Abstract
Global climate change is having a significant effect on the distributions of a wide variety of species, causing both range shifts and population extinctions. To date, however, no consensus has emerged on how these processes will affect the range-wide genetic diversity of impacted species. It has been suggested that species that recolonized from low-latitude refugia might harbour high levels of genetic variation in rear-edge populations, and that loss of these populations could cause a disproportionately large reduction in overall genetic diversity in such taxa. In the present study, we have examined the distribution of genetic diversity across the range of the seaweed Chondrus crispus, a species that has exhibited a northward shift in its southern limit in Europe over the last 40 years. Analysis of 19 populations from both sides of the North Atlantic using mitochondrial single nucleotide polymorphisms (SNPs), sequence data from two single-copy nuclear regions and allelic variation at eight microsatellite loci revealed unique genetic variation for all marker classes in the rear-edge populations in Iberia, but not in the rear-edge populations in North America. Palaeodistribution modelling and statistical testing of alternative phylogeographic scenarios indicate that the unique genetic diversity in Iberian populations is a result not only of persistence in the region during the last glacial maximum, but also because this refugium did not contribute substantially to the recolonization of Europe after the retreat of the ice. Consequently, loss of these rear-edge populations as a result of ongoing climate change will have a major effect on the overall genetic diversity of the species, particularly in Europe, and this could compromise the adaptive potential of the species as a whole in the face of future global warming.
Keywords: Chondrus crispus, distribution range, glacial refugia, phylogeography, population genetics
1. Introduction
Global climate change is having a significant effect on the distributions of a wide variety of taxa, with well-documented cases of range shifts [1,2] and even population extinctions [3,4]. Although it is now accepted that knowledge of the levels and patterns of genetic diversity within and between populations is crucial for formulating conservation strategies for potentially threatened species, no clear consensus has emerged on the relative importance of historical and contemporary factors in determining the distribution of genetic variation across species' ranges. It is generally assumed that population genetic parameters such as effective population size (Ne), genetic diversity and levels of gene flow would reflect the ‘abundant-centre’ or ‘central-marginal’ model of species' distributions [5,6]. In this model, optimal conditions at the centre of a species' range lead to greater abundance in central populations than in peripheral populations, which are likely to exist in sub-optimal conditions and thus be smaller and more isolated [7,8] (but see [9]). This should result in high levels of genetic diversity at the centre of the range, with a decrease in diversity coupled with an increase in population differentiation towards the range edge owing to elevated drift and/or selection. It has also been argued, however, that contemporary processes have less effect on the distribution of genetic diversity across species' ranges than longer-term processes, such as postglacial recolonization events associated with previous fluctuations in Earth's climate [10–13]. These have generally resulted in temperate terrestrial species showing greatest genetic diversity at lower latitudes owing to the ‘classic’ paradigm of recolonization from low-latitude refugia [11] (but see [13]). Thus, the conservation value of peripheral populations, particularly those at the rear edge, is of ongoing interest and debate since they could contain unique genetic variation. This would be especially important where one or more refugial populations have not contributed significantly to the recolonization of previously glaciated areas [12,14].
The impact of climate change on the distributions of intertidal species is of particular interest for several reasons. As well as increases in air and sea temperatures, coastal ecosystems are subject to additional effects of climate change, such as rises in sea level and a decrease in sea water pH associated with increased atmospheric carbon dioxide levels [15,16]. Many marine species are believed to exist at, or close to, their thermal tolerance limits [17], and thus are considered good early indicators of the overall effects of climate change on species' distributional ranges [18]. Unlike many terrestrial species, intertidal taxa generally lack the scope for altitudinal migration to counter the effects of warming climate and, on the whole, their ranges appear to be following the same patterns of poleward expansions in distribution as observed in terrestrial species [15,19]. A recent study on intertidal algae, however, highlighted the difficulties in making generalizations concerning range shifts, particularly when comparing warm- and cold-adapted species, since warm-water species that exhibited a range shift all expanded northwards, while cold-tolerant species displayed no consistent pattern [20].
Chondrus crispus Stackhouse is one species of seaweed that has exhibited a significant range shift in the last few decades, and is thus a potential indicator of the effects of climate change. Lima et al. [20] reported a northward shift in the southern limit of Portuguese populations of 180 km since 1971: these populations are morphologically distinct from all other populations of the species (figure S1 in the electronic supplementary material). Portugal is the southern limit of the species' distribution range in the eastern North Atlantic, where it occurs as far north as approximately 69° N in Norway and on the southern coast of Iceland. In the western Atlantic, it is found between New Jersey and southeastern Labrador (figure 1). Its congeners are Pacific species and, following a trans-Arctic migration, C. crispus is now endemic to the northern North Atlantic [21]. The species' present-day distribution is primarily determined by high temperatures, with spore mortality recorded at temperatures as low as 21°C [22]. Consequently, Iberian populations are probably on the cusp of their upper thermal tolerance limit.
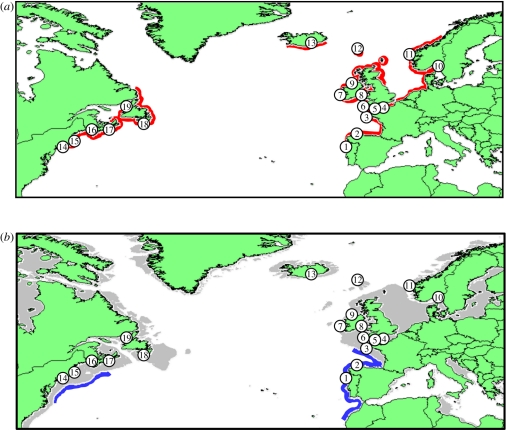
Figure 1.
Maps showing locations of C. crispus populations analysed in the present study. Numbers refer to locations in table S1 in the electronic supplementary material. (a) Red regions indicate the current distribution of C. crispus. (b) Blue regions show putative refugial areas identified by the palaeodistribution modelling, with grey shading indicating dry land at the LGM.
