Abstract
The toxin-producing microbial species Alexandrium minutum has a wide distribution in the Mediterranean Sea and causes high biomass blooms with consequences on the environment, human health and coastal-related economic activities. Comprehension of algal genetic differences and associated connectivity is fundamental to understand the geographical scale of adaptation and dispersal pathways of harmful microalgal species. In the present study, we combine A. minutum population genetic analyses based on microsatellites with indirect connectivity (Ci) estimations derived from a general circulation model of the Mediterranean sea. Our results show that four major clusters of genetically homogeneous groups can be identified, loosely corresponding to four regional seas: Adriatic, Ionian, Tyrrhenian and Catalan. Each of the four clusters included a small fraction of mixed and allochthonous genotypes from other Mediterranean areas, but the assignment to one of the four clusters was sufficiently robust as proved by the high ancestry coefficient values displayed by most of the individuals (>84%). The population structure of A. minutum on this scale can be explained by microalgal dispersion following the main regional circulation patterns over successive generations. We hypothesize that limited connectivity among the A. minutum populations results in low gene flow but not in the erosion of variability within the population, as indicated by the high gene diversity values. This study represents a first and new integrated approach, combining both genetic and numerical methods, to characterize and interpret the population structure of a toxic microalgal species. This approach of characterizing genetic population structure and connectivity at a regional scale holds promise for the control and management of the harmful algal bloom events in the Mediterranean Sea.
Keywords: connectivity, population structure, HAB (harmful algal blooms), microsatellite, genetic distance, dinoflagellate
1. Introduction
Management efforts against eutrophic events and high biomass proliferation of toxic microbial species in marine environments can benefit from information about how environmental factors influence genetic structure and population connectivity. Marine planktonic microbial species are generally thought to be dispersed throughout the ocean worldwide. Their ubiquity is attributed to huge population sizes [1,2] and their vast capacity for dispersal: they are passively transported and distributed homogeneously by different system currents, and there is an apparent absence of dispersal barriers [3]. This extensive dispersal capability has led to the assumption that microplanktonic populations are highly connected and they should therefore exhibit extensive gene flow preventing genetic isolation, fixation of genetic differences among populations and hence allopatric speciation [4,5]. As a consequence, these microbial species have been viewed as consisting of less genetically structured populations with a lower level of speciation than terrestrial animals and plants. However, molecular data have recently revealed a high degree of genetic diversity within microbial species with evident cryptic biodiversity that is well structured in the open ocean and benthic environment [6–9]. Together with other permanent and temporary components of the planktonic community, such as meroplankton, larval stages of animals and propaguli of seaweeds [10–14], micro-organisms show long-distance dispersal modulated by geographical and physical or ecological barriers [15,16]. The processes of gene flow within microbial species can result in different populations with different genetic structures that reflect the degree of connectivity of a geographical location with adjacent areas [17–19]. Gene flow may also be influenced by the duration of the vegetative planktonic form during the migration across the open ocean, as well as by local adaptation mediated by resting stages [20,21]. These circumstances are responsible for the successful dispersal and colonization of new areas by microbial micro-organisms [22].
The genetic seascape provides a scenario of connectivity and dispersal of both planktonic and benthic organisms through the coupling of sets of population genetic data and environmental descriptors within a spatial genetic structure [23,24]. Traditionally, estimates of dispersal have notoriously been difficult to obtain in marine environments owing to the high dispersal potential of many organisms and a variety of processes involved at different scales. Increasing efforts are underway in order to link population genetic structure with large-scale circulation patterns [25]. Most of these studies rely on isolation-by-distance analysis, but they very often show no linear relationship [26,27]. Alternatively, spatially explicit physical models, including dispersal kernels of different complexity, have been implemented in order to explain the genetic distances among populations within a species [28]. The combination of numerically modelling dispersion with population genetic structure analysis has notably improved the scenario of several marine species population dynamics and can also open new frontiers on the comprehension of the mechanisms for HAB species' dispersion and expansion.
