Abstract
The major histocompatibility complex (MHC) is a polymorphic gene family associated with immune defence, and it can play a role in mate choice. Under the genetic compatibility hypothesis, females choose mates that differ genetically from their own MHC genotypes, avoiding inbreeding and/or enhancing the immunocompetence of their offspring. We tested this hypothesis of disassortative mating based on MHC genotypes in a population of great frigatebirds (Fregata minor) by sequencing the second exon of MHC class II B. Extensive haploid cloning yielded two to four alleles per individual, suggesting the amplification of two genes. MHC similarity between mates was not significantly different between pairs that did (n = 4) or did not (n = 42) exhibit extra-pair paternity. Comparing all 46 mated pairs to a distribution based on randomized re-pairings, we observed the following (i): no evidence for mate choice based on maximal or intermediate levels of MHC allele sharing (ii), significantly disassortative mating based on similarity of MHC amino acid sequences, and (iii) no evidence for mate choice based on microsatellite alleles, as measured by either allele sharing or similarity in allele size. This suggests that females choose mates that differ genetically from themselves at MHC loci, but not as an inbreeding-avoidance mechanism.
Keywords: major histocompatibility complex, genetic compatibility, immunocompetence, inbreeding avoidance, microsatellites
1. Introduction
A fundamental goal in evolutionary biology is to gain a better understanding of forces that create and maintain genetic variation in natural populations. When natural selection is consistently directional, it can eliminate genetic variation over time. However, balancing selection can maintain genetic variation rather than eliminate it. Mounting evidence suggests that evolution at the major histocompatibility complex (MHC) might emerge as a taxonomically broad example of balancing selection acting to maintain genetic diversity [1]. The major histocompatibility complex is a highly polymorphic gene assemblage that plays a critical role in the development and activation of both the T-cell mediated and humoral arms of immune defence. Because of the fitness variation among MHC genotypes [2–7], and because MHC genotypes might be detectable by olfaction [8,9] or via condition-dependent traits [10,11], MHC could be an important target of mate choice [12,13].
From a ‘good genes’ perspective, selection might favour mate choice that targets particular MHC alleles [12,14]. If this type of pathogen-mediated selection is directional and constant, MHC diversity could decline over time. However, three types of balancing selection could maintain diversity of MHC alleles [1]: rare-allele advantage [15,16], fluctuating selection [17,18] and heterozygote advantage [6,7,19,20].
In this study, we use a seabird species to test an alternative process by which MHC diversity in offspring could shape mate choice—the genetic compatibility hypothesis [21]. Whereas a good genes process predicts that all females in the local breeding pool prefer either males with certain genotypes [12,14] or males that are heterozygous [22,23], the genetic compatibility mechanism is driven by whether a particular male's MHC genotype is a good fit for, or sufficiently different from, that of a particular female [24–26]. The choice of a genetically compatible mate should yield MHC-heterozygous offspring that can mount an effective immune response to a wider array of pathogens [19,20].
Importantly, dissimilar MHC genotypes between mates could arise from general inbreeding avoidance, rather than from mate choice based on MHC complementarity alone [27,28]. Some studies have found clear evidence that MHC per se is an important target of mate choice, with mates having dissimilar genotypes at MHC but not at neutral genomic markers [25,29,30]. Other datasets do not find clear distinctions between inbreeding avoidance and MHC complementarity [23,26,31,32], and additional studies have failed to find evidence for mate choice based on MHC dissimilarity at all (reviewed in [33]).
Testing for disassortative mating at MHC is complicated by several factors. First, there is uncertainty about the amount of gene duplication, concerted evolution and differential expression of duplicated MHC loci in non-model organisms. These phenomena seem prevalent in some species [34,35], but birds generally have a smaller, simpler MHC than mammals [36,37]. Among birds, gene duplication—and the presence of likely pseudogenes—appears to be more common in songbirds (Passeriformes) than in other groups [38]. Second, there is the potential for immune function to peak with an intermediate, rather than maximal, number of MHC alleles [39]. Accordingly, some studies have found mate choice favouring intermediate levels of MHC dissimilarity [40–42] rather than maximal levels [25,29,30], but this is an area of controversy [43]. Third, patterns of mate choice can be clouded by extra-pair parentage, particularly in birds [44]. For example, MHC does not influence social mate choice in Seychelles warblers, but females are more likely to seek extra-pair matings if socially paired to a male with low MHC diversity [22], and these extra-pair matings confer an MHC-mediated survival advantage upon offspring [7]. Thus, studies of MHC and mate choice need to be accompanied by parentage information.
Great frigatebirds Fregata minor are colony-breeding, sexually dimorphic seabirds. In our study population, there is serial monogamy and a male-biased operational sex ratio, such that females have many options as they choose mates for each breeding attempt [45,46]. We combined MHC sequencing with microsatellite genotyping of mated pairs to test whether great frigatebirds choose mates based on dissimilar MHC genotypes, or on genetic dissimilarity in a broader sense, or neither. We also used microsatellite genotypes of mates and their offspring to examine paternity. To assess MHC genotypes, we sequenced the second exon of MHC class II B genes, which codes for the most polymorphic segment of the peptide-binding region [47]. Disassortative mating at both MHC and microsatellite loci would suggest a genome-wide pattern of outbreeding. Disassortative mating at MHC but random mating at microsatellite loci would suggest, instead, that MHC dissimilarity is the actual target of mate choice.
2. Material and methods
(a). Study population
We studied great frigatebirds on Tern Island, a 14 ha island in French Frigate Shoals, Northwestern Hawaiian Islands (23°45′ N, 166°17′ W). Approximately 4000 great frigatebirds nest on Tern Island each year [46]. Egg laying ranges from February to May, with one egg per nest. In 2007, we monitored the breeding activities of 46 pairs. All 92 of these individuals, plus their 46 offspring, were captured by hand while at their nests, and approximately 50 µl of blood were collected from their brachial vein. Blood samples were stored in Longmire's lysis buffer at room temperature for the remainder of the field season.
