Segmentation analysis for both aCGH and exome log2 copy number ratios was performed through the use of the Circular Binary Segmentation Algorithm [27], as implemented in the DNAcopy package in R version 2.11.1. Default values for all parameters were used, except that consecutive segments were merged using the undo.splits = “sdundo” option with the undo.SD parameter set to 0.3/DLRS, where DLRS (derivative log ratio spread) represents the local SD in log ratio units, a well-known measure of local variability for aCGH microarrays. In this way, the segmentation algorithm was tuned to detect copy number changes of at least 0.3 in magnitude on the log2 scale. Segments were reported as amplified or deleted if the corresponding estimated copy number ratio was greater than 1.25 or less than 0.75, respectively; high-level amplifications and homozygous losses were called whenever the estimated copy number ratio was greater than 2.0 or less than 0.5. ROC analysis was performed using the ROC package in Bioconductor.
Informative Genes
The list of 2016 “informative” cancer genes used to quantify performance of copy number calls was generated by combining the list of 457 genes comprising the Sanger Institute's Cancer Gene Census together with a list of 1933 Protein Kinases, Tumor Suppressors, Tyrosine Kinases, and Oncogenes downloaded from Memorial Sloan-Kettering Cancer Center's Cancer Genes resource. This resulted in a list of 2217 unique genes, of which 2016 were targeted by the Agilent SureSelect Human All Exon Kit and did not map to the Y chromosome.
Results
Algorithm: Detecting Copy Number Alterations from Whole Exome Sequencing Data
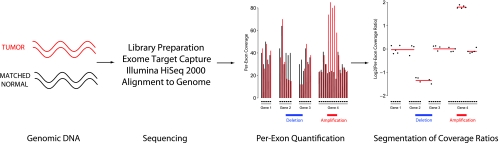
Figure 1 illustrates our approach to generating copy number data from whole exome sequencing data. Libraries are prepared from tumor and matched normal DNA, targeted exons are sequenced, and per-exon coverage is computed for both tumor and normal samples. In contrast to genomic sequencing, in which depth of coverage is approximately proportional to genomic copy number [11], exome sequencing involves varying capture efficiencies across the human exome, making the relationship between coverage and copy number less apparent. We accounted for variation in coverage across exons by performing per-exon comparisons between each tumor sample and its matched normal sample. This approach also corrects for variation in observed coverage across different regions of the human genome due to the presence of repetitive sequences and variation in GC content.
Figure 1.
Overview of copy number analysis by whole exome sequencing. Vertical bars represent per-exon coverage in the tumor (red) and matched normal (black) tissue. Log-transformed coverage ratios between tumor and normal tissues are computed for each exon (black dots) and altered regions are identified through segmentation analysis (red line segments).
More precisely, we generated copy number ratios for each tumor/normal matched pair through the following algorithm. First, exons containing fewer than 10 reads (i.e., 780 bp) worth of coverage in the matched normal sample were excluded from analysis. Second, coverage values were perturbed slightly by adding 780 bp of coverage to each exon's coverage quantification in both samples. Third, per-exon coverage in the tumor sample was divided by per-exon coverage in the matched normal sample, resulting in coverage ratios for each exon. These modified coverage ratios were then globally normalized by dividing each of them by the ratio of human mappings between the two samples (tumor/normal). After log2-transforming these normalized coverage ratios, the overall median value was subtracted, resulting in a set of log2-transformed coverage ratios with median zero for each tumor/normal matched pair. Ratios and logarithms were well defined throughout this process owing to the filtering of low-coverage exons in the benign sample, which ensured that division by zero did not occur, and the small perturbation of coverage values, which ensured that coverage ratios were always nonzero. The normalized log2-transformed coverage ratios were used for downstream segmentation analysis. The resulting data structure is analogous to that of a two-channel microarray with “probes” at each targeted exon and sequencing coverage replacing signal intensity.