This is the first reported study to explore population structure using microsatellite loci combined with a connectivity model of the toxic dinoflagellate Alexandrium minutum populations in the Mediterranean Sea. The study represents a novel advancement in the evaluation of unknown population genetic structure of a bloom-forming phytoplankton species by incorporating a model of indirect connectivity within the Mediterranean basin. Using a numerical simulation model of dispersion throughout different generations in the Mediterranean Sea, we constructed a hypothetical scenario of natural connectivity of this toxic microbial species that may explain some aspects of the observed genetic structure of A. minutum populations. Alexandrium minutum is responsible for harmful phenomena which negatively alter marine ecosystem functions and services with outbreaks of paralytic shellfish poisoning (PSP) [29,30]. In the Mediterranean Sea, the increase and expansion of A. minutum outbreaks have coincided with exploitation of the coastline that increasingly offers confined nutrient-enriched waters suitable for microalgal proliferation [31,32]. Despite the dinoflagellates' preference for settling in confined environments near shore, A. minutum has an enormous natural potential of dispersal because of its capacity to grow and produce resting cysts under a wide range of environmental conditions [33]. Our approach allowed characterizing the protist A. minutum population structure and connectivity at spatial scales for the control and management of the HAB events in the Mediterranean Sea.
2. Material and methods
(a). Origin and clonal cultures
A total of 116 clonal isolates of A. minutum were collected from different coastal areas of the Mediterranean Sea and the eastern Atlantic (electronic supplementary material, table S1). The Spanish Atlantic locality was sampled as it represented an external site to be compared with the Mediterranean sampling sites. The isolates were obtained from environmental samples which included both surface sea water and sediment, as described in Penna et al. [34].
(b). DNA extraction and microsatellite genotyping
Exponential growth phase of each isolate was collected by centrifugation for DNA extraction. Genomic DNA was purified using DNeasy Plant Mini Kit (Qiagen). The ribosomal gene 5.8S rDNA and internal transcribed spacer (ITS) regions of each microalgal isolate genomic DNA were amplified according to the protocol of Penna et al. [34] and the polymerase chain reaction (PCR)-amplified products were sequenced (electronic supplementary material, table S2).
After testing 12 microsatellite loci [35], seven loci were selected for genotyping the 116 isolates as the other five loci could not be confidently amplified in all the samples. PCR amplifications were carried out in a DNA Thermo Cycler 2720 (Applied Biosystems). PCR mixture contained 5 ng of template DNA, 200 µM of each dNTP, 0.5 µM of each primer, 2 mM MgCl2, 1 × reaction buffer and 0.6 U AmpliTaq Gold (Applied Biosystems) in a final volume of 25 µl. A concentration of 3 mM MgCl2 was used for the Aminu22 locus. For the Aminu11, Aminu29 and Aminu48 loci, only 0.8 µg µl−1 of BSA was added to the reaction mixture. The PCR conditions for Aminu22, Aminu41, Aminu43 and Aminu44 loci were as follows: an initial denaturation step of 10 min at 94°C, 38 cycles of 30 s at 94°C, 30 s at 50°C, 1 min at 72°C and a final extension step of 5 min at 72°C. The PCR conditions for Aminu11, Aminu29 and Aminu48 loci were as follows: an initial denaturation step of 10 min at 94°C, five cycles of 30 s at 94°C, 30 s at 58°C, 1 min at 72°C; five cycles of 30 s at 94°C, 30 s at 55°C, 1 min at 72°C; 28 cycles of 30 s at 94°C, 30 s at 50°C, 1 min at 72°C and a final extension step of 5 min at 72°C. Sizing of microsatellite loci was carried out by GeneMapper (Applied Biosystems).
(c). Population genetic analysis
The data were analysed in the following steps.
— Genetic diversity. The linkage disequilibrium (LD) test was performed with Fstat v. 2.9.3.2 (http://www2.unil.ch/popgen/softwares/fstat.htm). The number of alleles per locus was calculated using Arlequin v. 3.11 (http://cmpg.unibe.ch/software/arlequin3/), and gene diversity and allelic richness using Fstat.
— Population differentiation. Global and pairwise FST fixation indices and an analysis of molecular variance (AMOVA) were calculated using Arlequin v. 3.1. The significance of the FST values was assessed after 1000 permutations. A locus-by-locus AMOVA with FST calculation was carried out using Arlequin v. 3.1. Unrooted dendrogram of populations was constructed using the neighbour-joining (NJ) method, as implemented in the Phylip v. 3.6 (http://evolution.gs.washington.edu/phylip.html) using chord distance (Dc) [36] calculated with Microsat (http://hpgl.stanford.edu/projects/microsat/) v. 1.4d as genetic distance. The robustness of the tree was tested by 10 000 bootstrap pseudoreplicates. An isolation-by-distance analysis was carried out to correlate genetic distances, expressed as FST/(1 − FST), with geographical distances through the Isolation by Distance v. 3.16 (http://www.bio.sdsu.edu/pub/andy/IBD.html). Geographical distances were measured as the linear distances between pairs of locations. The significance of each test was examined by 10 000 permutations.