Phylogeographic studies can provide valuable insights into how species have previously responded to episodes of climatic change and the way in which these have shaped the distribution of genetic diversity across entire ranges. North Atlantic marine organisms exhibit complex patterns of species survival in various refugial areas during the last glacial maximum (LGM; ca 18–21 kyr ago), with different refugia contributing to postglacial recolonization to varying degrees [23]. Such complex scenarios have important implications for how species respond to climate change in two ways. Firstly, if species have also persisted in cryptic northern refugia as opposed to surviving solely in low-latitude regions, their dispersal capabilities in the face of previous phases of global warming may be overestimated [13,24,25]. The existence of these northerly refugia, if not recognized as such, would also confound to some degree the expectation of reduced genetic variation at the leading edge [12,13]. Secondly, where southern refugia have not contributed to postglacial recolonization, they may represent reservoirs of unique genetic variation that are under immediate threat from climate change [12,14]. Previous phylogeographic studies on North Atlantic seaweeds with similar distributions to C. crispus [26–28] have suggested that both scenarios are likely.
In the present study, we used a phylogeographic approach to test whether populations from the southern limits of the distribution range of C. crispus exhibit unique genetic variation that may be lost as a result of climate change. We have sampled across the entire range of the species on both sides of the Atlantic, since the relative importance of leading-edge versus rear-edge peripheral populations is still not fully understood [12,29,30]. Given that extinction of rear-edge populations is more likely than migration [1,29,31,32], they may merit special conservation effort in the face of global warming, particularly if they are genetically distinct.
2. Material and methods
(a). Sample collection and DNA extraction
A total of 354 samples were collected from 19 populations (14–23 individuals per population) across the range of C. crispus, including the southern edge of both western and eastern coasts of the North Atlantic (table S1 in the electronic supplementary material; figure 1). Haploid gametophytes were identified using the resorcinol staining test for κ-carrageenan [33] and total genomic DNA extracted using the Qiagen Plant DNeasy kit.
(b). Single nucleotide polymorphism analysis of mitochondrial DNA
The trnI intron of the C. crispus mitochondrial genome (EMBL accession no. Z47547) was analysed for SNP polymorphisms. A 498 bp fragment was amplified in an ascertainment set of six individuals from each population using the following primers: trnI-5—TGAGTCTAGATTGATTCGAACAATC and trnI-3—GAGTCTATAGCTTAAAGGTTAGAG. PCR was carried out on an MWG Primus thermal cycler using the following parameters: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 2 min and a final extension at 72°C for 5 min. Polymerase chain reaction (PCR) was carried out in a total volume of 20 µl containing 200 ng genomic DNA, 20 pmol of each primer, 1×PCR reaction buffer, 200 µM each dNTP, 2.5 mM MgCl2 and 0.5 U GoTaq Flexi DNA polymerase (Promega). 5 µl PCR product were resolved on 1.5 per cent agarose gels and visualized by ethidium bromide staining, and the remaining 15 µl sequenced commercially (Macrogen, Korea).
Mitochondrial SNPs identified in more than a single individual in the ascertainment set were screened in all 354 samples using allele-specific PCR (AS-PCR), which uses a competitive PCR approach to amplify either of two alleles at a specific position. AS-PCR primers were designed as described by Provan et al. [34] and are given in table S2 in the electronic supplementary material. The AS-PCR protocol was as follows: initial denaturation at 94°C for 3 min followed by 11 touchdown cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min (−0.7°C per cycle), extension at 72°C for 1 min followed by 24 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 1 min and a final extension at 72°C for 5 min. PCR was carried out in a total volume of 10 µl containing 100 ng genomic DNA, 10 pmol of each primer, 1×PCR reaction buffer, 200 µM each dNTP, 2.5 mM MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega). PCR products were resolved on 2 per cent agarose gels and visualized by ethidium bromide staining.
(c). Analysis of single-copy nuclear DNA sequences
Primers were designed to amplify 20 single-copy nuclear DNA (scnDNA) regions based on expressed sequence tag (EST) data from GenBank. Accession numbers and primers are given in table S3 in the electronic supplementary material. After an initial examination of levels of polymorphism and use of the Hudson & Kaplan [35] test in DNAsp (v. 4.90.1) [35,36] to eliminate regions exhibiting recombination (nine of the 20 regions), the two most variable loci (CO653303 and CO653370) were selected for analysis. PCR was carried out on eight individuals from each population as described for the mitochondrial DNA (mtDNA) except that an annealing temperature of 58°C was used, and the PCR products sequenced commercially (Macrogen, Korea).
(d). Microsatellite analysis
Microsatellite markers were developed by searching C. crispus EST sequences in the GenBank database for all trinucleotide repeat motifs, as described by Provan et al. [37]. Primer sequences are given in table S4 in the electronic supplementary material. All 354 individuals were genotyped. PCR was carried out in a total volume of 10 µl containing 100 ng genomic DNA, 10 pmol of dye-labelled M13 primer (6-FAM or HEX), 1 pmol of tailed forward primer, 10 pmol reverse primer, 1×PCR reaction buffer, 200 µM each dNTP, 2.5 mM MgCl2 and 0.25 U GoTaq Flexi DNA polymerase (Promega). PCR was carried out on an MWG Primus thermal cycler using the following parameters: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 2 min and a final extension at 72°C for 5 min. Genotyping was carried out on an AB3730xl capillary genotyping system. Allele sizes were scored using ROX-500 size standards and were checked by comparison with previously sized control samples. Scoring was carried out using the Genemapper software package (v. 4.1; Applied Biosystems).
