Abstract
Streptococcus suis, a major porcine pathogen, has been receiving growing attention not only for its role in severe and increasingly reported infections in humans, but also for its involvement in drug resistance. Recent studies and the analysis of sequenced genomes have been providing important insights into the S. suis resistome, and have resulted in the identification of resistance determinants for tetracyclines, macrolides, aminoglycosides, chloramphenicol, antifolate drugs, streptothricin, and cadmium salts. Resistance gene-carrying genetic elements described so far include integrative and conjugative elements, transposons, genomic islands, phages, and chimeric elements. Some of these elements are similar to those reported in major streptococcal pathogens such as Streptococcus pyogenes, Streptococcus pneumoniae, and Streptococcus agalactiae and share the same chromosomal insertion sites. The available information strongly suggests that S. suis is an important antibiotic resistance reservoir that can contribute to the spread of resistance genes to the above-mentioned streptococci. S. suis is thus a paradigmatic example of possible intersections between animal and human resistomes.
Keywords: Streptococcus suis, zoonotic pathogen, resistome, integrative and conjugative element, transposon, genomic island, phage, chimeric element
The massive use of antibiotics by the livestock industry, either for growth promotion (a practice recently banned in Europe) or for prophylaxis and therapy, contributed to the emergence, and spread of antibiotic resistance (McEwen and Fedorka-Cray, 2002). Antimicrobials for veterinary use are the same or belong to the same classes as those for human use and exert a constant pressure on the animal microflora, selecting for resistance (Witte, 1997). Streptococcus suis, an emerging zoonotic pathogen, has been receiving growing attention for its involvement not only in severe and increasingly reported human infections, but also in drug resistance. Here we outline the current knowledge about the S. suis resistome and assess its role as a possible reservoir of antibiotic resistance determinants for streptococcal pathogens.
S. suis and S. suis Infections
Streptococcus suis is a major porcine pathogen worldwide, endemic in all countries where intensive pig farming is practiced. Moreover, it is increasingly being isolated from mammalian species other than pigs, from birds, and from the environment (Gottschalk et al., 2010). In humans, S. suis is now considered as an emerging zoonotic pathogen, causing systemic infection: meningitis with possible residual deafness or vestibular dysfunctions is the most frequent clinical presentation; endocarditis, cellulitis, peritonitis, rhabdomyolysis, arthritis, spondylodiscitis, pneumonia, uveitis, and endophthalmitis have also been reported (Lun et al., 2007; Wertheim et al., 2009a). The human infection is mainly an occupational disease that may affect those who come into contact with animal infected blood or secretions or with pork-derived products. Thirty-three capsular serotypes are currently recognized (Staats et al., 1997; Hill et al., 2005). Serotype 2 is the most virulent and is responsible for severe infections in both swine and humans worldwide.
After the first reported human case of S. suis infection in Denmark in 1968 (Perch et al., 1968), sporadic cases (mainly of meningitis) have been reported in Europe and South-East Asia in the following decades (Lun et al., 2007; Wertheim et al., 2009a). In 2005, a severe epidemic caused by S. suis serotype 2 broke out in China’s Sichuan Province, preceded by a small outbreak in 1998 (Yu et al., 2006). From 2005 onward, an increasing number of S. suis human infections have been reported worldwide, also in countries where the infection had been rarely or never reported before (Gottschalk et al., 2010). Currently, the majority of cases occur in South-East Asia where S. suis is a leading cause of adult meningitis, in particular in Vietnam (Mai et al., 2008; Wertheim et al., 2009b). A multilocus sequence typing scheme (King et al., 2002) disclosed a high genetic diversity of S. suis isolates with over 250 sequence types (ST) identified (http://ssuis.mlst.net). Four major ST clonal complexes (CC; CC–ST1, CC–ST16, CC–ST25, and CC–ST27) dominate the population; CC–ST1, the most virulent, includes ST1, found throughout the world, and ST7, responsible for the Chinese epidemic (Ye et al., 2006). It has been suggested that the increase in human cases may also reflect the recent awareness of S. suis as an emerging agent of meningitis and the improvement of microbiological diagnostic techniques (Gottschalk et al., 2010). Actually, the fact that S. suis does share several characteristics with other bacteria causing meningitis can result in misdiagnosis. Indeed, many isolates that had originally been identified as Streptococcus pneumoniae, enterococci, Streptococcus bovis, viridans streptococci, or even Listeria spp. were re-identified as S. suis in retrospective studies. In addition, recent serologic data suggest that human infection occurs more frequently than previously believed (Smith et al., 2008). Altogether these findings suggest that S. suis disease has been under diagnosed in the past. The growing interest in this pathogen is reflected by recent (2007–2011) whole-genome sequencing studies. So far, eight strains have been sequenced (Chen et al., 2007; Holden et al., 2009; Ye et al., 2009; Hu et al., 2011a,b), showing that ~40% of the ~2 Mb genome is unique. This suggests that S. suis is phylogenetically distinct from other Streptococcus species whose genome sequences are currently available. Intraspecies genomic comparisons have shown high levels of sequence conservation. However, these data may be influenced by the clonal relatedness of the sequenced strains, six of eight being serotype 2 strains belonging to the CC–ST1.