(b). Major histocompatibility complex cloning
We digested blood samples using proteinase K. Genomic DNA was extracted by alcohol precipitation. To develop primers that would amplify the second exon of MHC class II B in great frigatebirds, we used a degenerate primer pair to amplify an approximately 1500 bp sequence encompassing part of exon 1, all of exon 2 and part of exon 3 (primers Exon1F-CTGGTGGCACTGSTGGYRCT and Exon3R-CCAGCANCACCAGCASCTGGTA; Colin Hughes 2009, unpublished data). Using this sequence, we designed a new primer pair that amplified a 557 bp sequence surrounding exon 2: 481F-CACACTGCCAGTCCTACCG and 1038R-AGGGACTCGTGTCCTCATGG. We then truncated this sequence to the 270 bp that comprise exon 2, the region that contributes the most polymorphism to the peptide-binding site. For all 92 adult birds, each DNA sample was PCR amplified three separate times (10 µl reactions each) to limit PCR artefacts, using the proof reading enzyme Pfx (Platinum Pfx, Invitrogen). After PCR, all three amplification products from an individual were combined and gel purified in a 2.5 per cent agarose gel (Wizard SV, Promega Corporation). To determine sequences from heterozygous individuals, purified DNA was transformed and cloned using a haploid cloning vector and high-efficiency cloning cells (Clone Jet, Fermentas; JM109, Promega). Cloning products were grown for 24 h on standard LB/amp plates and then used as template DNA for PCR screening. All clones were PCR and gel screened (primers 481F/1038R, 1.5% agarose gels) to ensure the presence of the proper insert. After gel screening, PCR products were cleaned and sequenced with BigDye Terminator Cycle Sequencing Kit (v. 3.1) and a 3130× genetic analyzer (Applied Biosystems, Inc.). For every individual, 24–28 positive clones were selected at random for sequencing.
(c). Sequence screening
Sequences were analysed using BioEdit v. 7.0.0 (Ibis Therapeutics) and MEGA4 [48]. MHC sequences occurring just once in the 24–28 clones from a single individual and differing by less than 3 bp from any other sequence found from that same individual were considered artefacts of PCR error and were removed from further analyses [47,49]. In addition, since the recombination of cloned PCR products can result in chimeras, all alleles were compared with direct sequences of uncloned PCR products to check for similarity of polymorphic sites [47]. Sequences that did not match polymorphic sites of their respective uncloned sequences were also removed from further analyses.
(d). Microsatellites
We used microsatellite genotypes to measure genetic similarity between mates and to conduct a paternity exclusion analysis. We genotyped the 92 adults and 46 offspring at 12 di- or tetra-nucleotide microsatellite loci [50]. We used three 10 µl multiplex PCR reactions of four loci each. All reactions used 1× GeneAmp Gold buffer (Applied Biosystems, Inc.), 0.2 mM each deoxyribonucleoside triphosphates (dNTP) and 10 ng of template DNA but differed in concentrations of other components. For loci Fmin04, Fmin06, Fmin13 and Fmin15, we used 1.5 mM MgCl2, 0.4 U AmpliTaq Gold polymerase (Applied Biosystems, Inc.), 0.5 µM of forward and reverse primers Fmin13, and 0.3 µM of forward and reverse primers for each of the other three loci (Fmin04, Fmin06 and Fmin15). Cycle parameters were 95°C for 7 min; 35 cycles of 95°C for 30 s, 53°C for 30 s, 70°C for 3 min and 70°C for 15 min. For loci Fmin01, Fmin03, Fmin11 and Fmin18, we used 2.0 mM MgCl2, 0.2 µM of forward and reverse primers Fmin11, 0.4 µM of forward and reverse primers for each of the other three loci (Fmin01, Fmin03 and Fmin18), 7.5 µM bovine serum albumin and 0.5 U AmpliTaq Gold polymerase. Cycle parameters were 95°C for 7 min; 35 cycles of 95°C for 30 s, 58°C for 30 s, 70°C for 3 min; and 70°C for 15 min. For loci Fmin02, Fmin10, Fmin14 and Fmin17, we used 2.0 mM MgCl2, 0.3 µM of forward and reverse primers for each locus, 7.5 µM bovine serum albumin and 0.5 U AmpliTaq Gold. Cycle parameters were 95°C for 7 min; 35 cycles of 95°C for 30 s, 56°C for 30 s, 70°C for 3 min; and 70°C for 15 min. Dye-labelled products were run on an ABI PRISM 3730XL, with manual verification of allele calls in GeneMapper v. 4.0 (Applied Biosystems, Inc.). To estimate the per-allele error rates of microsatellite genotyping [51], we blindly repeated the amplification and genotyping of 1236 single-locus microsatellite genotypes spread across 124 individuals.
(e). Data analysis
MHC nucleotide sequences were connected in a parsimony-based haplotype network using the genealogical software package TCS [52]. For all analyses of selection and mate choice, MHC sequences were converted from nucleotides to amino acids to better approximate functional differences between individuals. We used the modified Nei-Gojobori test in MEGA4 [48] to determine whether sequences were under positive selection.
Biologically speaking, two aspects of MHC complementarity were of interest to us: the extent to which mates had identical versus non-identical alleles, and the magnitude of the differences between non-identical alleles. On the one hand, it might be beneficial to have alleles that are even minimally different. On the other hand, two alleles differing by one amino acid could be less effective at responding to a diversity of pathogens than two alleles differing by eight amino acids. Thus, we chose two metrics to investigate whether mated pairs were less similar at MHC II B exon 2 than would be expected under random mating: an allele-sharing index, and an index that incorporates information on the magnitude of differences between alleles. All analyses were amenable to the likelihood that our primers amplified two loci containing MHC II B (see §3). The first analysis uses the simple proportion of alleles shared by mates [53]. We used this measure to compare the mean allele-sharing value of the 46 known pairs with the distribution of mean allele-sharing values generated from 10 000 simulations of 46 random male–female pairings selected from the same 92 individuals (e.g. [30]).
To analyse the magnitude of differences between alleles, we summed the number of amino acid differences from pairwise combinations of alleles in mated birds [25,40]. The average of this score for the 46 known pairs was then compared with the distribution of scores generated from 10 000 simulations of 46 random pairings selected from the same 92 individuals, just as with the allele-sharing analysis [25,40].
We used a randomization test of variance values [40] to look for evidence that females might select mates with intermediate, rather than maximal, levels of MHC dissimilarity. Under this hypothesis, the previously described randomization tests should not show evidence that the mean genetic similarity of true mates is in the extreme tail of the distribution of simulated random pairings. Instead, the mean value should be intermediate (and thus non-significant). However, there should be a low variance in the observed measures of genetic similarity between true mates (compared with the simulation of random pairings), such that mated pairs all tend to exhibit the same level of intermediate MHC dissimilarity.