(e). Palaeodistribution modelling
Ecological niche modelling was carried out to determine suitable climate envelopes for C. crispus at the LGM (ca 18 kyr ago) using the maximum entropy approach implemented in the Maxent software package (v. 3.2.1; [38]). Present-day species occurrence data for C. crispus were downloaded from the Global Biodiversity Information Facility data portal (www.gbif.org). A distribution model based on current-day sea-surface temperature (SST) at 1° resolution [39] was generated using Maxent with the default parameters for convergence threshold (10−5) and number of iterations (500), and projected onto reconstructed LGM data [39] to identify potential refugial areas. Duplicate records from the same locality were removed to reduce the effects of spatial autocorrelation. A presence threshold was determined using the sensitivity–specificity sum maximization approach [40] and the performance of the model was tested using 25 per cent of the occurrence data points to determine the area under the receiver operating characteristic (ROC) curve.
(f). Data analysis
Gene diversity (H) based on mtDNA haplotypes and microsatellites and numbers of unique haplotypes/alleles were calculated for each population. In addition, populations were also grouped into regions based on putative refugia and recolonized areas representing Iberia (population nos 1 and 2), English Channel (population nos 3–6), Ireland (population nos 7–9), Scandinavia (population nos 10 and 13), Northern Europe (population nos 11 and 12) and North America (population nos 14–19). Levels of within-population and within-region gene diversity were calculated using the Arlequin software package (v. 3.01) [41]. Owing to small sample sizes (n = 8 in each case), diversity statistics based on the two scnDNA regions were not calculated.
To evaluate alternative recolonization scenarios for Europe, we employed a model testing approach as described by Dépraz et al. [42]. Putative refugial regions were identified based on a combination of the palaeodistribution modelling, the presence of private haplotypes and previously published phylogeographic evidence (summarized in table S5 in the electronic supplementary material). For computational feasibility, populations were pooled into groups representing refugia (Iberia and English Channel—two separate groups as defined previously) and recolonized areas (Ireland, Northern Europe and Scandinavia—three separate groups), and maximum-likelihood migration rates between groups and values of Θ were calculated from the two-locus scnDNA data using Migrate-N (v. 3.0.3) [43]. For each recolonization model, runs were started using values of Θ and M (migration rate) calculated from FST values and consisted of 10 short chains of 50 000 generations from which trees were sampled at 50 generation intervals (1000 trees sampled) followed by three long chains of 1 million generations from which trees were sampled at 100 generation intervals (10 000 trees sampled). The first 20 000 generations were discarded as a burn-in period and the results of the final chains were combined to generate values of Θ and M. The analysis was then re-run based on an unconstrained migration model to carry out likelihood ratio tests relative to the various recolonization models. These were then used to calculate the Akaike information criterion (AIC) [44] to obtain relative measures of support for each model. The equivalent analysis was not carried out for North America owing to the difficulty in identifying precisely the location of refugial areas.
3. Results
(a). Mitochondrial single nucleotide polymorphism analysis
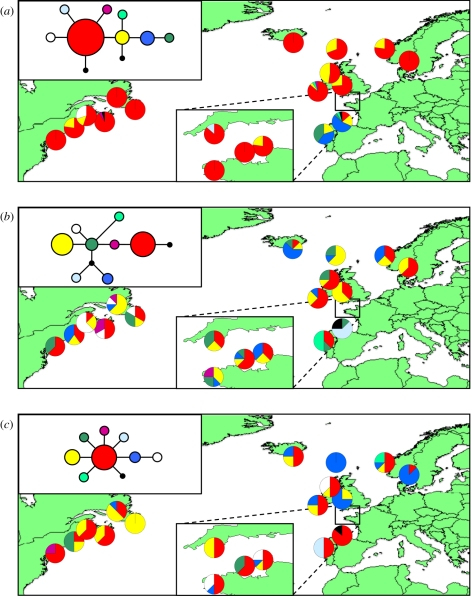
Seven SNPs were detected in the ascertainment set and AS-PCR of these SNPs in 356 individuals from 19 populations revealed a total of 10 haplotypes (figure 2; table S6a,b in the electronic supplementary material). The dominant haplotype (shown in red) was found in 18 of the 19 populations studied, the exception being the Portuguese population. Three of the remaining haplotypes were endemic to Iberia, and were found in approximately 75 per cent of the individuals studied from the Portugal (no. 1) and Spain (no. 2) populations. A further two haplotypes were endemic to North America, and a single haplotype was found only in the Torquay (no. 6) population in the English Channel. All populations from the recolonized areas of Europe displayed one or both of the two most common haplotypes with the exception of the Galway (no. 7) population, where two individuals each possessed a haplotype found in the Iberian populations. Gene diversity values based on mtDNA SNP haplotypes (table S6a,b in the electronic supplementary material) by population ranged from zero (several populations) to 0.692 (Spain), and by region ranged from 0.214 (Northern Europe) to 0.714 (Iberia).
Figure 2.
Maps showing distribution of haplotypes revealed by (a) mitochondrial SNP analysis (mtDNA) and (b,c) sequencing of two single-copy nuclear loci (CO653303 and CO653370). Insets show genetic relationships between haplotypes.
(b). Single-copy nuclear DNA analysis
The two scnDNA loci, CO653303 and CO653370, exhibited 10 and 9 haplotypes, respectively (figure 2; table S6a in the electronic supplementary material). The Iberian populations possessed four and two private haplotypes at the two loci, while North American populations contained two and one endemic haplotypes, respectively (figure 2; table S6b in the electronic supplementary material). For both loci, recolonized European populations shared all their haplotypes with English Channel populations, with the exception of a single private haplotype in the Øygarden (no. 11) population at locus CO653370.
(c). Microsatellite analysis
Analysis of eight microsatellite loci revealed between 2 and 12 alleles per locus (average 5.25; table S6a,b in the electronic supplementary material). Gene diversity values by population ranged from 0.105 (Øygarden) to 0.436 (Portugal), and by region ranged from 0.181 (Northern Europe) to 0.343 (Iberia). Six populations exhibited private alleles, ranging from one in the Portugal and New Brunswick (no. 17) populations to three in the Roscoff (no. 4) population. Based on regions, two private alleles were found in Ireland, Northern Europe and North America, three in the English Channel and four in Iberia.