Benchmarking: Concordance with Copy Number Alterations Generated Using aCGH
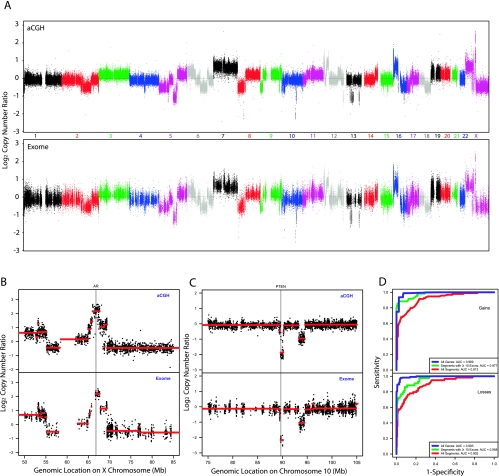
We investigated the ability of these normalized exome capture coverage ratios to yield accurate copy number assessments by comparing them against the corresponding copy number ratios from Agilent 244K CGH microarray data. We used three matched metastatic castration-resistant prostate tumor/normal pairs for this comparison, using the 38-Mb Agilent SureSelect Human All Exon Kit to target the human exome and performing next-generation sequencing on the Illumina HiSeq 2000 platform. Representative assessments for one sample WA54 are shown in Figure 2 with assessments for the other two samples in Figures W1 and W2. Genome-wide copy number ratios are highly concordant between the two technologies (Figure 2A) with large-scale amplifications agreeing in magnitude. Large regions of gain and loss spanning whole chromosomes and chromosomal arms, such as chromosome 7 gain, 8p loss, 8q gain, 16p gain, 16q loss, 18q loss, and Xp gain, are easily visible by both technologies. We verified this concordance more formally by comparing copy number ratios on disjoint windows covering the genome. Specifically, we partitioned the genome into windows containing at least five targeted exons and five aCGH probes and computed mean log ratios by each technology on each window. The resulting quantifications exhibit strong correlations for each of the three samples (minimum Pearson correlation coefficient = 0.92, P < .001; Figure W3). This genome-wide comparison illustrates that copy number ratios from exome capture sequencing exhibit strong concordance with and are on the same scale as those from aCGH microarrays.
Figure 2.
Comparison of exome sequencing to array CGH in detecting CNAs. (A) Overall copy number across the genome for metastatic prostate tumor sample WA54 by aCGH (upper panel) and exome sequencing (lower panel). Log2(copy number ratio) between tumor and matched normal is shown on the vertical axis; each point represents the log-transformed ratio for each aCGH probe or targeted exon, ordered by genomic coordinates. (B) Copy number assessment for sample WA54 by aCGH and exome sequencing in a 35-Mb region containing the AR gene. Red line segments represent segmented copy number data. (C) Copy number assessment for sample WA54 by aCGH and exome sequencing in a 30-Mb region containing the PTEN gene. Red line segments represent segmented copy number data. (D) Classification performance of exome capture sequencing relative to aCGH for sample WA54. ROC curves are shown, using aCGH copy number assessments as a criterion standard. ROC curves are presented for classifying all aCGH segments (red), segments containing at least 10 targeted exons (green), and all targeted genes (blue).
In addition, we examined copy number at genes well known to be gained or lost in prostate cancer and found that copy number assessments for these genes were concordant as well. Both technologies reveal a focal amplification of the AR gene in sample WA54 (Figure 2B), and the patterns of copy number changes in that region are strikingly similar. The estimated number of copies in the segment overlapping the AR gene was similar by each approach (4.50 copies by aCGH, 4.57 copies by exome capture), revealing that exome capture coverage ratios exhibit sufficient dynamic range to capture high-level amplifications. Similarly, both aCGH and exome sequencing reveal a focal region of two-copy loss at the PTEN gene in sample WA54 (Figure 2C), and the two technologies agree on the approximate number of copies: 0.27 copies of PTEN by aCGH and 0.22 copies of PTEN by exome capture.