To test for disassortative mating at non-MHC loci, we calculated the genetic similarity of mated pairs based on the 12 microsatellite loci. To parallel our analysis of MHC alleles, we used two approaches—one based on allelic identity (i.e. allele sharing) and one based on the extent of the differences between the alleles. As a measure of allelic identity, we calculated Queller & Goodnight's r [54]. As a measure of allelic similarity, we calculated Streiff's I' [55], which incorporates information on allele size: microsatellite mutations most commonly occur in a stepwise manner, driven primarily by replication slippage [56,57], in which case there can be useful evolutionary information in the size difference between alleles (e.g. [58,59]; but see [60]). The mean Streiff's I' was calculated for all 46 pairs and was then compared with the distribution of the mean Streiff's I' scores generated from 10 000 simulations of 46 random pairings of the same 92 individuals.
Microsatellite-based measures of genetic similarity between mates were computed with SPAGeDi v. 1.3 [61]. Paternity exclusion analysis was conducted with the 12 microsatellite loci using Cervus v. 3.0 [62] and incorporating our calculated rate of genotyping error. The chick-brooding female was assumed to be the mother [45], and likelihood ratios were used to assess the competing hypotheses that the chick-brooding male either was or was not the genetic sire. All statistical tests were two-tailed.
3. Results
(a). Major histocompatibility complex
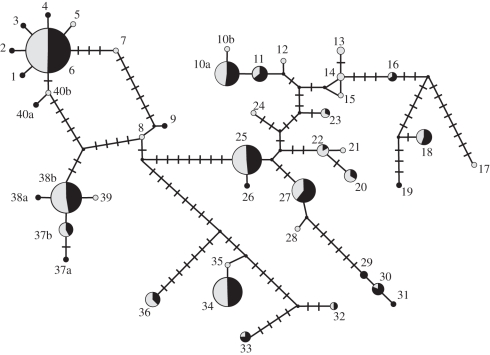
Multiple clones of the second exon of the MHC class II B gene(s) were sequenced for all 92 adults. After removal of sequences resulting from PCR error, a total of 2025 sequenced clones remained (range 17–26 sequenced clones per individual, mean = 22.01 ± 0.20 s.e.), yielding 44 unique exon 2 nucleotide sequences (GenBank accession numbers JF916727–JF916770; electronic supplementary material, S1). Sequences differed by 1–36 bp, with no indels, and comprised a large number of moderate-frequency haplotypes (figure 1). All remaining results are based on conversion to amino acid sequences. In 92 birds, there were 40 unique exon 2 amino acid sequences, differing by 1–23 amino acids. Each individual possessed two to four unique alleles (two alleles in 16 individuals (17.4%), three alleles in 38 individuals (41.3%), four alleles in 38 individuals (41.3%); electronic supplementary material, S2), suggesting that two loci were being amplified. There were no signs of pseudogenes (no stop codons or frame shift mutations) and the sequences were consistent with expressed loci subject to positive selection (modified Nei-Gojobori Z = 2.33, p = 0.011).
Figure 1.
Network of nucleic acid haplotypes, based on statistical parsimony. The area of each circle is proportional to the number of copies of that haplotype in our sample, with males shaded black and females shaded grey. Each connecting line segment represents a change of 1 bp. Haplotypes delineated with letters (e.g. 10a and 10b) yield identical amino acid sequences.
(b). Microsatellites
Individuals were genotyped at 12 loci (89% of birds) or 11 loci (11% of birds; electronic supplementary material, S2). Based on blind repeat PCR and repeat sizing of 1236 single-locus genotypes, the genotyping error rate was 0.00202 per allele. Queller & Goodnight's r for allele sharing between mated pairs ranged from −0.305778 to 0.559283 (mean = −0.004073, variance = 0.03422, n = 46 pairs). Streiff's I' for genetic similarity between mated pairs ranged from −0.605 to 0.523 (mean = 0.0121, variance = 0.0454, n = 46 pairs). Paternity exclusion analysis in these 46 single-chick families identified four likely cases of extra-pair paternity, with offspring mismatching their social fathers at three, three, four and six loci, respectively. Assuming that the female is a genetic parent (0 mismatches observed; and see [45]), the log of the odds (LOD) score for a parent–offspring relationship between the social father and the offspring was less than −5.5 for these four pairs versus LOD scores ranging from +2.6 to 17.1 among the remaining 42 pairs.
(c). Disassortative mating
There was no significant difference in allele sharing (i.e. allelic identity) between actual mated pairs and randomly assigned pairs (allele sharingmated = 0.345 ± 0.033 s.e., allele sharingrandom = 0.360 ± 0.00033 s.e., p = 0.672; figure 2a and electronic supplementary material, S3). Because the allele-sharing value of actual pairs was slightly, but not significantly, lower than the mean from the randomizations, we used a variance test to assess the possibility that females choose mates for an intermediate level of MHC dissimilarity. Contrary to this intermediate-similarity hypothesis, the variance in the allele-sharing scores of actual mates was not significantly smaller than expected under a model of random pairing (p = 0.264; figure 2b).
Figure 2.
Measures of MHC allele sharing in 46 known breeding pairs (arrow and bold line) compared with the distribution of values generated from 10 000 simulations of 46 random male–female pairings selected from the same 92 individuals. Dashed lines indicate cut-offs for significant departures from random mating. (a) Mean allele-sharing values. (b) Variance in allele-sharing values.
To test for disassortative mating based on MHC allele sequence similarity (rather than simple identity), we tested whether the sum of pairwise amino acid differences between mated pairs was extremely large (or small) relative to randomized pairings. Mates had significantly greater pairwise amino acid differences compared with the randomly assigned pairs (AAdistancemated = 130.85 ± 8.02 s.e., AAdistancerandom = 126.31 ± 0.02 s.e., p = 0.0352; figure 3 and electronic supplementary material, S3).
Figure 3.
Mean of the summed pairwise differences in MHC amino acid sequences between alleles of 46 known pairs (arrow and bold line) compared with the distribution of values generated from 10 000 simulations of 46 random male–female pairings selected from the same 92 individuals. Dashed lines indicate cut-offs for significant departures from random mating.