(d). Palaeodistribution modelling
For the ecological niche model generated by Maxent under current SSTs, the value of 0.823 for the area under the ROC curve indicated a prediction that was better than random. A binomial test of omission also indicated that the model predictions were significantly better than random (p < 0.001) for all typical threshold values. Palaeodistribution modelling based on reconstructed SSTs suggested the presence of refugial areas for C. crispus on either side of the North Atlantic during the LGM (figure 1). On the western side of the North Atlantic, the potential distribution was limited to latitudes between approximately 37° N and approximately 44° N, which includes the area around Georges Bank and extends northwards to the Nova Scotian shelf. These latitudes correspond to those inhabited by the present-day Long Island Sound, New Hampshire, Nova Scotia and New Brunswick (nos 14–17) populations, although their current locations would have been dry land at the LGM owing to the decrease in sea levels. The modelled LGM distribution on the eastern side of the North Atlantic was more extensive, ranging from approximately 30° N to approximately 50° N, and included the present-day Iberian populations. It should be borne in mind, however, that the relatively low resolution (1°) of the SST grids used means that they might not completely reflect true intertidal temperatures, either in the current day or at the LGM. Nevertheless, they do provide a broad-scale indication of species occurrence.
(e). Postglacial recolonization model selection
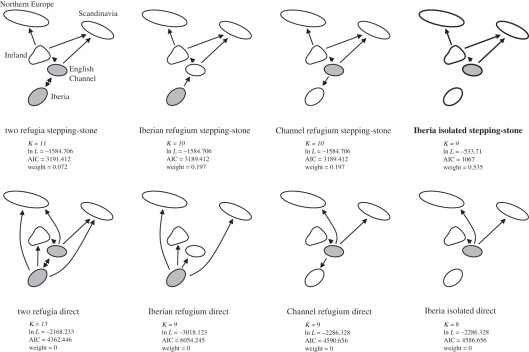
Based on the results of the palaeodistribution modelling, the occurrence of private haplotypes/alleles (table S6a,b in the electronic supplementary material) and evidence from previous phylogeographic studies (see §2), we identified three putative refugia: Iberia and the English Channel in the northeastern North Atlantic, and a further refugium in the northwestern North Atlantic. The precise location of the refugial area(s) on the western side of the Atlantic could not be determined based on the same criteria, but the occurrence of private haplotypes/alleles for all three marker types analysed suggests in situ persistence of C. crispus during the LGM. For Europe, the model testing (figure 3) indicated that a stepping-stone pattern of recolonization of Europe from the English Channel refugium with the Iberian populations remaining isolated was the most likely scenario (AIC = 1067; weight = 0.535). All scenarios involving stepping-stone recolonization were deemed more probable than any of those based on direct recolonization from refugia.
Figure 3.
Results of the model selection analysis. For each model, areas in grey indicate refugia from which recolonization took place and arrows indicate postglacial gene flow. ln L: log-likelihood of parameter set; K: number of free parameters for each model; AIC: Akaike information criterion; weight: relative support for each model. The most likely model is shown in bold.
4. Discussion
The findings of the present study suggest that while rear-edge populations of C. crispus in the European North Atlantic are reservoirs of unique genetic variation, this is not the case for the corresponding rear-edge populations in North America. Because the Iberian refugium did not contribute to the postglacial recolonization of the northern North Atlantic by C. crispus to the same extent as the English Channel populations, probably owing to the general lack of suitable substrate in the Bay of Biscay, populations from the area possess a particularly high proportion of endemic haplotypes and alleles across all the marker types analysed (mtDNA SNPs, scnDNA and microsatellites). A similar scenario in the brown seaweed Fucus serratus based on mtDNA sequences led Hoarau et al. [27] to conclude that Iberian populations of the species represent ‘unstable remnants’ of a glacial refugium. Populations of both species in the region frequently suffer stochastic extinction events during hot summers, and although C. crispus thalli exhibit an upper thermal tolerance of 28–29°C (water temperature) [45], these temperatures may be exceeded in intertidal rock pools. Given that predictions of future climate indicate a rise in both air and SSTs [46], there is thus a real danger that the unique genetic diversity maintained in southern-edge populations will be lost. Range-edge populations, particularly those at low latitudes, have been previously highlighted as possibly requiring special conservation efforts [12,14,47]. They often encompass a disproportionate amount of the total genetic diversity across a species' range [48,49]. As these populations frequently comprise the most evolutionarily divergent lineages (e.g. the light green haplotype at locus CO653303), they are often active areas of speciation [50,51]. Consequently, loss of the Iberian populations of C. crispus might have a major detrimental effect on the evolutionary potential of the species. This might also apply to other marine organisms that have recently been shown to possess unique genetic variation in Iberian populations, such as the seaweeds Palmaria palmata [27] and F. serratus [28], the ascidian Botryllus schlosseri [52], the bryozoan Celleporella hyalina [46], the sand goby Pomatoschistus minutus [53], and the mysid Neomysis integer [54].
For sessile species with limited dispersal capabilities in particular, such as seaweeds and bryozoans, the inability to track changes in climate may further compound the risk of extinction and subsequent loss of the unique genetic diversity found at the rear edge. The statistical phylogeographic analysis indicated that Europe was recolonized by C. crispus via a stepping-stone model, rather than by direct long-distance dispersal from refugial areas. Fertilization in the genus Chondrus occurs in situ and dispersal is via diploid carpospores or haploid tetraspores, and occasionally via vegetative fragments [55]; but spore viability in seaweeds is generally limited to 24–48 h [56], and a previous population genetic study in C. crispus indicated limited gene flow and isolation by distance [57]. Furthermore, surveys of the volcanic island of Surtsey [58] at intervals of 5 years had not recorded the species as late as 1997, 32 years after the arrival of the first recorded seaweed, Urospora penicilliformis. Given the occurrence of suitable substrate on Surtsey and the fact that C. crispus is found on the Vestmannaeyjar archipelago, 2.7 nautical miles from Surtsey, this further suggests that the species has a limited dispersal capacity.