— Inference of population structure. A principal coordinates analysis (PCoA), based on the matrix of genetic distances among alleles, was performed using genalex v. 6.1. Population structure was investigated with a Bayesian clustering method implemented in structure v. 2.2 [37]. The program implements a model-based clustering method to infer population structure and assign individuals to populations using multilocus genotype data (microsatellite data). Using the estimated allele frequencies for each individuals also based on a modest number of loci, it is possible to compute the likelihood of origin of a given genotype from a population. After inferring the most likely number of different clusters, this method estimates the ancestry coefficient of each individual, which represents the likelihood of it belonging to one of the identified clusters. In this study, the model for the assignment of ancestry of individuals was the admixture model, where each individual draws some fraction of his genome from each of the K populations. The most likely number of K cluster was identified through three independent runs. Each run was based on 1 000 000 MCMC simulations with an initial ‘burn-in’ period of 250 000; 10 iterations were run for each K value ranging from 1 to 10. The second-order statistics ΔK [38] was calculated to better refine estimation of K. The Structure analysis was also run hierarchically with the following scheme: a first run (same settings as above) was performed setting K = 2. Each of the two identified groups was then subjected to a canonical analysis in order to find the most likely number of inferred group. Finally, the estimated cluster membership coefficient (Q) matrices of multiple runs generated by Structure were analysed using CLUster Matching and Permutation Program (CLUMPP v. 1.1.2; [39]). The program evaluates the similarity of outcomes between populations structure estimates [40] identifying the run that produces the highest average pairwise similiarity (H-value). The input of CLUMPP is represented by the output files generated by Structure for each of the K-values tested. After running the program, determination of the best configuration (in terms of K) is accomplished by taking the highest H-value among those estimated for different K.
(d). Population connectivity estimation
The dispersal of A. minutum in the Mediterranean Sea was investigated with a coupled biophysical algorithm, which included a high-resolution ocean circulation model and an individual-based model (IBM). The ocean circulation model, described by Jordi and Wang [41], reproduced the major characteristics of the observed mean circulation and mesoscale variability in the Mediterranean Sea. The IBM, which incorporated the free-living planktonic stage of A. minutum, was embedded in the ocean circulation model. At each time-step (Δt = 1 h), the final position of each particle representing a large number of A. minutum cells was estimated from particle speed at the initial position. Speed was determined by interpolating the model currents with the particle position. In addition, a random walk movement (zero mean and variance of 2KhΔt, where Kh is the horizontal diffusivity set to 1000 m2 s−1) was superimposed onto this speed to simulate particle dispersion owing to sub-grid scale processes that cannot be resolved by the model currents. A total of 10 particles were released at each model grid point adjacent to the coast every 10 days over the last 8 years of model currents. Each release was carried forward for 30 days (considered to represent the vegetative cell phase) without inclusion of cell mortality. The probability that a particle is transported from one coastal grid point to arrive at another in 30 days is the direct connectivity (Cd), calculated as the average percentage of particles released from one model grid point that arrived to another grid point adjacent to the coast at the end of the 30 days. Indirect connectivity (Ci) is the probability of a vegetative cell being transported from one coastal grid point to another along indirect routes of migration by means of island stepping stones and coastal-zone diffusion and provides information on dispersal over several life cycles (or cell phases). According to graph theory, Ci is defined as:
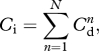 |
where N is the number of life cycles, Cd identifies the direct routes and Cd2 includes all the routes requiring one step (or two direct routes), etc. [42]. Ci reached an approximate steady state within 200 life cycles. Therefore, the life cycle of A. minutum limits the geographical range over which vegetative cells can advect or mix, even taking possible indirect routes of migration into account. Despite this limit, Ci was calculated over 500 life cycles to ensure that the steady state was reached.