Current Knowledge of the S. suis Resistome
High rates of S. suis resistance to tetracyclines (up to >90%) and macrolides (up to >70%) have been reported in pig isolates worldwide (Wisselink et al., 2006; Hendriksen et al., 2008; Zhang et al., 2008; Princivalli et al., 2009). In the 1990s, a retrospective study of historic pig isolates in Denmark demonstrated that the increase in tetracycline and macrolide resistance had begun in the early 1980s (Aarestrup et al., 1998). Resistances to tetracyclines and macrolides in human strains were first reported in the second half of the first decade of 2000, but retrospective studies showed that they were already widespread in previous decades (Ye et al., 2008; Chu et al., 2009; Hoa et al., 2011). The genetic basis of tetracycline and macrolide resistance in S. suis has been extensively investigated.
Tetracycline resistance in streptococci is mainly due to ribosomal protection genes tet(M) and tet(O), and less frequently tet(Q), tet(T), and tet(W), and to efflux genes tet(K) and tet(L) (Chopra and Roberts, 2001; Roberts, 2005). Until a few years ago, tet(M) and tet(O) were the only tetracycline resistance determinants reported in S. suis; further determinants have lately been detected, such as ribosomal protection genes tet(W) and mosaic tet(O/W/32/O) and efflux genes tet(L), tet(B), and tet(40). Of these, tet(O/W/32/O), tet(B), and tet(40) had never been described in the genus Streptococcus before. tet(W) is an emerging tetracycline resistance determinant whose host range, including Gram-positive and Gram-negative, aerobic and anaerobic bacteria, is second only to that of tet(M) among ribosomal protection tet genes (Roberts, 2005). In S. suis, tet(W) was first detected in 2008, in an isolate from a case of meningitis in Italy (ST1 strain SsCA) (Manzin et al., 2008); it was subsequently described in other Italian strains (ST1 human strain SSUD and three pig isolates) (Princivalli et al., 2009), in the sequenced genome of the Chinese ST1 human strain GZ1 (Ye et al., 2009), and in two Vietnamese human isolates (Hoa et al., 2011). tet(O/W/32/O) is a new mosaic gene reported in 2009 in clonally unrelated pig isolates of S. suis (Princivalli et al., 2009). Mosaic tet genes are a recently discovered class of hybrids of ribosomal protection genes (Thaker et al., 2010). Mosaic derivatives of tet(O) and tet(W) were first detected in 2003 in anaerobic Gram-negative Megasphaera elsdenii from swine intestine (Stanton and Humphrey, 2003). Other mosaic genes, also including portions of tet(32), were later detected in Clostridium difficile (Patterson et al., 2007) and Clostridium saccharolyticum (Kazimierczak et al., 2008). tet(L), commonly carried in streptococci by small transmissible plasmids (Chopra and Roberts, 2001), has recently been detected in a sequenced genome of S. suis, where it is carried by a Tn916-like element (Holden et al., 2009), and lately, still outside plasmids, in Vietnamese human isolates (Hoa et al., 2011). tet(B), which had never been described in Gram-positive bacteria, was detected in 2010 in Chinese pig isolates of S. suis (Chander et al., 2011). tet(40) is a novel efflux gene recently detected in C. saccharolyticum in tandem with a mosaic tet gene [tet(O/32/O)] (Kazimierczak et al., 2008).
Macrolide resistance in streptococci is mainly due to methylase-mediated target site modification by erm genes and to active efflux by mef genes. erm(B), the old-established erm determinant in streptococci, can be expressed either constitutively or inducibly and is usually associated with high-level resistance; erm(TR), an erm(A) subclass, is normally inducible and is widely distributed in S. pyogenes isolates; erm(T) was detected in inducibly erythromycin-resistant isolates of group D streptococci and in S. pyogenes (Varaldo et al., 2009), and more recently in Streptococcus dysgalactiae subsp. equisimilis (Palmieri et al., 2011b). mef-class genes, which include some variants, are associated to a low-level resistance pattern affecting, among MLS antibiotics, only 14- and 15-membered macrolides (M phenotype) (Sutcliffe et al., 1996). mef(A) and mef(E) are widespread in S. pyogenes and S. pneumoniae, respectively, but they are also common in other streptococcal species (Varaldo et al., 2009). In S. suis, while mef(A) (Martel et al., 2003; Chu et al., 2009) and mef(E) (Hu et al., 2011a,c) have only occasionally been reported, erm(B) is found in >90% of macrolide-resistant pig isolates (Martel et al., 2001, 2003; Princivalli et al., 2009; Hoa et al., 2011); it has recently been reported also in human isolates (Manzin et al., 2008; Holden et al., 2009; Princivalli et al., 2009; Hoa et al., 2011). Although no further macrolide resistance determinants have been documented, the isolation of macrolide-resistant strains negative for the above genes (Martel et al., 2003; Princivalli et al., 2009; Hoa et al., 2011) suggests that other genetic determinants or target site mutations may occur.