The occurrence of unique haplotypes in southern European populations was not paralleled in the North American samples analysed. Although the high number of endemic haplotypes and alleles observed in these populations suggests recolonization from an in situ refugium, the precise location of the refugial area or areas is difficult to identify. The area south of the southern limit of the Laurentide ice sheet (south of Long Island Sound) lacks suitable rocky substrate for intertidal species [59,60] and it was generally assumed that the present-day occurrence of such taxa on Northwest Atlantic coastlines was the result of postglacial recolonization from Europe following extirpation of local populations during the LGM [60]. Recent geological reconstructions and modelling studies, however, have indicated that the area around the Grand Banks remained unglaciated at the LGM [61–63]. The exposure of the continental shelf owing to the drop in sea level may have provided suitable habitat for rocky intertidal species [60], and this idea is supported by phylogeographic evidence for persistence of several intertidal taxa around the northeastern coast of North America throughout the LGM [27,29,60,64–66]. The occurrence of suitable refugial habitats around the George's Bank and the southern Nova Scotian Shelf area as indicated by the palaeodistribution modelling is less likely, since most reconstructions suggest that the Laurentide ice sheet extended to the edge of the continental shelf in the Gulf of Maine [62,63,67]. Nevertheless, our findings based upon multiple molecular markers support the idea of a northeastern North American refugium for C. crispus, rather than recolonization from Europe, although this American refugium did not include present-day rear-edge populations. The most probable location for the refugium, based on the balance of evidence across the different markers studied, would appear to be in the area of the Canadian Maritime provinces, or further north in Newfoundland. This region has previously been proposed as a refugium for the hermit crab Pagurus longicarpus [64], the hydrozoan Obelia geniculata [65] and the periwinkle Littorina littorea [66].
This fact that the genetic signatures of glacial refugia were not obvious in North America, unlike Europe, might also reflect the prediction that ‘stable’ rear-edge populations (i.e. those that have persisted in the same areas throughout glacial and postglacial periods) should preserve a higher proportion of genetic variation than ‘trailing’ rear edges, which have migrated since the LGM [12]. The palaeodistribution modelling indicated that the present-day Iberian populations lie within the putative refugial area on the eastern side of the North Atlantic, and thus represent a stable rear edge. Current-day populations of C. crispus in the western North Atlantic, however, are situated north of the putative shoreline at the LGM and must have recolonized after the post-LGM rise in sea levels, which would class them as trailing rear edges. Likewise, populations that persisted in the Hurd Deep refugium in the English Channel [26] must have migrated to their current locations since the LGM. Under the two scenarios proposed by Hampe & Petit [12] (stable versus trailing), this would explain the fact that the Portuguese population exhibits by far the highest level of microsatellite diversity in Europe, whereas levels of microsatellite diversity are more consistent across the North American populations. The Iberian populations also exhibited far higher levels of mtDNA diversity than those found elsewhere in Europe. As many of these rear-edge populations are under threat of extinction from climate change, our findings suggest that such populations merit special attention in order to preserve the range-wide evolutionary potential of the species, and in particular those that might represent stable rear edges. These findings also further confirm the importance of historical forces in shaping present-day patterns of genetic variation across species ranges, particularly for species with limited potential for dispersal.
Acknowledgements
The authors are extremely grateful to everybody who provided samples for this study. We would also like to thank Ruth Kelly for assistance in the laboratory, Marcus Pfenninger for advice on data analysis, Grant Bigg for providing the GIS layers used in the distribution modelling and four anonymous referees for valuable comments on an original version of the manuscript.
References
- 1.Parmesan C., et al. 1999. Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature 399, 579–583 10.1038/21181 (doi:10.1038/21181) [DOI] [Google Scholar]
- 2.Parmesan C., Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 10.1038/nature01286 (doi:10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 3.Thomas C. D., et al. 2004. Extinction risk from climate change. Nature 427, 145–148 10.1038/nature02121 (doi:10.1038/nature02121) [DOI] [PubMed] [Google Scholar]
- 4.Parmesan C. 2006. Ecological and evolutionary response to recent climate change. Ann. Rev. Ecol. Evol. Syst. 37, 637–669 10.1146/annurev.ecolsys.37.091305.110100 (doi:10.1146/annurev.ecolsys.37.091305.110100) [DOI] [Google Scholar]