3. Results
Most of the A. minutum isolates were sequenced for species-specific identification. The length of the 5.8S rDNA gene and ITS1–ITS2 regions of A. minutum isolates was 520 bp. Alignment analyses of sequences of different dinoflagellate species in GenBank showed complete identity (100%) with the Mediterranean A. minutum confirming the species assignment of our isolates.
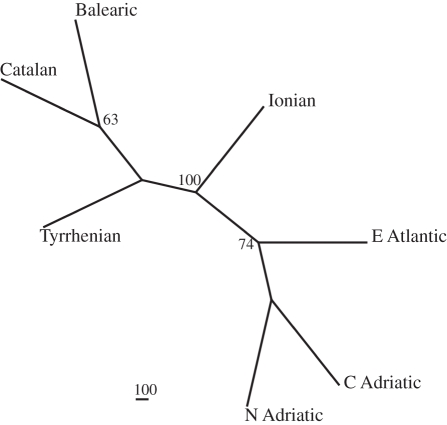
Of the 12 polymorphic microsatellites tested for the amplification of the A. minutum isolates, seven microsatellites gave robust and repeatable amplification products, namely Aminu10, Aminu11, Aminu22, Aminu29, Aminu41, Aminu43, Aminu44 and Aminu48. The remaining five microsatellites were discarded, as PCR products were obtained in less than 50 per cent of the A. minutum isolates. As expected, all seven loci showed a single allele in each isolate owing to the haploid phase of the vegetative cells of this species. Statistical analyses of the seven microsatellites gave no signs of linkage disequilibrium (data not shown). The seven loci were polymorphic in all sampled localities except for the Aminu48 locus in the Catalan population, where only one allele was found. An NJ tree was constructed based on the assumption that each A. minutum sampling site corresponded to a distinct genetic group. The NJ tree identified six distinct groups corresponding to the different regional seas: north-western Adriatic, Ionian, Tyrrhenian, Catalan, Balearic and Atlantic (figure 1). The topology of the NJ tree was strongly supported by high bootstrap values.
Figure 1.
Neighbour-joining unrooted tree of seven Alexandrium minutum sampling sites in the Mediterranean Sea and Atlantic based on a chord distance matrix. Bootstrap values (>60%) are placed at each node.
The main indices of genetic diversity for each group are shown in the electronic supplementary material, table S3. The Adriatic emerged as the group with the highest genetic diversity, having the highest values of both gene diversity and allelic richness. The AMOVA based on allele frequencies showed that most of the variation was owing to differences between individuals within groups (81%), and yet a remarkable 19 per cent of variation was owing to differences between groups; the global FST value was 0.189 (p < 0.001) (electronic supplementary material, table S4). A pairwise comparison between the six A. minutum groups provided FST values ranging from 0.113 to 0.302. All 15 pairwise FST values were statistically highly significant (p < 0.001, after false discovery rate correction for multiple tests) demonstrating a highly structured pattern mainly based on the geographical distribution of the different groups (electronic supplementary material, table S5). The two most distant sampling sites, Adriatic and Atlantic, showed the lowest pairwise FST values (0.113, p < 0.001), while none of the other values correlated with the geographical distance between the sampling sites. An AMOVA was also performed independently for each locus, as locus-by-locus AMOVA (data not shown). The global variance components could be different because the degree of freedom varies from locus to locus, and therefore the FST estimates could also vary. In fact, the Aminu29 locus had a low percentage of variation (7.92%) owing to differences among the populations. Even though the FST value for this locus was the lowest when compared with the other loci, it was statistically significant (p < 0.05, after correction for multiple tests). This means that the differentiation measured with the global FST was not owing to the predominating effect of few loci.
No evidence of isolation-by-distance was found after correlating genetic and geographical distances (r = 0.0346, p = 0.5419) with a Mantel test.