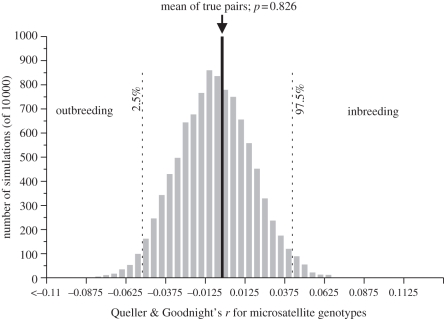
To test whether MHC itself is a target of mate choice, we also compared mates' genotypes at 12 microsatellite loci. Based on allele sharing, as measured by Queller & Goodnight's r, the microsatellite similarity between mated pairs was not significantly small (or large) compared with randomly assigned pairs (Queller & Goodnight's rmated = −0.004073 ± 0.0273 s.e.; rrandom =−0.009204 ± 0.000237 s.e., p = 0.826; figure 4 and electronic supplementary material, S3). Similar results (i.e. no evidence for non-random mating at microsatellite loci) were obtained when using Wang's r [63] as a measure of allele sharing (p = 0.564). When incorporating information on allele size, the microsatellite similarity between mated pairs was likewise not significantly small (or large) compared with randomized pairs (Streiff's I′mated = 0.0121 ± 0.000299 s.e., Streiff's I′random = 0.00300 ± 000 299 s.e., p = 0.758; figure 5 and electronic supplementary material, S3).
Figure 4.
Mean coefficient of relatedness for 46 pairs (arrow and bold line) compared with the distribution of the mean relatedness coefficients generated from 10 000 simulations of 46 random pairings selected from the same 92 individuals. Relatedness coefficient is Queller & Goodnight's r, based on allelic identity at 12 microsatellite loci. Dashed lines indicate cut-offs for significant departures from random mating.
Figure 5.
Mean of the microsatellite allelic similarity for 46 pairs (arrow and bold line) compared with the distribution of the mean allelic similarity generated from 10 000 simulations of 46 random pairings selected from the same 92 individuals. Allelic similarity is Streiff's I′, based on 12 microsatellite loci. Dashed lines indicate cut-offs for significant departures from random mating.
In the small sample of four social pairs with extra-pair young (EPY), the mean MHC allele-sharing score between social mates was 0.4125 (±0.0901 s.e.) compared with 0.3389 (±0.0351 s.e.) in the remaining 42 pairs (t44 = 0.626, p = 0.535). The MHC allelic dissimilarity score (based on the sum of pairwise differences between alleles in the male and the female of a pair) between the four social pairs with EPY was 116.5 (±24.30 s.e.) compared with 132.2 (±55.23 s.e.) in the remaining 42 social pairs (t44 = 0.610, p = 0.586). Thus, by both measures of MHC similarity, parents of extra-pair young were slightly but not significantly more similar in their MHC alleles than parents of within-pair young. The true genetic sires of extra-pair offspring were not identified, given that this breeding population may contain more than 10 000 mate-seeking males [46].
4. Discussion
Our data on MHC and microsatellite genotypes suggest that mate choice favours partners with divergent MHC amino acid sequences, in a manner beyond simple inbreeding avoidance. Specifically, at exon 2 of two MHC class II B loci, mated pairs had alleles that differed by more amino acids than expected by chance. This pattern of MHC-disassortative mating was not detected when simply categorizing MHC alleles as identical versus non-identical. And in contrast to our finding of mate choice for divergent MHC alleles, pairs did not exhibit significantly disassortative mating at microsatellite loci. This lack of evidence for inbreeding avoidance in our microsatellite data is perhaps not unexpected, given that previous work in this population found evidence of inbreeding [64]. Together, the combination of MHC and microsatellite results suggests that MHC complementarity is an actual target of mate choice in this system. Similar evidence for disassortative mating based on MHC genotypes but not on microsatellites has been found in a growing number of species, including tuataras [30], lemurs [24], salmon [25] and trout [40] (but see [32]). Importantly, the contrasting results of our two MHC analyses suggest the further possibility that offspring might benefit from complex MHC genotypes as defined not simply by heterozygosity but by having alleles that are very different from one another and thus more likely to differ in pathogen-binding function.
Our data were consistent with the hypothesis of mate choice for maximal, rather than intermediate, MHC dissimilarity between mates. Previous studies have been mixed in this regard, with evidence of mate choice for maximal [25,29,30], intermediate [12,40–42] or random [33] levels of MHC dissimilarity. The observed population-level pattern of mate choice might depend upon how much dissimilarity exists in the available pool of mates—i.e. mate choice for intermediate differences might be seen in populations with enormous variation in MHC, whereas mate choice for maximal differences might be seen in populations with lower amounts of MHC variation. Such a hypothesis would predict that MHC variation is lower in our frigatebird population than in populations with mate choice for intermediate dissimilarity, though testing this prediction with existing data would be difficult owing to variation between studies in MHC sampling methods.
The parentage data allowed us to consider whether MHC similarity between mates might drive extra-pair matings, as seen in Seychelles warblers [7,22]. Previous work has shown that extra-pair fertilizations are rare in seabirds generally [65] and in this population of frigatebirds, in particular [45], so the scope for MHC to influence paternity is somewhat limited here. Our microsatellite data revealed that four of 46 offspring were likely to have extra-pair sires. We did find a trend towards greater MHC similarity in these four social pairs compared with the 42 genetically monogamous pairs, but this trend was not significant.
A possible caveat to our conclusions about MHC-disassortative mate choice is that our primers appeared to amplify two loci, as we found two, three or four alleles per bird. Our approach of haploid cloning and sequencing provides some confidence that we uncovered all unique alleles within each individual. However, because of the presumed similar flanking sequence of these two MHC loci, we were unable to assign alleles to a particular locus and thus could not analyse mate choice based on each locus separately. Our study also did not examine gene expression; therefore, we cannot say definitively that all MHC sequences presented here produce proteins that are important in the immune repertoire [35,66]. Nonetheless, several pieces of indirect evidence suggest that both loci are functional: analyses of all unique sequences revealed no stop codons or frameshifts, and significant positive selection was detected on the entire dataset. Furthermore, including a non-functional locus in our analysis would weaken our ability to detect a pattern of disassortative mating. Our finding of a significant result suggests either that both loci were functional genes or that the strength of mating preferences on one of the loci was sufficient to allow for detection of a disassortative pattern.
The findings on mate choice predict that offspring of MHC-disassortative pairs would experience immune-dependent fitness benefits. Future work could test whether MHC diversity is predictive of survival, as has recently been shown in several other avian systems [6,7]. Great frigatebirds, like many marine birds, are colonial nesters on islands, and dense breeding colonies on islands are likely to facilitate transmission of pathogens and parasites [67]. The seabird breeding colony on Tern Island includes approximately 4000 frigatebirds and 200 000 seabirds of 14 other species, all nesting in only 10 ha of habitat. Furthermore, Tern Island includes haematophagous flies (Hippoboscidae) that can serve as vectors of disease [68,69]. Whether vector-transmitted diseases have strong fitness consequences in frigatebirds is still unresolved, with Haemosporidian parasites causing severe health problems in some host–parasite systems but not others [70].