To quantify the ability of exome capture sequencing to identify regions of gain and loss, we performed ROC analysis of exome capture quantifications, using the matched aCGH data as a criterion standard (Figure 2D). First, we performed segmentation analysis (Materials and Methods) on both aCGH and exome capture log-transformed copy number ratios, and using copy number ratio cutoffs of 1.25 and 0.75 to define regions of gain and loss, respectively, we identified a set of altered regions by aCGH. The segmented exome capture copy number ratios were computed on these altered segments; performance of this classifier relative to the aCGH calls was quantified using the area under the ROC curve (AUC). For each of the three samples, exome copy number analysis performed well in classifying these aCGH segments, with AUCs of at least 0.89 across the three samples (Table W1). As expected, restricting attention to segments that contain targeted exons improved classification performance; for segments containing at least two targeted exons, the minimum AUC across the three samples was 0.94. Finally, we did a gene-centric analysis, comparing copy number calls for each of the 18,090 genes targeted by the exome capture kit, and performance was even better, with a minimum AUC of 0.989 across the three samples. We repeated this analysis for a smaller set of 2016 “informative” genes (Materials and Methods) that have been implicated in cancer (e.g., kinases, oncogenes, and tumor suppressors) and verified that the strong performance persists when restricted to genes that are likely to be relevant to cancer progression.
Application: Copy Number Alterations in Prostate Cancer Tissues
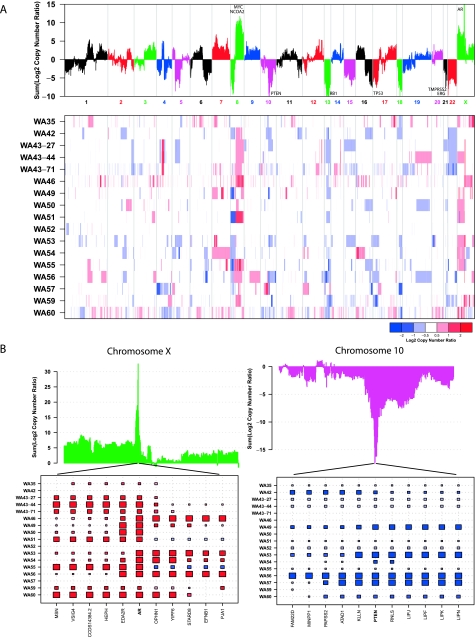
Next, we sought to demonstrate that our CNA detection method can be used to generate results qualitatively equivalent to aCGH-based methods by applying it to the exomes of 17 lethal metastatic castration-resistant prostate cancers (Table W2), including the three samples used for benchmarking, and matched normal tissues from the same patients. In total we generated 395,489,506,152 bases, with 105.27 average fold coverage of each targeted base per tissue sample (Table W3). Using copy number ratio cutoffs of 1.25 to define regions of gain and 0.75 to define regions of loss, we identified a median of 93 gained regions and 79 lost regions across the 17 samples (Table W4). The median total length of these altered regions across samples was 407.5 Mb (gains) and 406.2 Mb (losses). Using more stringent copy number ratio cutoffs of 2.0 to define high-level gain and 0.5 to define homozygous loss identified a median of 19 high-level gains and 17 homozygous losses covering 23.3 and 25 Mb, respectively. Three of the cancer samples were derived from different metastatic sites from the same patient; these three samples had multiple amplifications and deletions in common, including focal amplifications on chromosomes 4 and 14, the broad 8q amplification, loss of chromosome 22, and a focal loss on the end of 2q, reflecting the likely clonal origin of the tumor (Figure 3A).
Figure 3.