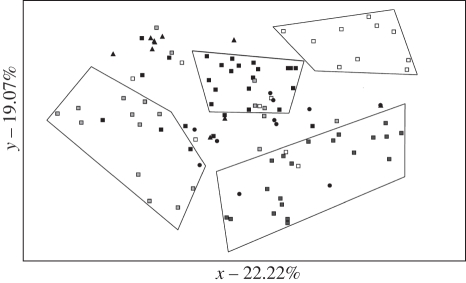
When considering the 116 individuals instead of sampling localities, that is, not assuming any a priori knowledge, a PCoA identified four main clusters corresponding to four different seas—Adriatic, Ionian, Tyrrhenian and Catalan (figure 2). The two PCoA axes explained 22.22 and 19.07 per cent of the variance among genotypes, respectively. Furthermore, it was evident that each group also comprised some allochthonous genotypes derived from other regional seas. A Bayesian clustering method (implemented in Structure) was used on single genotypes to infer the most likely number of homogeneous genetic groups, K. Comparison of the ln-likelihood of the different runs for the different K tested, revealed no peak, although a slight but constant increase in ln-likelihood up to the highest number of K tested (detailed analyses of Structure were also shown in a paragraph of the electronic supplementary material). At the same time, the second-order statistic devised to infer the most likely number of K, namely ΔK, did not yield unambiguous results either, there being a major peak at K = 2 and another peak at K = 4. To distinguish between K = 2 and K = 4, we estimated the highest average pairwise similarity index (H-value) according to the method described in Nordborg et al. [40] and implemented in the software CLUMPP. We inferred H for K = 2, K = 3 and K = 4: the results clearly showed that the highest H-value is that associated with K = 4 (electronic supplementary material, table S6). Furthermore, adopting a hierarchical Structure analysis, we recovered the same four distinct genetically homogeneous groups identified by the standard Structure analysis and by CLUMPP. Not only did these four clusters correspond to those identified by PCoA, but with K = 4 we recorded the highest ancestry coefficient for each genotype. The overall genetic structure of the seven sampling sites can be therefore assigned to four distinct homogeneous clusters or populations (table 1). Taking K = 4, we found that the two sampled Adriatic sites could be assigned to cluster 1, the Ionian site to cluster 2, the Tyrrhenian site to cluster 3 and the Catalan site to cluster 4. The Balearic and Atlantic sampled sites could not be assigned to any of the four clusters as they consisted either of individuals with mixed genotypic components or of individuals with components from two or more clusters. It was also evident from the individual ancestry coefficient that each population contained allochthonous individuals as well as individuals with mixed genotypes. At any rate, it is also relevant that a very high percentage of individuals (84.4%) showed ancestry coefficients higher than 75 per cent highly indicative of assignment to one of the four inferred clusters (figure 3).
Figure 2.
Principal coordinate analysis (PCoA) based on 116 isolates of Alexandrium minutum typed at seven microsatellite loci. Black square, N Adriatic; dark grey square, C Adriatic; white square, Ionian; light grey square, Tyrrhenian; grey square, Catalan; black circle, Balearic; black triangle, E Atlantic.
Table 1.
Average coefficients of ancestry obtained from Structure at K = 4. The highest values of each samples are indicated in bold.
| Regional seas | Cluster (K) |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| N Adriatic | 0.659 | 0.025 | 0.299 | 0.017 |
| C Adriatic | 0.719 | 0.064 | 0.147 | 0.070 |
| Ionian | 0.008 | 0.833 | 0.056 | 0.104 |
| Tyrrhenian | 0.043 | 0.083 | 0.749 | 0.125 |
| Catalan | 0.048 | 0.023 | 0.007 | 0.922 |
| Balearic | 0.103 | 0.338 | 0.078 | 0.481 |
| Atlantic | 0.538 | 0.145 | 0.213 | 0.104 |
Figure 3.
Population structure based on seven microsatellite loci of Alexandrium minutum as estimated by genotypic clustering in Structure. Assignment of 116 individuals to K = 4 genetically distinct groups. Each individual is represented by a vertical bar coloured according to the assigned cluster.