- 5.Vucetich J. A., Waite T. A. 2003. Spatial patterns of demography and genetic processes across the species' range: null hypotheses for landscape conservation genetics. Conserv. Genet. 4, 639–645 10.1023/A:1025671831349 (doi:10.1023/A:1025671831349) [DOI] [Google Scholar]
- 6.Eckert C. G., Samis K. E., Lougheed S. C. 2008. Genetic variation across species' geographical ranges: the central-marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188 10.1111/j.1365-294X.2007.03659.x (doi:10.1111/j.1365-294X.2007.03659.x) [DOI] [PubMed] [Google Scholar]
- 7.Lawton J. H. 1993. Range, population abundance and conservation. Trends Ecol. Evol. 8, 409–413 10.1016/0169-5347(93)90043-O (doi:10.1016/0169-5347(93)90043-O) [DOI] [PubMed] [Google Scholar]
- 8.Gaston K. J. 2003. The structure and dynamics of geographic ranges. Oxford, UK: Oxford University Press [Google Scholar]
- 9.Sagarin R. D., Gaines S. D. 2002. The ‘abundant centre’ distribution: to what extent is it a biogeographical rule? Ecol. Lett. 5, 137–147 10.1046/j.1461-0248.2002.00297.x (doi:10.1046/j.1461-0248.2002.00297.x) [DOI] [Google Scholar]
- 10.Taberlet P., Fumagalli L., Wust-Saucy A. G., Cosson J. F. 1998. Comparative phylogeography and postglacial recolonization routes in Europe. Mol. Ecol. 7, 453–464 10.1046/j.1365-294x.1998.00289.x (doi:10.1046/j.1365-294x.1998.00289.x) [DOI] [PubMed] [Google Scholar]
- 11.Hewitt G. M. 1999. Post-glacial recolonisation of European biota. Biol. J. Linn. Soc. 68, 87–112 10.1111/j.1095-8312.1999.tb01160.x (doi:10.1111/j.1095-8312.1999.tb01160.x) [DOI] [Google Scholar]
- 12.Hampe A., Petit R. J. 2005. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 8, 461–467 10.1111/j.1461-0248.2005.00739.x (doi:10.1111/j.1461-0248.2005.00739.x) [DOI] [PubMed] [Google Scholar]
- 13.Provan J., Bennett K. D. 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571 10.1016/j.tree.2008.06.010 (doi:10.1016/j.tree.2008.06.010) [DOI] [PubMed] [Google Scholar]
- 14.Lesica P., Allendorf F. W. 1995. When are peripheral populations valuable for conservation? Conserv. Biol. 9, 753–760 10.1046/j.1523-1739.1995.09040753.x (doi:10.1046/j.1523-1739.1995.09040753.x) [DOI] [Google Scholar]
- 15.Harley C. D. G., Hughes A. R., Hultgren K. M., Miner B. G., Sorte C. J. B., Thornber C. S., Rodriguez L. F., Tomanek L., Williams S. L. 2006. The impacts of climate change in coastal marine systems. Ecol. Lett. 9, 228–241 10.1111/j.1461-0248.2005.00871.x (doi:10.1111/j.1461-0248.2005.00871.x) [DOI] [PubMed] [Google Scholar]
- 16.Brierley A. S., Kingsford M. J. 2009. Impacts of climate change on marine organisms and ecosystems. Curr. Biol. 19, 602–614 10.1016/j.cub.2009.05.046 (doi:10.1016/j.cub.2009.05.046) [DOI] [PubMed] [Google Scholar]
- 17.Helmuth B., Harley C. D. G., Halpin P. M., O'Donnell M., Hofmann G. E., Blanchette C. A. 2002. Climate change and latitudinal patterns of intertidal thermal stress. Science 298, 1015–1017 10.1126/science.1076814 (doi:10.1126/science.1076814) [DOI] [PubMed] [Google Scholar]
- 18.Southward A. J., et al. 2005. Long-term oceanographic and ecological research in the western English Channel. Adv. Mar. Biol. 47, 1–105 10.1016/S0065-2881(04)47001-1 (doi:10.1016/S0065-2881(04)47001-1) [DOI] [PubMed] [Google Scholar]
- 19.Helmuth B., Mieszkowska N., Moore P., Hawkins S. J. 2006. Living on the edge of two changing worlds: forecasting the responses of rocky intertidal ecosystems to climate change. Ann. Rev. Ecol. Evol. Syst. 37, 373–404 10.1146/annurev.ecolsys.37.091305.110149 (doi:10.1146/annurev.ecolsys.37.091305.110149) [DOI] [Google Scholar]
- 20.Lima F. P., Ribeiro P. A., Queiroz N., Hawkins S. J., Santos A. M. 2007. Do distributional shifts of northern and southern species of algae match the warming pattern? Global Change Biol. 13, 2592–2604 10.1111/j.1365-2486.2007.01451.x (doi:10.1111/j.1365-2486.2007.01451.x) [DOI] [Google Scholar]
- 21.Hu Z., Critchley A. T., Gao T. X., Zeng X. Q., Morrell S. L., Duan D. L. 2007. Delineation of Chondrus (Gigartinales, Florideophyceae) in China and the origin of C. crispus inferred from molecular data. Mar. Biol. Res. 3, 145–154 10.1080/17451000701335679 (doi:10.1080/17451000701335679) [DOI] [Google Scholar]
- 22.Prince J. S., Kingsbury J. M. 1973. The ecology of Chondrus crispus at Plymouth, Massachusetts. III. Effect of elevated temperature on growth and survival. Biol. Bull. 145, 580–588 10.2307/1540638 (doi:10.2307/1540638) [DOI] [Google Scholar]
- 23.Maggs C. A., et al. 2008. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122 10.1890/08-0257.1 (doi:10.1890/08-0257.1) [DOI] [PubMed] [Google Scholar]
- 24.Stewart J. R., Lister A. M. 2001. Cryptic northern refugia and the origins of the modern biota. Trends Ecol. Evol. 16, 608–613 10.1016/S0169-5347(01)02338-2 (doi:10.1016/S0169-5347(01)02338-2) [DOI] [Google Scholar]
- 25.Bennett K. D., Provan J. 2008. What do we mean by ‘refugia’? Quaternary Sci. Rev. 27, 2449–2455 10.1016/j.quascirev.2008.08.019 (doi:10.1016/j.quascirev.2008.08.019) [DOI] [Google Scholar]
- 26.Provan J., Wattier R. A., Maggs C. A. 2005. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol. Ecol. 14, 793–804 10.1111/j.1365-294X.2005.02447.x (doi:10.1111/j.1365-294X.2005.02447.x) [DOI] [PubMed] [Google Scholar]