Last, we consider the broader process of mate choice for a combination of traits. Male great frigatebirds are known for secondary sexual characters and elaborate courtship displays. These types of traits are thought to signal good genes to potential mates [71], though to date there is limited evidence for such a role in this species [72]. Even if males with good ornaments possess good genes, they may not possess the most suitable genes for females attempting to maximize their genetic compatibility [73,74]. Good genes and compatible genes are not necessarily mutually exclusive, and females could use nested rule-based preferences [75,76] where they initially select a subset of males with attractive ornaments, then from this subset choose the most genetically compatible male. In our population of frigatebirds, the observed behaviour of mate-choosing females is consistent with this possibility. Females initially assess potential mates by repeatedly flying over displaying males; during this stage, females might be relying on visual or auditory cues. Next, a female lands beside a suitor, and the potential pair engages in mutual preening, during which females may have the opportunity to use olfactory cues. In mammals and fish, peptides derived from MHC proteins can contribute to individual olfactory profiles that subsequently play a role in mate choice decisions [8,77,78]. Birds appear also to have sophisticated olfactory capabilities [79], and there has been an explosion of studies from a wide range of avian orders showing a link between olfaction and mate choice [80–82] or other behaviours [83–85]. Although nothing is known of frigatebird olfaction, female great frigatebirds reject males at both stages of mate choice—at the initial stage of flying over displaying males and also at the subsequent stage of perching in physical contact (Dearborn & Juola 2005, personal observation). If mate choice in this and other species is a multi-stage process that emphasizes different sensory modalities at different stages, it could be possible to maintain directional selection for elaborate morphological or behavioural ornaments while still favouring mate selection for MHC-complementary partners [76]. As MHC sequencing becomes feasible in additional species, we may soon have an even broader taxonomic view of the evolutionary role of MHC complementarity in mate choice.
Acknowledgements
We thank Bill Searcy for extensive support and advice. Colin Hughes kindly provided the degenerate primers used for initial MHC amplification. Scott Edwards, Miguel Alcaide, Carla Hurt and Colin Hughes gave helpful suggestions on avian MHC and laboratory work. We thank Morgan Gilmour for assistance in the field, Lyndsey Kiss and Taylor Gibbons for assistance in the laboratory, and Mike Robinson and Frank Hailer for discussions about data analysis. Bill Searcy, Tommaso Pizzari, Andrea Gager and two anonymous reviewers provided excellent feedback on the manuscript. The U.S. Fish and Wildlife Service provided logistical support and access to Tern Island. Funding for this research was provided by the University of Miami's Department of Biology, College of Liberal Arts and Sciences, the Maytag Endowment, the Cushland Fund and NSF IOS-0717976.
References
- 1.Spurgin L. G., Richardson D. S. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988 10.1098/rspb.2009.2084 (doi:10.1098/rspb.2009.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paterson S., Wilson K., Pemberton J. M. 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proc. Natl Acad. Sci. USA 95, 3714–3719 10.1073/pnas.95.7.3714 (doi:10.1073/pnas.95.7.3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrington M., et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283, 1748–1752 10.1126/science.283.5408.1748 (doi:10.1126/science.283.5408.1748) [DOI] [PubMed] [Google Scholar]
- 4.Kurtz J., Kalbe M., Aeschlimann P. B., Haberli M. A., Wegner K. M., Reusch T. B. H., Milinski M. 2004. Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc. R. Soc. Lond. B 271, 197–204 10.1098/rspb.2003.2567 (doi:10.1098/rspb.2003.2567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madsen T., Ujvari B. 2006. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 19, 1973–1978 10.1111/j.1420-9101.2006.01158.x (doi:10.1111/j.1420-9101.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 6.Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x) [DOI] [PubMed] [Google Scholar]
- 7.Brouwer L., Barr I., Van De Pol M., Burke T., Komdeur J., Richardson D. S. 2010. MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Mol. Ecol. 19, 3444–3455 10.1111/j.1365-294X.2010.04750.x (doi:10.1111/j.1365-294X.2010.04750.x) [DOI] [PubMed] [Google Scholar]
- 8.Boehm T., Zufall F. 2006. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 29, 100–107 10.1016/j.tins.2005.11.006 (doi:10.1016/j.tins.2005.11.006) [DOI] [PubMed] [Google Scholar]
- 9.Brennan P. A., Zufall F. 2006. Pheromonal communication in vertebrates. Nature 444, 308–315 10.1038/nature05404 (doi:10.1038/nature05404) [DOI] [PubMed] [Google Scholar]
- 10.Von Schantz T., Wittzell H., Göransson G., Grahn M. 1997. Mate choice, male condition-dependent ornamentation and MHC in the pheasant. Hereditas 127, 133–140 10.1111/j.1601-5223.1997.t01-1-00133.x (doi:10.1111/j.1601-5223.1997.t01-1-00133.x) [DOI] [Google Scholar]
- 11.Jäger I., Eizaguirre C., Griffiths S. W., Kalbe M., Krobbach C. K., Reusch T. B. H., Schaschl H., Milinski M. 2007. Individual MHC class I and MHC class IIB diversities are associated with male and female reproductive traits in the three-spined stickleback. J. Evol. Biol. 20, 2005–2015 10.1111/j.1420-9101.2007.01366.x (doi:10.1111/j.1420-9101.2007.01366.x) [DOI] [PubMed] [Google Scholar]