Strains resistant to other antibiotics, such as β-lactams, aminoglycosides, trimethoprim–sulfamethoxazole, chloramphenicol, and fluoroquinolones, have been reported. The genetic basis of resistance to these antibiotics has only occasionally been investigated. Penicillin resistance was first reported in a human isolate in UK in 1980 (Shneerson et al., 1980) and more recently among pig isolates (Marie et al., 2002; Higgins and Gottschalk, 2005; Huang et al., 2005; Zhang et al., 2008). Penicillin-binding protein modifications (altered molecular weight and/or decreased affinity for penicillin) are involved in the resistance mechanism (Cain et al., 1995). Quite recently, resistance also to third-generation cephalosporins has been reported (Hu et al., 2011c). Aminoglycoside resistance has frequently been reported in S. suis (Touil et al., 1988; Wasteson et al., 1994; Marie et al., 2002; Tian et al., 2004; Wisselink et al., 2006; Hendriksen et al., 2008). Recently, genes coding for resistance to kanamycin [aminoglycoside-3′-phosphotransferase (aphA)] and to streptomycin [aminoglycoside-6′-adenyltransferase (aadE)] have been detected in multiresistant strains (Chen et al., 2007; Holden et al., 2009; Hu et al., 2011a; Palmieri et al., 2011a). Resistance to antifolate drugs has frequently been reported (Mengelers et al., 1989; Wisselink et al., 2006; Zhang et al., 2008). A dihydrofolate reductase (dhfr) resistance gene has been detected in the sequenced strain BM407 (Holden et al., 2009). Chloramphenicol resistance has rarely been reported (Takamatsu et al., 2003). However, a recent increase in chloramphenicol-resistant strains has been noted among human isolates in Vietnam (Hoa et al., 2011). Fluoroquinolone resistance has occasionally been described (Aarestrup et al., 1998; Escudero et al., 2007, 2011; Hendriksen et al., 2008; Hu et al., 2011c). Resistance is associated with single point mutations in the quinolone resistance-determining regions of ParC and GyrA (Escudero et al., 2007; Hu et al., 2011c), but a novel efflux pump has recently been described (Escudero et al., 2011).
Contribution of Exogenous Genetic Elements to the S. suis Resistome
Recent studies (Li et al., 2011; Palmieri et al., 2011a,c) and the analysis of sequenced genomes (Chen et al., 2007; Holden et al., 2009; Ye et al., 2009; Hu et al., 2011a,b,c) have provided significant insights into the S. suis resistome, leading to the identification of several genetic elements carrying resistance determinants for tetracyclines, macrolides, aminoglycosides, chloramphenicol, antifolate drugs, streptothricin, and cadmium salts. These elements, that include integrative and conjugative elements (ICEs), transposons, genomic islands (GIs), phages, and chimeric elements, are illustrated in Figure 1. A scheme of their integration sites into the S. suis core chromosome is shown in Figure 2.
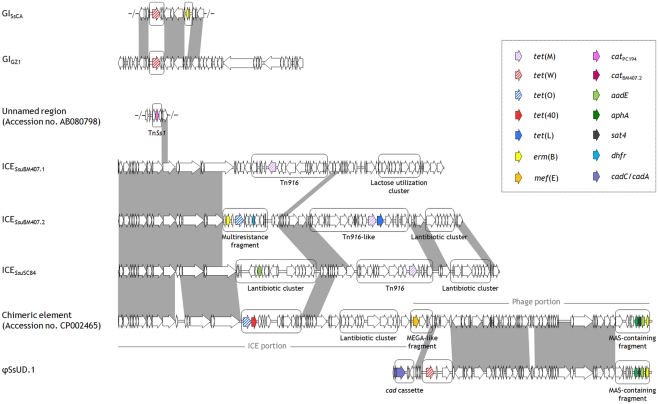
Figure 1.
ORF maps and genome organization of recognized S. suis genetic elements carrying resistance genes. ICESsuSC84 was chosen as a representative of the ICEs of ST7 strains. The ORFs, indicated as‘arrows pointing in the direction of transcription, are depicted as white arrows, except for resistance genes which are specified in the box. Cargo regions are enclosed in rounded rectangles. Gray areas between ORF maps denote >90% DNA identity (identities between cargo regions are not shown).