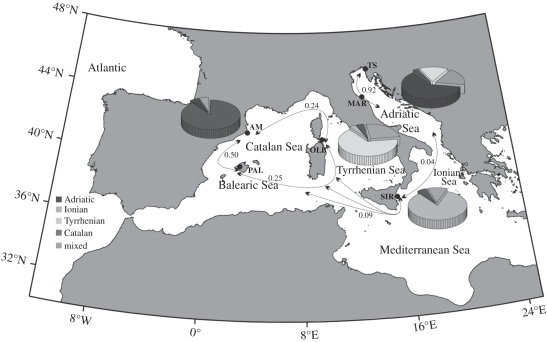
Hydrodynamic model simulations indicated that direct cell exchange (Cd) on a regional scale, even between the geographically closest locations, is highly unlikely on the timescale considered for vegetative cell survival. This is because of the typically low current intensities in the open ocean (<30 cm s−1) and to the ocean circulation patterns that favour alongshore transport. Indirect connectivity (Ci) should therefore be considered the main dispersal pathway on this spatial scale. The Ci pattern revealed two large regions of high internal connectivity: the western Adriatic and western Mediterranean sites, linked by the Ionian Sea. The highest Ci was obtained for the Adriatic sites of Trieste and Marotta (0.92). Particles tended to recirculate within this sea and there was relatively little exchange with other areas (<0.05). Low renewal through the Strait of Otranto was responsible for the natural isolation of this geographical area. Indirect connectivity between sites in the western Mediterranean was also relatively high. In particular, Arenys de Mar and Palma (Ci = 0.5) were well connected, and transport to Olbia, although weaker, was also significant (Ci = 0.25 and 0.24 from Palma and Arenys, respectively). The Ionian was better connected to Gibraltar, because of the strong Algerian Current, than to geographically closer regions such as the Adriatic Sea (figure 4).
Figure 4.
Distribution of different populations of Alexandrium minutum and indirect connectivity estimated from different sampling sites in the Mediterranean Sea. The pie graph for each site shows the percentage of individuals, typed at seven microsatellites, assigned to one of the four clusters identified by PCoA and the Bayesian method (see also electronic supplementary material, table S7).
4. Discussion
Recent phylogenetic studies on the A. minutum on macrogeographical scale using both ribosomal genes and microsatellites revealed that this species is geographically differentiated, although the isolates of Mediterranean origin do not appear to be geographically structured but rather dispersed in different clusters [43,44]. This lack of structure is not supported by our data that demonstrate the existence of regional scale genetic population structure of the toxin-producing phytoplankton species A. minutum in the Mediterranean Sea. Moreover, we suggest that the genetic population differentiation is related to the basin scale transport patterns through successive generations of the vegetative microalgal cells. By combining indirect connectivity and genetic models, we were able to add new insights into microbial species connectivity patterns relevant to oceanic scenarios and useful for understanding the distribution, dispersal and evolution for the so defined cosmopolitan marine micro-organism. The diversity detected with microsatellite markers significantly exceeds that of ribosomal DNA markers revealing enough allelic variation to investigate the genetic structure of A. minutum in the Mediterranean Sea. The A. minutum isolates were collected from seven different geographical areas. We started from the strong assumption that each sampling area corresponded to a distinct, genetically homogeneous A. minutum population. The NJ dendrogram confirmed that six of the seven sampling areas were well differentiated, while the two Adriatic sites merged into a single cluster. A PCoA of individual genotypes, with no a priori assumptions regarding their origin, identified four major groups of isolates. The geographical origin of most of the isolates in each group showed that these four groups correspond to four different regional seas: Adriatic, Ionian, Tyrrhenian and Catalan. Allochthonous individuals were present within each group. The Structure and subsequent CLUMPP analyses highlighted the best configuration was that with the setting K = 4. Most of the isolates in the Mediterranean Sea were assigned to one of the four inferred clusters with a very high probability. Each of the four clusters included a small fraction of mixed and allochthonous genotypes from other Mediterranean areas, but the assignment to one of the four clusters was sufficiently robust, as proved by the high ancestry coefficient values which most of the individuals (>84%) showed. The strong population structure was also confirmed by the global FST value, while the pairwise FST values among the four Mediterranean clusters were also highly significant. Within each cluster, high gene diversity values were encountered. This unexpected within-population variability that was not related to any gene flow among sampling areas can be explained by the biological and genetic role of the seed bank in marine habitats. It has been proved that resting cysts settled in the bottom sediment can remain viable for many decades and accumulate over several years constituting a reservoir of potential genetic diversity and resulting in high values of gene diversity. It is therefore expected that A. minutum local cyst banks in each examined area in the Mediterranean Sea also potentially host high gene diversity. The role of resting cysts in promoting genetic diversity has been demonstrated in other geographical areas, such as the northern European Gullmar Fjord, where the diatom population of Skeletonema marinoi is genetically differentiated from the open sea population. Furthermore, the genotypes in this fjord represent a continuous gradient of genetic differentiation that permits the recruitment of different clones with different live-permanence [45]. Plankton and benthic environments are genetically linked with the marine ecosystem, as benthic stages of organisms are continuously resuspended from a large gene pool in the bottom to the surface where the propagules germinate and persist in the vegetative form. Populations or clones are probably selected by environmental conditions to grow and dominate the water column [46]. Like other phytoplankton species, the dinoflagellate A. minutum alternates between planktonic (vegetative cells) and benthic (resting cysts) stages as a response to endogenous and/or environmental factors [47,48].