- 27.Hoarau G., Coyer J. A., Veldsink J. H., Stam W. T., Olsen J. L. 2007. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol. Ecol. 16, 3606–3616 10.1111/j.1365-294X.2007.03408.x (doi:10.1111/j.1365-294X.2007.03408.x) [DOI] [PubMed] [Google Scholar]
- 28.Olsen J. L., Zechman F. W., Hoarau G., Coyer J. A., Stam W. T., Valero M., Aberg P. 2010. The phylogeographic architecture of the fucoid seaweed Ascophyllum nodosum: an intertidal ‘marine tree’ and survivor of more than one glacial–interglacial cycle. J. Biogeogr. 37, 842–856 10.1111/j.1365-2699.2009.02262.x (doi:10.1111/j.1365-2699.2009.02262.x) [DOI] [Google Scholar]
- 29.Gibson S. Y., van der Marel R. C., Starzomski B. M. 2009. Climate change and conservation of leading-edge peripheral populations. Conserv. Biol. 23, 1369–1373 10.1111/j.1523-1739.2009.01375.x (doi:10.1111/j.1523-1739.2009.01375.x) [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer M., Passalacqua N. G., Bartram S., Schatz B., Croce A., Carey P. D., Kraudelt H., Jeltsch F. 2010. Conservation priorities differ at opposing species borders of a European orchid. Biol. Conserv. 143, 2207–2220 10.1016/j.biocon.2010.06.005 (doi:10.1016/j.biocon.2010.06.005) [DOI] [Google Scholar]
- 31.Thomas C. D. 2005. Recent evolutionary effects of climate change. In Climate change and biodiversity (eds Lovejoy T. E., Hannah L.), pp. 75–88 New Haven, CT: Yale University Press [Google Scholar]
- 32.Aitken S. N., Yeaman S., Holliday J. A., Wang T., Curtis-McLane S. 2008. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95–111 10.1111/j.1752-4571.2007.00013.x (doi:10.1111/j.1752-4571.2007.00013.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garbary D. J., de Wreede R. E. 1988. Life history phases in natural populations of Gigartinaceae (Rhodophyta): quantification using resorcinol. In Experimental phycology: a laboratory manual (eds Lobban C. S., Chapman D. J., Kremer B. P.), pp. 174–178 Cambridge, UK: Cambridge University Press [Google Scholar]
- 34.Provan J., Beatty G. E., Hunter A. M., McDonald R. A., McLaughlin E., Preston S. J., Wilson S. 2008. Restricted gene flow in fragmented populations of a wind-pollinated tree. Conserv. Genet. 9, 1521–1532 10.1007/s10592-007-9484-y (doi:10.1007/s10592-007-9484-y) [DOI] [Google Scholar]
- 35.Hudson R. R., Kaplan N. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111, 47–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozas J., Rozas R. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175 10.1093/bioinformatics/15.2.174 (doi:10.1093/bioinformatics/15.2.174) [DOI] [PubMed] [Google Scholar]
- 37.Provan J., Beatty G. E., Maggs C. A., Savidge G. 2007. Expressed sequence tag-derived microsatellites for the cool-water marine copepod Calanus finmarchicus. Mol. Ecol. Notes 7, 1369–1371 10.1111/j.1471-8286.2007.01889.x (doi:10.1111/j.1471-8286.2007.01889.x) [DOI] [Google Scholar]
- 38.Phillips S. J., Anderson R. P., Schapire R. E. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 10.1016/j.ecolmodel.2005.03.026 (doi:10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 39.Bigg G. R., Cunningham C. W., Ottersen G., Pogson G. H., Wadley M. R., Williamson P. 2008. Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proc. R. Soc. B 275, 163–172 10.1098/rspb.2007.1153 (doi:10.1098/rspb.2007.1153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantor S. B., Sun C. C., Tortolero-Luna G., Richards-Kortum R., Follen M. 1999. A comparison of C/B ratios from studies using receiver operating characteristic curve analysis. J. Clin. Epidemiol. 52, 885–892 10.1016/S0895-4356(99)00075-X (doi:10.1016/S0895-4356(99)00075-X) [DOI] [PubMed] [Google Scholar]
- 41.Excoffier L., Laval L. G., Schneider S. 2005. Arlequin, version 3.0: an integrated software package for population genetic data analysis. Evol. Bioinformatics Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 42.Dépraz A., Cordellier M., Hausser J., Pfenninger M. 2008. Postglacial recolonization at a snail's pace (Trochulus villosus): confronting competing refugia hypotheses using model selection. Mol. Ecol. 17, 2449–2462 10.1111/j.1365-294X.2008.03760.x (doi:10.1111/j.1365-294X.2008.03760.x) [DOI] [PubMed] [Google Scholar]
- 43.Beerli P., Felsenstein J. 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl Acad. Sci. USA 98, 4563–4568 10.1073/pnas.081068098 (doi:10.1073/pnas.081068098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automatic Control 19, 716–723 10.1109/TAC.1974.1100705 (doi:10.1109/TAC.1974.1100705) [DOI] [Google Scholar]
- 45.Lüning K., Guiry M. D., Masuda M. 1986. Upper temperature tolerance of North Atlantic and North Pacific geographical isolates of Chondrus species (Rhodophyta). Helgol. Meeresunters. 41, 297–306 10.1007/BF02366194 (doi:10.1007/BF02366194) [DOI] [Google Scholar]
- 46.Gómez A., Hughes R. N., Wright P. J., Carvalho G. R., Lunt D. H. 2007. Mitochondrial DNA phylogeography and mating compatibility reveal marked genetic structuring and speciation in the NE Atlantic bryozoan Celleporella hyalina.. Mol. Ecol. 16, 2173–2188 10.1111/j.1365-294X.2007.03308.x (doi:10.1111/j.1365-294X.2007.03308.x) [DOI] [PubMed] [Google Scholar]