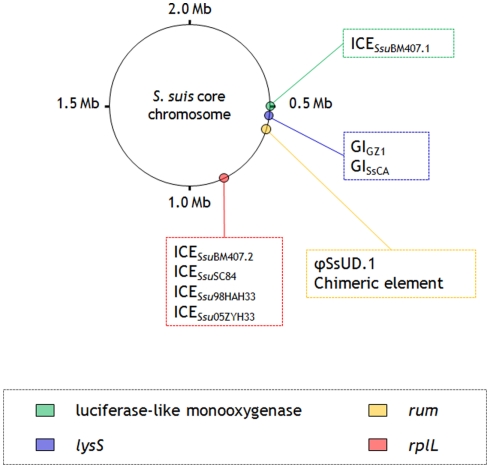
Figure 2.
Schematic illustration showing the S. suis core chromosome with the insertion sites of recognized genetic elements carrying resistance genes.
Five ICEs have been described in the sequenced genomes of ST7 (98HAH33, 05ZYH33, and SC84) and ST1 (BM407) human strains (Chen et al., 2007; Holden et al., 2009). Of these, four – ICESsu98HAH33, ICESsu05ZYH33, and ICESsuSC84 (~90 kb, three virtually identical elements), and ICESsuBM407.2 (~80 kb) – are closely related, except for cargo genes, to ICESde3396 of S. dysgalactiae subsp. equisimilis (Davies et al., 2009): they are integrated immediately downstream of the 50S ribosomal gene L7/L12 (rplL), they harbor a tyrosine family integrase, and share an almost identical set of genes for the conjugative machinery. ICESsu98HAH33, ICESsu05ZYH33, and ICESsuSC84 bear three distinct cargo regions: (i) a putative bacteriocin biosynthesis cluster that is disrupted by a putative integron containing the aadE gene; (ii) a Tn916 transposon, regularly carrying tet(M); and (iii) a cluster of genes associated with lantibiotic export/resistance. ICESsuBM407.2 contains three distinct cargo regions: (i) a Tn916-like element carrying, besides tet(M), tet(L) and a novel chloramphenicol acetyltransferase (cat) gene; (ii) a multiresistance fragment bearing tet(O), erm(B), and a dhfr gene; and (iii) a lantibiotic cluster similar to the one mentioned above. The fifth ICE (ICESsuBM407.1) displays a similar scaffold but contains a different integrase, belonging to serine recombinase family, which directs the insertion of this element into a luciferase-like monooxygenase gene (Holden et al., 2009). ICESsuBM407.1 contains two cargo regions: (i) a Tn916 transposon, regularly carrying tet(M), and (ii) a lactose utilization cluster. Structural and compositional analysis strongly suggests that all five ICEs have the potential to undergo excision and transfer. In particular, ICESsu05ZYH33 was transferred at high frequency to S. suis recipients in conjugation assays (Li et al., 2011). Another ICE (ICESsu32457, ~56 kb), recently identified in a pig isolate in Italy and shown to carry tet(O/W/32/O), is the first reported genetic support for a mosaic tet gene in streptococci (Palmieri et al., 2011c). ICESsu32457 is transferable to S. suis, S. pyogenes, S. agalactiae, and S. pneumoniae, and similar to ICESsu98HAH33, ICESsu05ZYH33, ICESsuSC84, and ICESsuBM407.2 mentioned above, it is closely related to ICESde3396 of S. dysgalactiae subsp. equisimilis except for cargo genes (Palmieri et al., submitted).
A tet(W)-carrying ~47 kb GI (here designated GIGZ1), located immediately downstream of the lysyl-tRNA synthetase chromosomal gene (lysS), has been identified in the sequenced genome of the Chinese strain GZ1 (Ye et al., 2009). An almost identical, non-transferable, tet(W)-carrying GI (GISsCA), differing from GIGZ1 only for the presence of an erm(B)-containing insertion, has recently been detected in an Italian human strain (Palmieri et al., 2011a).
In another Italian human strain, the presence of tet(W) has been reported in a phage (φSsUD.1, ~61 kb) (Palmieri et al., 2011a). φSsUD.1 carries a unique combination of antibiotic and heavy metal resistance genes resulting from the presence, besides tet(W), of an erm(B)-containing MAS (macrolide–aminoglycoside–streptothricin) – like fragment and a cadC/cadA cadmium efflux cassette. The MAS-like fragment closely resembles the one recently described in the pneumococcal transposons Tn6003 and Tn1545 (Cochetti et al., 2007, 2008). The resistance genes fitting in the φSsUD.1 phage scaffold differ from, but are in the same position as, the cargo genes carried by S. pyogenes phages such as φ10394.4 (Banks et al., 2003) and φm46.1 (Brenciani et al., 2010). φSsUD.1 is integrated at the 3′ end of a conserved RNA uracil methyltransferase (rum) gene and is transferable to S. pyogenes.