- 12.Eizaguirre C., Yeates S. E., Lenz T. L., Kalbe M., Milinski M. 2009. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329 10.1111/j.1365-294X.2009.04243.x (doi:10.1111/j.1365-294X.2009.04243.x) [DOI] [PubMed] [Google Scholar]
- 13.Zelano B., Edwards S. V. 2002. An MHC component to kin recognition and mate choice in birds: predictions, progress, and prospects. Am. Nat. 160, S225–S237 10.1086/342897 (doi:10.1086/342897) [DOI] [PubMed] [Google Scholar]
- 14.Ekblom R., Saether S. A., Fiske P., Käläs J. A., Höglund J. 2010. Balancing selection, sexual selection and geographic structure in MHC genes of Great Snipe. Genetica 138, 453–461 10.1007/s10709-008-9335-x (doi:10.1007/s10709-008-9335-x) [DOI] [PubMed] [Google Scholar]
- 15.Ejsmond M. J., Babik W., Radwan J. 2010. MHC allele frequency distributions under parasite-driven selection: a simulation model. BMC Evol. Biol. 10, 332. 10.1186/1471-2148-10-332 (doi:10.1186/1471-2148-10-332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westerdahl H., Waldenstrom J., Hansson B., Hasselquist D., von Schantz T., Bensch S. 2005. Associations between malaria and MHC genes in a migratory songbird. Proc. R. Soc. B 272, 1511–1518 10.1098/rspb.2005.3113 (doi:10.1098/rspb.2005.3113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekblom R., Saether S. A., Grahn M., Fiske P., Kålås J. A., Höglund J. 2004. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media). Mol. Ecol. 13, 3821–3828 10.1111/j.1365-294X.2004.02361.x (doi:10.1111/j.1365-294X.2004.02361.x) [DOI] [PubMed] [Google Scholar]
- 18.Hedrick P. W. 2002. Pathogen resistance and genetic variation at MHC loci. Evolution 56, 1902–1908 10.1111/j.0014-3820.2002.tb00116.x (doi:10.1111/j.0014-3820.2002.tb00116.x) [DOI] [PubMed] [Google Scholar]
- 19.Penn D. J., Damjanovich K., Potts W. K. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11 260–11 264 10.1073/pnas.162006499 (doi:10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver M. K., Telfer S., Piertney S. B. 2009. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. R. Soc. B 276, 1119–1128 10.1098/rspb.2008.1525 (doi:10.1098/rspb.2008.1525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neff B. D., Pitcher T. E. 2005. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol. Ecol. 14, 19–38 10.1111/j.1365-294X.2004.02395.x (doi:10.1111/j.1365-294X.2004.02395.x) [DOI] [PubMed] [Google Scholar]
- 22.Richardson D. S., Komdeur J., Burke T., von Schantz T. 2005. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B 272, 759–767 10.1098/rspb.2004.3028 (doi:10.1098/rspb.2004.3028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwensow N., Fietz J., Dausmann K., Sommer S. 2008. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evol. Ecol. 22, 617–636 10.1007/s10682-007-9186-4 (doi:10.1007/s10682-007-9186-4) [DOI] [Google Scholar]
- 24.Schwensow N., Eberle M., Sommer S. 2008. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc. R. Soc. B 275, 555–564 10.1098/rspb.2007.1433 (doi:10.1098/rspb.2007.1433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landry C., Garant D., Duchesne P., Bernatchez L. 2001. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc. R. Soc. Lond. B 268, 1279–1285 10.1098/rspb.2001.1659 (doi:10.1098/rspb.2001.1659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson M., Madsen T., Nordby J., Wapstra E., Ujvari B., Wittsell H. 2003. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. Lond. B 270, (doi:10.1098/rsbl.2003.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grob B., Knapp L. A., Martin R. D., Anzenberger G. 1998. The major histocompatibility complex and mate choice: inbreeding avoidance and selection of good genes. Exp. Clin. Immunogenet. 15, 119–129 10.1159/000019063 (doi:10.1159/000019063) [DOI] [PubMed] [Google Scholar]
- 28.Jordan W. C., Bruford M. W. 1998. New perspectives on mate choice and the MHC. Heredity 81, 127–133 10.1038/sj.hdy.6884281 (doi:10.1038/sj.hdy.6884281) [DOI] [PubMed] [Google Scholar]
- 29.Neff B. D., Garner S. R., Heath J. W., Heath D. D. 2008. The MHC and non-random mating in a captive population of Chinook salmon. Heredity 101, 175–185 10.1038/hdy.2008.43 (doi:10.1038/hdy.2008.43) [DOI] [PubMed] [Google Scholar]
- 30.Miller H. C., Moore J. A., Nelson N. J., Daugherty C. H. 2009. Influence of major histocompatibility complex genotype on mating success in a free-ranging reptile population. Proc. R. Soc. B 276, 1695–1704 10.1098/rspb.2008.1840 (doi:10.1098/rspb.2008.1840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherborne A. L., Thom M. D., Paterson S., Jury F., Ollier W. E. R., Stockley P., Beynon R. J., Hurst J. L. 2007. The genetic basis of inbreeding avoidance in house mice. Curr. Biol. 17, 2061–2066 10.1016/j.cub.2007.10.041 (doi:10.1016/j.cub.2007.10.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setchell J. M., Charpentier M. J. E., Abbott K. M., Wickings E. J., Knapp L. A. 2010. Opposites attract: MHC-associated mate choice in a polygynous primate. J. Evol. Biol. 23, 136–148 10.1111/j.1420-9101.2009.01880.x (doi:10.1111/j.1420-9101.2009.01880.x) [DOI] [PubMed] [Google Scholar]
- 33.Huchard E., Knapp L. A., Wang J., Raymond M., Cowlishaw G. 2010. MHC, mate choice and heterozygote advantage in a wild social primate. Mol. Ecol. 19, 2545–2561 10.1111/j.1365-294X.2010.04644.x (doi:10.1111/j.1365-294X.2010.04644.x) [DOI] [PubMed] [Google Scholar]
- 34.Edwards S. V., Hedrick P. W. 1998. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol. Evol. 13, 305–311 10.1016/S0169-5347(98)01416-5 (doi:10.1016/S0169-5347(98)01416-5) [DOI] [PubMed] [Google Scholar]
- 35.Wallny H. J., et al. 2006. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc. Natl Acad. Sci. USA 103, 1434–1439 10.1073/pnas.0507386103 (doi:10.1073/pnas.0507386103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess C. M., Edwards S. V. 2002. The evolution of the major histocompatibility complex in birds. Bioscience 52, 423–431 10.1641/0006-3568(2002)052[0423:TEOTMH]2.0.CO;2 (doi:10.1641/0006-3568(2002)052[0423:TEOTMH]2.0.CO;2) [DOI] [Google Scholar]