Qualitative comparison to prior CNAs observed in prostate cancer. (A) Overall summary of copy number across 17 lethal metastatic castration-resistant prostate cancers. Summed segmented log2 copy number ratios (top panel) for all targeted genes across the 17 samples are shown. Genes exhibiting recurrent amplifications or deletions across the cohort will have large positive or negative values, respectively. Regions of copy number gain and loss for all 17 samples are shown in a heat map (bottom panel). Red represents amplification; white, copy number neutral; blue, deletion. Three samples are derived from three different metastatic foci from a man with lethal castrate-resistant prostate cancer: celiac lymph node metastatic site (WA43-27), lung metastatic site (WA43-71), and bladder metastatic site (WA43-44). (B) Focal amplifications of the AR gene and deletions of the PTEN gene in this cohort. AR has the largest positive summed log copy number ratio across the 17 samples, with a total sum of 32.6, whereas PTEN has the largest negative summed log copy number ratio, with a total sum of -17.5. A plot of this sum over the entire chromosome (top) is shown; a large positive peak is present at AR and a large negative peak is present at PTEN. Segmented copy number ratios are represented by boxes, with the area (absolute log2 ratio) and color intensity (log2 ratio; copy number gain in red; loss in blue) of each box proportional to mean copy number across that gene. Missing boxes indicate that the gene is neither amplified nor deleted in that sample.
Global analysis of copy number profiles of all 17 prostate cancers (Figure 3A) identified recurrent aberrations previously associated with prostate cancer development and progression, including broad losses of 1p, 8p, 6q, and losses of large regions of chromosome 13, 15, 18 and 22, as well as gains of 1q, 3q, 7q, and 8q, containing two prostate cancer oncogenes, MYC and NCOA2 [24,28–30]. We also identified previously reported deletions between TMPRSS2 and ERG in cases with TMPRSS2:ERG gene fusions [24,28–30]. In addition, we identified recurrent focal amplifications of AR (Figure 3B) and recurrent homozygous focal deletions of PTEN (Figure 3C), consistent with prior observations [24,28–30]. Examination of each sample's copy number ratio in these regions shows that the pattern of nearby amplifications around AR can be different for each sample; however, the region of gain always includes AR itself. The same is true with respect to the region of loss including PTEN. We also detected specific disruptions of RB1 and TP53 (Figure 3A), two genes previously associated with focal losses in prostate cancer [24,28,30].
Discussion
There are a number of benefits of using targeted exome sequencing for assessing CNAs. One benefit of the exome-based approach to identifying CNAs, over using evenly spaced genomic hybridization probes in aCGH, is the possibility of using the data to gain exon-level resolution of the genomic rearrangements associated with copy number changes. Another benefit is that one has the potential to leverage a vast amount of data that is already being generated as part of large genome sequencing projects, such as The Cancer Genome Atlas (TCGA). For example, at the time of writing, the TCGA project had just released 316 whole exomes from ovarian cancer [22], more than 500 whole exomes had just been published, and sequencing for thousands more was underway. In essence, point mutations (such as BRAF V600E) and amplifications/deletions (such as amplification of ERBB2 or loss of PTEN) can be monitored from the same whole exome sequencing data set. This type of assessment will be powerful for integrative mutational studies in the context of cancer and toward personalized medicine.
Sequencing-based approaches to copy number detection have the advantage of being able to not only assess CNAs using depth of coverage, like aCGH, but also using SNPs or somatic point mutation to assess for shifts in zygosity indicative of copy number changes, as in SNP array approaches. In this study, we have presented a depth of coverage approach to detect CNAs using exome data, but an approach using SNPs or somatic point mutation is equally feasible in theory. Moreover, a combined approach using both depth of coverage and SNPs has the potential to be even more effective, especially compared with both aCGH and SNP arrays.