A ~124 kb chimeric element constituted of two portions, an ICE (~69 kb) and a phage (~55 kb), has recently been detected in the sequenced genome of JS14 (Hu et al., 2011a), integrated at the 3′ end of the rum gene. The ICE portion harbors a site-specific integrase, it displays a scaffold similar to the S. suis ICEs mentioned above, and contains two cargo regions, one bearing tet(O) in tandem with tet(40) and the other a bacteriocin gene cluster. The phage portion displays the typical modular organization of tailed phages, and closely resembles φSsUD.1 (Palmieri et al., 2011a). The right end of the phage portion bears an erm(B)-containing MAS-like fragment, like φSsUD.1; the left end bears a mega-like genetic structure, similar to the mef(E)-carrying mega element originally described in S. pneumoniae (Gay and Stephens, 2001), in the same position as the cadC/cadA cassette in φSsUD.1.
Beside the Tn916-family elements, only another transposon carrying resistance genes, TnSs1, has been described in S. suis (Takamatsu et al., 2003). It contains a cat gene showing 97% identity with catPC194 (Widdowson et al., 2000) flanked by direct repeats of an IS6-family element.
Concluding Remarks
Although the emergence of S. suis as a human pathogen has caused a flurry of research, current knowledge on the S. suis resistome is still fairly sketchy. This is mainly due to the so far limited number of studies of the genetic basis of resistance and to the redundancy of data from the strains sequenced, many of which are clonally related. Next-generation DNA sequencing will probably provide a wealth of new data in the next few years. The available information suggests that S. suis may contribute to the spread of antibiotic resistance genes to streptococcal human pathogens such as S. pyogenes, S. pneumoniae, and S. agalactiae, acting as a resistance reservoir. The notion is supported by studies demonstrating that S. suis harbors mobile resistance genetic elements, similar to those of the above-mentioned streptococci, that share the same conserved chromosomal insertion sites. Thus, S. suis is a paradigmatic example of possible intersections between animal and human resistomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partly supported by the Italian Ministry of Education, University, and Research.
References
- Aarestrup F. M., Rasmussen S. R., Artursson K., Jensen N. E. (1998). Trends in the resistance to antimicrobial agents of Streptococcus suis isolates from Denmark and Sweden. Vet. Microbiol. 63, 71–80 10.1016/S0378-1135(98)00228-4 [DOI] [PubMed] [Google Scholar]
- Banks D. J., Porcella S. F., Barbian K. D., Martin J. M., Musser J. M. (2003). Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J. Infect. Dis. 188, 1898–1908 10.1086/379897 [DOI] [PubMed] [Google Scholar]
- Brenciani A., Bacciaglia A., Vignaroli C., Pugnaloni A., Varaldo P. E., Giovanetti E. (2010). Characterization of φm46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54, 221–229 10.1128/AAC.00499-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain D., Malouin F., Dargis M., Harel J., Gottschalk M. (1995). Alterations in penicillin-binding proteins in strains of Streptococcus suis possessing moderate and high levels of resistance to penicillin. FEMS Microbiol. Lett. 130, 12–17 10.1111/j.1574-6968.1995.tb07708.x [DOI] [PubMed] [Google Scholar]
- Chander Y., Oliveira S. R., Goyal S. M. (2011). Identification of the tet(B) resistance gene in Streptococcus suis. Vet. J. 189, 359–360 10.1016/j.tvjl.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Chen C., Tang J., Dong W., Wang C., Feng Y., Wang J., Zheng F., Pan X., Liu D., Li M., Song Y., Zhu X., Sun H., Feng T., Guo Z., Ju A., Ge J., Dong Y., Sun W., Jiang Y., Wang J., Yan J., Yang H., Wang X., Gao G. F., Yang R., Wang J., Yu J. (2007). A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE 2, e315. 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Roberts M. C. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. W., Cheung T. K. M., Chu M. Y., Tsang V. Y. M., Fung J. T. L., Kam K. M., Lo J. Y. C. (2009). Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int. J. Antimicrob. Agents 34, 181–182 10.1016/j.ijantimicag.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Cochetti I., Tili E., Mingoia M., Varaldo P. E., Montanari M. P. (2008). erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob. Agents Chemother. 52, 1285–1290 10.1128/AAC.01457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochetti I., Tili E., Vecchi M., Manzin A., Mingoia M., Varaldo P. E., Montanari M. P. (2007). New Tn916-related elements causing erm(B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. J. Antimicrob. Chemother. 60, 127–131 10.1093/jac/dkm120 [DOI] [PubMed] [Google Scholar]