- 37.Kaufman J., Milne S., Göbel T. W. F., Walker B. A., Jacob J. P., Auffray C., Zoorob R., Beck S. 1999. The chicken B locus is a minimal essential major histocompatibility complex. Nature 401, 923–925 10.1038/44856 (doi:10.1038/44856) [DOI] [PubMed] [Google Scholar]
- 38.Bollmer J. L., Dunn P. O., Whittingham L. A., Wimpee C. 2010. Extensive MHC class II B gene duplication in a passerine, the common yellowthroat (Geothlypis trichas). J. Hered. 101, 448–460 10.1093/jhered/esq018 (doi:10.1093/jhered/esq018) [DOI] [PubMed] [Google Scholar]
- 39.Kalbe M., Eizaguirre C., Dankert I., Reusch T. B., Sommerfeld R. D., Wegner K. M., Milinski M. 2009. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc. R. Soc. B 276, 925–934 10.1098/rspb.2008.1466 (doi:10.1098/rspb.2008.1466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forsberg L. A., Dannewitz J., Petersson E., Grahn M. 2007. Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout—females fishing for optimal MHC dissimilarity. J. Evol. Biol. 20, 1859–1869 10.1111/j.1420-9101.2007.01380.x (doi:10.1111/j.1420-9101.2007.01380.x) [DOI] [PubMed] [Google Scholar]
- 41.Wegner K. M., Kalbe M., Kurtz J., Reusch T. B. H., Milinski M. 2003. Parasite selection for immunogenetic optimality. Science 301, 1343. 10.1126/science.1088293 (doi:10.1126/science.1088293) [DOI] [PubMed] [Google Scholar]
- 42.Lenz T. L., Eizaguirre C., Scharsack J. P., Kalbe M., Milinski M. 2009. Disentangling the role of MHC-dependent ‘good genes’ and ‘compatible genes’ in mate-choice decisions of three-spined sticklebacks Gasterosteus aculeatus under semi-natural conditions. J. Fish Biol. 75, 2122–2142 10.1111/j.1095-8649.2009.02410.x (doi:10.1111/j.1095-8649.2009.02410.x) [DOI] [PubMed] [Google Scholar]
- 43.Hedrick P. W. 2004. Comment on ‘Parasite selection for immunogenetic optimality’. Science 303, 957. 10.1126/science.1092163 (doi:10.1126/science.1092163) [DOI] [PubMed] [Google Scholar]
- 44.Westneat D. F., Stewart I. R. K. 2003. Extra-pair paternity in birds: causes, correlates, and conflict. Annu. Rev. Ecol. Evol. Syst. 34, 365–396 10.1146/annurev.ecolsys.34.011802.132439 (doi:10.1146/annurev.ecolsys.34.011802.132439) [DOI] [Google Scholar]
- 45.Dearborn D. C., Anders A. D., Parker P. G. 2001. Sexual dimorphism, extrapair fertilizations, and operational sex ratio in great frigatebirds (Fregata minor). Behav. Ecol. 12, 746–752 10.1093/beheco/12.6.746 (doi:10.1093/beheco/12.6.746) [DOI] [Google Scholar]
- 46.Dearborn D. C., Anders A. D. 2006. Demography and reproductive ecology of great frigatebirds. Atoll Res. Bull. 543, 159–171 [Google Scholar]
- 47.Alcaide M., Edwards S. V., Negro J. J. 2007. Characterization, polymorphism, and evolution of MHC class II B genes in birds of prey. J. Mol. Evol. 65, 541–554 10.1007/s00239-007-9033-9 (doi:10.1007/s00239-007-9033-9) [DOI] [PubMed] [Google Scholar]
- 48.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 49.Edwards S. V., Grahn M., Pott W. K. 1995. Dynamics of MHC evolution in birds and crocodilians: amplification of Class-II genes with degenerate primers. Mol. Ecol. 4, 719–729 10.1111/j.1365-294X.1995.tb00272.x (doi:10.1111/j.1365-294X.1995.tb00272.x) [DOI] [PubMed] [Google Scholar]
- 50.Dearborn D. C., Hailer F., Fleischer R. C. 2008. Microsatellite primers for relatedness and population structure in great frigatebirds (Pelecaniformes: Fregatidae). Mol. Ecol. Resour. 8, 1399–1401 10.1111/j.1755-0998.2008.02351.x (doi:10.1111/j.1755-0998.2008.02351.x) [DOI] [PubMed] [Google Scholar]
- 51.Hoffman J. I., Amos W. 2005. Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol. Ecol. 14, 599–612 10.1111/j.1365-294X.2004.02419.x (doi:10.1111/j.1365-294X.2004.02419.x) [DOI] [PubMed] [Google Scholar]
- 52.Clement M., Posada D., Crandall K. A. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 10.1046/j.1365-294x.2000.01020.x (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 53.Wetton J. H., Carter R. E., Parkin D. T., Walters D. 1987. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature 327, 147–149 10.1038/327147a0 (doi:10.1038/327147a0) [DOI] [PubMed] [Google Scholar]
- 54.Queller D. C., Goodnight K. F. 1989. Estimating relatedness using genetic markers. Evolution 43, 258–275 10.2307/2409206 (doi:10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- 55.Streiff R., Labbe T., Bacilieri R., Steinkellner H., Glössl J., Kremer A. 1998. Within-population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. assessed with isozymes and microsatellites. Mol. Ecol. 7, 317–328 10.1046/j.1365-294X.1998.00360.x (doi:10.1046/j.1365-294X.1998.00360.x) [DOI] [Google Scholar]
- 56.Ellegren H. 2000. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 16, 551–558 10.1016/S0168-9525(00)02139-9 (doi:10.1016/S0168-9525(00)02139-9) [DOI] [PubMed] [Google Scholar]
- 57.Caliebe A., Jochens A., Krawczak M., Rösler U. 2010. A Markov chain description of the stepwise mutation model: local and global behaviour of the allele process. J. Theor. Biol. 266, 336–342 10.1016/j.jtbi.2010.06.033 (doi:10.1016/j.jtbi.2010.06.033) [DOI] [PubMed] [Google Scholar]
- 58.Hardy O. J., Charbonnel N., Fréville H., Heuertz M. 2003. Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163, 1467–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coulson T. N., Pemberton J. M., Albon S. D., Beaumont M., Marshall T. C., Slate J., Guinness F. E., Clutton-Brock T. H. 1998. Microsatellites reveal heterosis in red deer. Proc. R. Soc. Lond. B 265, 489–495 10.1098/rspb.1998.0321 (doi:10.1098/rspb.1998.0321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansson B. 2010. The use (or misuse) of microsatellite allelic distances in the context of inbreeding and conservation genetics. Mol. Ecol. 19, 1082–1090 10.1111/j.1365-294X.2010.04556.x (doi:10.1111/j.1365-294X.2010.04556.x) [DOI] [PubMed] [Google Scholar]