As intimated, the exome approach is limited by mapping issues, making genes containing highly repetitive sequence difficult to target for exome sequencing and therefore difficult to assess for CNAs using this method. For example, the second exon of FOXA1, a two-exon gene, has two large gaps in coverage in both the 38- and the 50-Mb Agilent SureSelect All Human Exon design, resulting from repetitive sequence, so that the computed coverage of the exon is always a gross underestimate of the actual coverage. These sorts of coverage issues make detection of focal CNAs of certain genes difficult. A second limitation is that exome capture copy number analysis will clearly fail to detect genomic aberrations that occur in regions containing no nearby genes. If exome capture was deliberately used for assessing CNAs, both limitations could be overcome by optimizing the exome capture design to improve detection of these alterations using the excess sequencing capacity afforded by deep sequencing of only the coding regions of the genome (which represent <1% of the genome). The results of this analysis suggest that additional effort should also be put toward designing exon capture platforms that add additional targets to improve detection of CNAs. This could be done by placing additional targets throughout the genome and near genes with highly repetitive regions, even if they are not directly sequencing a region of interest. Personalized medicine approaches that emphasize somatic mutations in informative coding genes would clearly benefit from an exon capture platform and could efficiently assess genes of interest for both somatic point mutations and for somatic CNAs.
Supplementary Material
Acknowledgments
The authors thank Javed Siddiqui and Rohit Mehra for assisting with sample acquisition, Terrence Barrette for assisting with sequence data generation using the Illumina pipeline, and Jyoti Athanikar for assistance with manuscript preparation.
Footnotes
This work was supported in part by the National Institutes of Health (NIH) Specialized Program of Research Excellence (P50 CA69568) and the Early Detection Research Network (U01 CA111275). A.M.C. is supported by the Howard Hughes Medical Institute, the Prostate Cancer Foundation, the Taubman Research Institute, the Doris Duke Foundation, and the American Cancer Society as a clinical research professor. K.J.P. is supported by the Prostate Cancer Foundation, the Taubman Research Institute, and the American Cancer Society as a clinical research professor (NIH 1 PO1 CA093900 and 1 U01CA143055).
This article refers to supplementary materials, which are designated by Figures W1 to W3 and Tables W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Pinkel D, Seagraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 2.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 3.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Seagraves R, et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, deBakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 6.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 8.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 9.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, Dallaire S, Gabriel SB, Lee C, Daly MJ, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 10.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Landers ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nord AS, Lee M, King MC, Walsh T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genomics. 2011;12:184. doi: 10.1186/1471-2164-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magi A, Benelli M, Yoon S, Roviello F, Torricelli F. Detecting common copy number variants in high-throughput sequencing data by using JointSLM algorithm. Nucleic Acids Res. 2011;39:e65. doi: 10.1093/nar/gkr068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TM, Luquette LJ, Xi R, Park PJ. rSW-seq: algorithm for detection of copy number alterations in deep sequencing data. BMC Bioinformatics. 2010;11:432. doi: 10.1186/1471-2105-11-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medvedev P, Fiume M, Dzamba M, Smith T, Brudno M. Detecting copy number variation with mated short reads. Genome Res. 2010;20:1613–1622. doi: 10.1101/gr.106344.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeva V, Zinovyev A, Bleakley K, Vert JP, Janoueix-Lerosey I, Delattre O, Barillot E. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics. 2011;27:268–269. doi: 10.1093/bioinformatics/btq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 19.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones S, Wang TL, Shihle M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Research Network, author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Byseck R, Siemers NO. Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines. PLoS One. 2011;6:e21097. doi: 10.1371/journal.pone.0021097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 26.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 28.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberg MA, Carducci MA, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holcomb IN, Young JM, Coleman IM, Salari K, Grove DI, Hsu L, True LD, Roudier MP, Morrissey CM, Higano CS, et al. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer Res. 2009;69:7793–7802. doi: 10.1158/0008-5472.CAN-08-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demichelis F, Setlur SR, Beroukhim R, Perner S, Korbel JO, Lafargue CJ, Pflueger D, Pina C, Hofer MD, Sboner A, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.