- Davies M. R., Shera J., Van Domselaar G. H., Sriprakash K. S., McMillan D. J. (2009). A novel integrative conjugative element mediates genetic transfer from group G streptococcus to other β-hemolytic streptococci. J. Bacteriol. 191, 2257–2265 10.1128/JB.01624-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero J. A., San Millan A., Catalan A., de la Campa A. G., Rivero E., Lopez G., Dominguez L., Moreno M. A., Gonzalez-Zorn B. (2007). First characterization of fluoroquinolone resistance in Streptococcus suis. Antimicrob. Agents Chemother. 51, 777–782 10.1128/AAC.00972-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero J. A., San Millan A., Gutierrez B., Hidalgo L., La Ragione R. M., Abuoun M., Galimand M., Ferrandiz M. J., Dominguez L., De La Campa A. G., Gonzalez-Zorn B. (2011). Fluoroquinolone efflux in Streptococcus suis is mediated by SatAB and not by SmrA. Antimicrob. Agents Chemother. 55, 5850–5860 10.1128/AAC.00498-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay K., Stephens D. S. (2001). Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184, 56–65 10.1086/321001 [DOI] [PubMed] [Google Scholar]
- Gottschalk M., Xu J., Calzas C., Segura M. (2010). Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5, 371–391 10.2217/fmb.10.2 [DOI] [PubMed] [Google Scholar]
- Hendriksen R. S., Mevius D. J., Schroeter A., Teale C., Jouy E., Butaye P., Franco A., Utinane A., Amado A., Moreno M., Greko C., Stärk K. D., Berghold C., Myllyniemi A. L., Hoszowski A., Sunde M., Aarestrup F. M. (2008). Occurrence of antimicrobial resistance among bacterial pathogens and indicator bacteria in pigs in different European countries from year 2002–2004: the ARBAO-II study. Acta Vet. Scand. 50, 19. 10.1186/1751-0147-50-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R., Gottschalk M. (2005). “Streptococcal diseases,” in Diseases of Swine, eds Straw B. E., D’Allaire S., Mengeling W. L., Taylor D. J. (Ames: Iowa State University Press; ), 769–783 [Google Scholar]
- Hill J. E., Gottschalk M., Brousseau R., Harel J., Hemmingsen S. M., Goh S. H. (2005). Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 107, 63–69 10.1016/j.vetmic.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Hoa N. T., Chieu T. T., Nghia H. D., Mai N. T., Anh P. H., Wolbers M., Baker S., Campbell J. I., Chau N. V., Hien T. T., Farrar J., Schultsz C. (2011). The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997–2008. BMC Infect. Dis. 6, 11–16 10.1186/1471-2334-11-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T., Hauser H., Sanders M., Ngo T. H., Cherevach I., Cronin A., Goodhead I., Mungall K., Quail M. A., Price C., Rabbinowitsch E., Sharp S., Croucher N. J., Chieu T. B., Mai N. T., Diep T. S., Chinh N. T., Kehoe M., Leigh J. A., Ward P. N., Dowson C. G., Whatmore A. M., Chanter N., Iversen P., Gottschalk M., Slater J. D., Smith H. E., Spratt B. G., Xu J., Ye C., Bentley S., Barrell B. G., Schultsz C., Maskell D. J., Parkhill J. (2009). Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE 4, e6072. 10.1371/journal.pone.0006072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Yang M., Zhang A., Wu J., Chen B., Hua Y., Yu J., Xiao J., Jin M. (2011a). Complete genome sequence of Streptococcus suis serotype 14 strain JS14. J. Bacteriol. 193, 2375–2376 10.1128/JB.00083-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Yang M., Zhang A., Wu J., Chen B., Hua Y., Yu J., Chen H., Xiao J., Jin M. (2011b). Complete genome sequence of Streptococcus suis serotype 3 strain ST3. J. Bacteriol. 193, 3428–3429 10.1128/JB.00083-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Yang M., Zhang A., Wu J., Chen B., Hua Y., Yu J., Xiao J., Jin M. (2011c). Comparative genomics study of multi-drug-resistance mechanisms in the antibiotic-resistant Streptococcus suis R61 strain. PLoS ONE 6, e24988. 10.1371/journal.pone.0024988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. T., Teng L. J., Ho S. W., Hsueh P. R. (2005). Streptococcus suis infection. J. Microbiol. Immunol. Infect. 38, 306–313 [PubMed] [Google Scholar]