- 61.Hardy O. J., Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620 10.1046/j.1471-8286.2002.00305.x (doi:10.1046/j.1471-8286.2002.00305.x) [DOI] [Google Scholar]
- 62.Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106 10.1111/j.1365-294X.2007.03089.x (doi:10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 63.Wang J. L. 2002. An estimator for pairwise relatedness using molecular markers. Genetics 160, 1203–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen L. B., Dearborn D. C. 2004. Great frigatebirds (Fregata minor) choose mates that are genetically similar. Anim. Behav. 68, 1229–1236 10.1016/j.anbehav.2003.12.021 (doi:10.1016/j.anbehav.2003.12.021) [DOI] [Google Scholar]
- 65.Baião P. C., Parker P. G. 2009. No evidence of extra-pair fertilization in Red-footed Boobies (Sula sula). Waterbirds 32, 179–182 10.1675/063.032.0122 (doi:10.1675/063.032.0122) [DOI] [Google Scholar]
- 66.Moon D. A., Veniamin S. M., Parks-Dely J. A., Magor K. E. 2005. The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J. Immunol. 175, 6702–6712 [DOI] [PubMed] [Google Scholar]
- 67.Muzaffar S. B., Jones I. L. 2004. Parasites and diseases of the auks (Alcidae) of the world and their ecology: a review. Mar. Ornithol. 32, 121–146 [Google Scholar]
- 68.Work T. M., Rameyer R. A. 1996. Haemoproteus iwa n. sp. in great frigatebirds (Fregata minor [Gmelin]) from Hawaii: parasite morphology and prevalence. J. Parasitol. 82, 489–491 10.2307/3284091 (doi:10.2307/3284091) [DOI] [PubMed] [Google Scholar]
- 69.Levin I. I., et al. In press Hippoboscid-transmitted Haemoproteus infecting Galapagos Pelecaniform birds: evidence from mitochondrial DNA and morphological description of blood stages of H. iwa. Int. J. Parasitol.. [DOI] [PubMed] [Google Scholar]
- 70.Beadell J. S., et al. 2006. Global phylogeographic limits of Hawaii's avian malaria. Proc. R. Soc. B 273, 2935–2944 10.1098/rspb.2006.3671 (doi:10.1098/rspb.2006.3671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Andersson M. 1994. Sexual selection. Princeton, NJ: Princeton University Press [Google Scholar]
- 72.Wright S. G., Dearborn D. C. 2009. Male ornament variation in a sexually dimorphic seabird with variable male mating success. Evol. Ecol. Res. 11, 759–770 [Google Scholar]
- 73.Foerster K., Delhey K., Johnsen A., Lifjeld J. T., Kempenaers B. 2003. Females increase offspring heterozygosity and fitness through extra-pair matings. Nature 425, 714–717 10.1038/nature01969 (doi:10.1038/nature01969) [DOI] [PubMed] [Google Scholar]
- 74.Stapleton M. K., Kleven O., Lifjeld J. T., Robertson R. J. 2007. Female tree swallows (Tachycineta bicolor) increase offspring heterozygosity through extrapair mating. Behav. Ecol. Sociobiol. 61, 1725–1733 10.1007/s00265-007-0404-4 (doi:10.1007/s00265-007-0404-4) [DOI] [Google Scholar]
- 75.Candolin U. 2003. The use of multiple cues in mate choice. Biol. Rev. Camb. Phil. Soc. 78, 575–595 10.1017/S1464793103006158 (doi:10.1017/S1464793103006158) [DOI] [PubMed] [Google Scholar]
- 76.Oh K. P., Badyaev A. V. 2006. Adaptive genetic complementarity in mate choice coexists with selection for elaborate sexual traits. Proc. R. Soc. B 273, 1913–1919 10.1098/rspb.2006.3528 (doi:10.1098/rspb.2006.3528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leinders-Zufall T., et al. 2004. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306, 1033–1037 10.1126/science.1102818 (doi:10.1126/science.1102818) [DOI] [PubMed] [Google Scholar]
- 78.Milinski M., Griffiths S., Wegner K. M., Reusch T. B. H., Haas-Assenbaum A., Boehm T. 2005. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA 102, 4414–4418 10.1073/pnas.0408264102 (doi:10.1073/pnas.0408264102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balthazart J., Taziaux M. 2009. The underestimated role of olfaction in avian reproduction? Behav. Brain Res. 200, 248–259 10.1016/j.bbr.2008.08.036 (doi:10.1016/j.bbr.2008.08.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonadonna F., Nevitt G. A. 2004. Partner-specific odor recognition in an antarctic seabird. Science 306, 835. 10.1126/science.1103001 (doi:10.1126/science.1103001) [DOI] [PubMed] [Google Scholar]
- 81.Hagelin J. C., Jones I. L., Rasmussen L. E. L. 2003. A tangerine-scented social odour in a monogamous seabird. Proc. R. Soc. Lond. B 270, 1323–1329 10.1098/rspb.2003.2379 (doi:10.1098/rspb.2003.2379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soini H. A., Schrock S. E., Bruce K. E., Wiesler D., Ketterson E. D., Novotny M. V. 2007. Seasonal variation in volatile compound profiles of preen gland secretions of the dark-eyed junco (Junco hyemalis). J. Chem. Ecol. 33, 183–198 10.1007/s10886-006-9210-0 (doi:10.1007/s10886-006-9210-0) [DOI] [PubMed] [Google Scholar]
- 83.Bonadonna F., Cunningham G. B., Jouventin P., Hesters F., Nevitt G. A. 2003. Evidence for nest-odour recognition in two species of diving petrel. J. Exp. Biol. 206, 3719–3722 10.1242/jeb.00610 (doi:10.1242/jeb.00610) [DOI] [PubMed] [Google Scholar]
- 84.Holland R. A., Thorup K., Gagliardo A., Bisson I. A., Knecht E., Mizrahi D., Wikelski M. 2009. Testing the role of sensory systems in the migratory heading of a songbird. J. Exp. Biol. 212, 4065–4071 10.1242/jeb.034504 (doi:10.1242/jeb.034504) [DOI] [PubMed] [Google Scholar]
- 85.Nevitt G. A., Losekoot M., Weimerskirch H. 2008. Evidence for olfactory search in wandering albatross, Diomedea exulans. Proc. Natl Acad. Sci. USA 105, 4576–4581 10.1073/pnas.0709047105 (doi:10.1073/pnas.0709047105) [DOI] [PMC free article] [PubMed] [Google Scholar]