- 47.Beatty G. E., McEvoy P. M., Sweeney O., Provan J. 2008. Range-edge effects promote clonal growth in peripheral populations of the one-sided wintergreen (Orthilia secunda). Divers. Distrib. 14, 546–555 10.1111/j.1472-4642.2008.00469.x (doi:10.1111/j.1472-4642.2008.00469.x) [DOI] [Google Scholar]
- 48.Millar C. I., Libby W. J. 1991. Strategies for conserving clinal, ecotypic and disjunct population diversity in widespread species. In Genetics and conservation of rare plants (eds Falk D. A., Holsinger K. E.), pp. 149–170 New York, NY: Oxford University Press [Google Scholar]
- 49.Beatty G. E., Provan J. 2011. Comparative phylogeography of two related plant species with overlapping ranges in Europe, and the potential effects of climate change on their intraspecific genetic diversity. BMC Evol. Biol. 11, 29. 10.1186/1471-2148-11-29 (doi:10.1186/1471-2148-11-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stebbins G. L., Major J. 1965. Endemism and speciation in the Californian flora. Ecol. Monogr. 35, 231–251 10.2307/1942216 (doi:10.2307/1942216) [DOI] [Google Scholar]
- 51.Levin D. A. 1970. Developmental instability and evolution in peripheral isolates. Am. Nat. 104, 343–353 10.1086/282668 (doi:10.1086/282668) [DOI] [Google Scholar]
- 52.Ben-Shlomo R., Paz G., Rinkevich B. 2006. Postglacial-period and recent invasions shape the population genetics of botryllid ascidians along European coasts. Ecosystems 9, 1118–1127 10.1007/s10021-006-0141-y (doi:10.1007/s10021-006-0141-y) [DOI] [Google Scholar]
- 53.Larmuseau M. H. D., van Houdt J. K. J., Guelinckx J., Hellemans B., Volckaert F. A. M. 2009. Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J. Biogeogr. 36, 1138–1151 10.1111/j.1365-2699.2008.02072.x (doi:10.1111/j.1365-2699.2008.02072.x) [DOI] [Google Scholar]
- 54.Remerie T., Vierstraete A., Weekers P. H. H., Vanfleteren J. R., Vanreusel A. 2009. Phylogeography of an estuarine mysid, Neomysis integer (Crustacea, Mysida), along the north-east Atlantic coasts. J. Biogeogr. 36, 39–54 10.1111/j.1365-2699.2008.01970.x (doi:10.1111/j.1365-2699.2008.01970.x) [DOI] [Google Scholar]
- 55.Taylor A. R. A., Chen L. C. M. 1994. Chondrus Stackhouse. In Biology of Economic Algae (ed. Akatsuka I.), pp. 35–76 The Hague, The Netherlands: SPB Academic Publishing [Google Scholar]
- 56.Santelices B. 1990. Patterns of reproduction, dispersal and recruitment in seaweeds. Oceanogr. Mar. Biol. 28, 177–276 [Google Scholar]
- 57.Wang X., Zhao F., Hu Z., Critchley A. T., Morrell S. L., Duan D. 2008. Inter-simple sequence repeat (ISSR) analysis of genetic variation of Chondrus crispus populations from North Atlantic. Aquat. Bot. 88, 154–159 10.1016/j.aquabot.2007.10.001 (doi:10.1016/j.aquabot.2007.10.001) [DOI] [Google Scholar]
- 58.Jónsson S., Gunnarsson K. 2000. Seaweed colonisation at Surtsey, the volcanic island south of Iceland. Surtsey Res. 11, 59–68 [Google Scholar]
- 59.Riggs S. R., Snyder S. W., Hine A. C., Mearns D. L. 1996. Hardbottom morphology and relationship to the geologic framework: mid-Atlantic continental shelf. J. Sediment Res. 66, 830–846 [Google Scholar]
- 60.Wares J. P., Cunningham C. W. 2001. Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55, 2455–2469 [DOI] [PubMed] [Google Scholar]
- 61.Dyke A. S., Andrews J. T., Clark P. U., England J. H., Miller G. H., Shaw J., Veillette J. J. 2002. The Laurentide and Innutian ice sheets during the last glacial maximum. Quaternary Sci. Rev. 21, 9–31 10.1016/S0277-3791(01)00095-6 (doi:10.1016/S0277-3791(01)00095-6) [DOI] [Google Scholar]
- 62.Miller G. H., Wolfe A. P., Steig E. J., Sauer P. E., Kaplan M. R., Briner J. P. 2002. The Goldilocks dilemma: big ice, little ice or ‘just-right’ ice in the Eastern Canadian Arctic. Quaternary Sci. Rev. 21, 33–48 10.1016/S0277-3791(01)00085-3 (doi:10.1016/S0277-3791(01)00085-3) [DOI] [Google Scholar]
- 63.Charbit S., Ritz C., Philippon G., Peyaud V., Kageyama M. 2007. Numerical reconstruction of the Northern Hemisphere ice sheets through the last glacial–interglacial cycle. Clim. Past 3, 15–37 10.5194/cp-3-15-2007 (doi:10.5194/cp-3-15-2007) [DOI] [Google Scholar]
- 64.Young A. M., Torres C., Mack J. E., Cunningham C. W. 2002. Morphological and genetic evidence for vicariance and refugium in Atlantic and Gulf of Mexico populations of the hermit crab Pagurus longicarpus. Mar. Biol. 140, 1059–1066 10.1007/s00227-002-0780-2 (doi:10.1007/s00227-002-0780-2) [DOI] [Google Scholar]
- 65.Govindarajan A. F., Halanych K. M., Cunningham C. W. 2005. Mitochondrial evolution and phylogeography in the hydrozoans Obelia geniculata (Cnidaria). Mar. Biol. 146, 213–222 10.1007/s00227-004-1434-3 (doi:10.1007/s00227-004-1434-3) [DOI] [Google Scholar]
- 66.Cunningham C. W. 2008. How to use genetic data to distinguish between natural and human-mediated introduction of Littorina littorea to North America. Biol. Invasions 10, 1–6 10.1007/s10530-007-9099-8 (doi:10.1007/s10530-007-9099-8) [DOI] [Google Scholar]
- 67.Uchupi E., Bolmer S. T. 2008. Geologic evolution of the Gulf of Maine region. Earth Sci. Rev. 91, 27–76 10.1016/j.earscirev.2008.09.002 (doi:10.1016/j.earscirev.2008.09.002) [DOI] [Google Scholar]