- Kazimierczak K. A., Rincon M. T., Patterson A. J., Martin J. C., Young P., Flint H. J., Scott K. P. (2008). A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob. Agents Chemother. 52, 4001–4009 10.1128/AAC.00308-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Leigh J. A., Heath P. J., Luque I., Tarradas C., Dowson C. G., Whatmore A. M. (2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40, 3671–3680 10.1128/JCM.40.10.3671-3680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Shen X., Yan J., Han H., Zheng B., Liu D., Cheng H., Zhao Y., Rao X., Wang C., Tang J., Hu F., Gao G. F. (2011). GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79, 1670–1683 10.1111/j.1365-2958.2011.07553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun Z. R., Wang Q. P., Chen X. G., Li A. X., Zhu X. Q. (2007). Streptococcus suis: an emerging zoonotic pathogen. Lancet Infect. Dis. 7, 201–209 10.1016/S1473-3099(07)70001-4 [DOI] [PubMed] [Google Scholar]
- Mai N. T. H., Hoa N. T., Nga T. V. T., Linh L. D., Chau T. T. H., Sinh D. X., Phu N. H., Chuong L. V., Diep T. S., Campbell J., Nghia H. D. T., Minh T. N., Chau N. V. V., de Jong M. D., Chinh N. T., Hien T. T., Farrar J., Schultsz C. (2008). Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46, 659–667 10.1086/527385 [DOI] [PubMed] [Google Scholar]
- Manzin A., Palmieri C., Serra C., Saddi B., Princivalli M. S., Loi G., Angioni G., Tiddia F., Varaldo P. E., Facinelli B. (2008). Streptococcus suis meningitis with no evidence of animal contact. Emerging Infect. Dis. 14, 1946–1948 10.3201/eid1412.080679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie J., Morvan H., Berthelot-Hérault F., Sanders P., Kempf I., Gautier-Bouchardon A. V., Jouy E., Kobisch M. (2002). Antimicrobial susceptibility of Streptococcus suis isolated from swine in France and from humans in different countries between 1996 and 2000. J. Antimicrob. Chemother. 50, 201–209 10.1093/jac/dkf099 [DOI] [PubMed] [Google Scholar]
- Martel A., Baele M., Devriese L. A., Goossens H., Wisselink H. J., Decostere A., Haesebrouck F. (2001). Prevalence and mechanism of resistance against macrolides and lincosamides in Streptococcus suis isolates. Vet. Microbiol. 83, 287–297 10.1016/S0378-1135(01)00426-6 [DOI] [PubMed] [Google Scholar]
- Martel A., Meulenaere V., Devriese L. A., Decostere A., Haesebrouck F. (2003). Macrolide and lincosamide resistance in the gram-positive nasal and tonsillar flora of pigs. Microb. Drug Resist. 84, 27–32 [DOI] [PubMed] [Google Scholar]
- McEwen S. A., Fedorka-Cray P. J. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3), S93–S106 10.1086/340246 [DOI] [PubMed] [Google Scholar]
- Mengelers M. J., van Klingeren B., van Miert A. S. (1989). In vitro antimicrobial activity of sulfonamides against some porcine pathogens. Am. J. Vet. Res. 50, 1022–1028 [PubMed] [Google Scholar]
- Palmieri C., Princivalli M. S., Brenciani A., Varaldo P. E., Facinelli B. (2011a). Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55, 631–636 10.1128/AAC.00965-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri C., Creti R., Imperi M., Gherardi G., Baldassarri L., Magi G., Bagnarelli P., Facinelli B. (2011b). “erm(T)-carrying pRW35 can be mobilized by a co-resident integrative conjugative element (ICESde5580) in Streptococcus dysgalactiae subsp. equisimilis,” in XVIII Lancefield International Symposium (Palermo: EAC srl; ), 259 [Google Scholar]
- Palmieri C., Magi G., Mingoia M., Varaldo P. E., Facinelli B. (2011c). “Characterization of a tet(O/W/32/O)-carrying integrative conjugative element (ICESsu32457) in Streptococcus suis,” in XVIII Lancefield International Symposium (Palermo: EAC srl; ), 231 [Google Scholar]
- Patterson A. J., Rincon M. T., Flint H. J., Scott K. P. (2007). Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob. Agents Chemother. 51, 1115–1118 10.1128/AAC.00725-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perch B., Kristjansen P., Skadhauge K. (1968). Group R streptococci pathogenic for man: two cases of meningitis and one fatal case of sepsis. Acta Pathol. Microbiol. Scand. 74, 69–76 10.1111/j.1699-0463.1968.tb03456.x [DOI] [PubMed] [Google Scholar]
- Princivalli M. S., Palmieri C., Magi G., Vignaroli C., Manzin A., Camporese A., Barocci S., Magistrali C., Facinelli B. (2009). Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003–2007). Euro Surveill. 14 pii, 19310. [DOI] [PubMed] [Google Scholar]
- Roberts M. C. (2005). Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203 10.1016/j.femsle.2005.02.034 [DOI] [PubMed] [Google Scholar]
- Shneerson J. M., Chattopadhyay B., Murphy M. F., Fawcett I. W. (1980). Permanent perceptive deafness due to Streptococcus suis type II infection. J. Laryngol. Otol. 94, 425–427 10.1017/S0022215100089040 [DOI] [PubMed] [Google Scholar]
- Smith T. C., Capuano A. W., Boese B., Myers K. P., Gray G. C. (2008). Exposure to Streptococcus suis among US swine workers. Emerging Infect. Dis. 14, 1925–1927 10.3201/eid1412.080162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats J. J., Feder I., Okwumabua O., Chengappa M. M. (1997). Streptococcus suis: past and present. Vet. Res. Commun. 21, 381–407 10.1023/A:1005870317757 [DOI] [PubMed] [Google Scholar]
- Stanton T. B., Humphrey S. B. (2003). Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69, 3874–3882 10.1128/AEM.69.7.3874-3882.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J., Tait-Kamradt A., Wondrack L. (1996). Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D., Osaki M., Sekizaki T. (2003). Chloramphenicol resistance transposable element TnSs1 of Streptococcus suis, a transposon flanked by IS6-family elements. Plasmid 49, 143–151 10.1016/S0147-619X(02)00149-X [DOI] [PubMed] [Google Scholar]
- Thaker M., Spanogiannopoulos P., Wright G. D. (2010). The tetracycline resistome. Cell. Mol. Life Sci. 67, 419–431 10.1007/s00018-009-0172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Aarestrup F. M., Lu C. P. (2004). Characterization of Streptococcus suis serotype 7 isolates from diseased pigs in Denmark. Vet. Microbiol. 103, 55–62 10.1016/j.vetmic.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Touil F., Higgins R., Nadeau M. (1988). Isolation of Streptococcus suis from diseased pigs in Canada. Vet. Microbiol. 17, 171–177 10.1016/0378-1135(88)90008-9 [DOI] [PubMed] [Google Scholar]
- Varaldo P. E., Montanari M. P., Giovanetti E. (2009). Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53, 343–353 10.1128/AAC.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteson Y., Høie S., Roberts M. C. (1994). Characterization of antibiotic resistance in Streptococcus suis. Vet. Microbiol. 41, 41–49 10.1016/0378-1135(94)90134-1 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Nghia H. D., Taylor W., Schultsz C. (2009a). Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48, 617–625 10.1086/596763 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Nguyen H. N., Taylor W., Lien T. T., Ngo H. T., Nguyen T. Q., Nguyen B. N., Nguyen H. H., Nguyen H. M., Nguyen C. T., Dao T. T., Nguyen T. V., Fox A., Farrar J., Schultsz C., Nguyen H. D., Nguyen K. V., Horby P. (2009b). Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS ONE 4, e5973. 10.1371/journal.pone.0005973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdowson C. A., Adrian P. V., Klugman K. P. (2000). Acquisition of chloramphenicol resistance by the linearization and integration of the entire staphylococcal plasmid pC194 into the chromosome of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 393–395 10.1128/AAC.44.2.393-395.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink H. J., Veldman K. T., Van den Eede C., Salmon S. A., Mevius D. J. (2006). Quantitative susceptibility of Streptococcus suis strains isolated from diseased pigs in seven European countries to antimicrobial agents licensed in veterinary medicine. Vet. Microbiol. 113, 73–82 10.1016/j.vetmic.2005.10.035 [DOI] [PubMed] [Google Scholar]
- Witte W. (1997). Impact of antibiotic use in animal feeding on resistance of bacterial pathogens in humans. Ciba Found. Symp. 207, 61–71 [DOI] [PubMed] [Google Scholar]
- Ye C., Bai X., Zhang J., Jing H., Zheng H., Du H., Cui Z., Zhang S., Jin D., Xu Y., Xiong Y., Zhao A., Luo X., Sun Q., Gottschalk M., Xu J. (2008). Spread of Streptococcus suis sequence type 7, China. Emerging Infect. Dis. 14, 787–791 10.3201/eid1405.070437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Zheng H., Zhang J., Jing H., Wang L., Xiong Y., Wang W., Zhou Z., Sun Q., Luo X., Du H., Gottschalk M., Xu J. (2009). Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 199, 97–107 10.1086/594370 [DOI] [PubMed] [Google Scholar]
- Ye C., Zhu X., Jing H., Du H., Segura M., Zheng H., Kan B., Wang L., Bai X., Zhou Y., Cui Z., Zhang S., Jin D., Sun N., Luo X., Zhang J., Gong Z., Wang X., Wang L., Sun H., Li Z., Sun Q., Liu H., Dong B., Ke C., Yuan H., Wang H., Tian K., Wang Y., Gottschalk M., Xu J. (2006). Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerging Infect. Dis. 12, 1203–1212 10.3201/eid1708.060232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Jing H., Chen Z., Zheng H., Zhu X., Wang H., Wang S., Liu L., Zu R., Luo L., Xiang N., Liu H., Liu X., Shu Y., Lee S. S., Chuang S. K., Wang Y., Xu J., Yang W., the Streptococcus suis Study Groups (2006). Human Streptococcus suis outbreak, Sichuan, China. Emerging Infect. Dis. 12, 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Ning Y., Zhang Z., Song L., Qiu H., Gao H. (2008). In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet. Microbiol. 131, 386–392 10.1016/j.vetmic.2008.04.005 [DOI] [PubMed] [Google Scholar]