Calculation of oceanographic distance in relation to the population genetic structure based on FST was applied to tentatively explain the genetic pattern of A. minutum in the Mediterranean basin. The population structure of A. minutum was not correlated to Euclidean distance as expected under an isolation-by-distance model; in other terms, the probabilities of A. minutum gene flow among sites have little to do with the physical distance between them [49,50]. The decoupling between the genetic structure and Euclidean distances between the sampled sites is probably due to the complex of geography and hydrodynamics of the Mediterranean regions as already depicted for other organisms. Marine biotic communities exhibit patterns of genetic structure that are more in accordance with asymmetric dispersal processes than to geographical distances [51,52]. More recent genetic studies have demonstrated an asymmetric gene flow under the Antarctic Circumpolar Current (ACC) in fish population [53], as well as an asymmetrical dispersal of bull-kelp in southern New Zealand [54] or multiple events of introgression of allochthonous genotypes and rapid dispersal into the Mediterranean Sea that explain the population genetic variability of the seaweed Asparagopsis taxiformis [55].
Straight geographical distances are not good indicators for inferring dispersal patterns in oceanic systems, particularly in the Mediterranean Sea, which is characterized by closed circulation patterns produced by coastal topography. In our study it seems, therefore, that the four major distinct genetic clusters reflect a situation of rather well-established geographical isolation with very little, if any, gene flow between them. Although the situation regarding the Balearic Sea and the eastern Atlantic deserves further investigation, mainly by additional sampling, some preliminary hypotheses can be advanced with respect to the four major clusters identified.
We considered the indirect connectivity model based on the Mediterranean basin circulation at different regime scales. Further, as this study demonstrates, direct connectivity at a regional level is highly unlikely. This may be attributed essentially to the relatively short vegetative stage that precludes dispersal over long distances. Alexandrium minutum vegetative cells are known to spend probably less than one month in the water column, although they are present throughout the year [56]. Moreover, long-distance propagation of A. minutum in open ocean conditions is unlikely because of the severe nutrient limitations [57]. Instead, a scenario of the indirect connectivity provides a more likely explanation of the main features of genetic differentiation among the four populations of A. minutum examined. The Ci allowed two areas with relatively high internal connections to be identified: the western Adriatic and Mediterranean Sea. This is consistent with the previously described ecoregions [58] and with the general concept that the Mediterranean Sea consists of two relatively independent basins communicating through the Strait of Sicily, a situation which constrains basin-scale circulation and has potential consequences for the distribution of planktonic organisms [59]. Within each region, the Ci was relatively high. In particular, there was considerable internal connectivity in the Adriatic Sea (Ci = 0.92) attributed to its closed circulation pattern. Stable cyclonic circulation in the Adriatic Sea not only favours alongshelf transportation, but also acts as a frontal structure reducing offshore transportation of coastal phytoplankton [60]. Furthermore, the phytoplanktonic cell survival in the northern Adriatic Sea can be sustained by outflow from the river Po, which markedly influences the hydrology of the western side of the northern Adriatic Basin [61,62]. This Ci scenario in the Adriatic Sea is consistent with the genetic structure of the two sampling sites, Trieste and Marotta identified by the NJ and PCoA configurations, and Structure assignment to belong to the same cluster. In the western Mediterranean Sea, connectivity estimates revealed the close link between Palma and Arenys de Mar (Ci = 0.50), which are connected by the anti-clockwise-circulating northern current [63]. Less robust were the connections between Olbia, which appeared genetically well-differentiated. The presence of a semi-permanent sub-basin gyre in the northern Tyrrhenian Sea [64] may play a key role in promoting genetic differentiation between the Arenys de Mar and Olbia sites. Finally, the Ionian site, Siracusa, had the highest FST pairwise values when compared with the other Mediterranean clusters, making this site highly differentiated and isolated, and apparently uninfluenced by any gene flow. In fact, there was little connectivity between Siracusa and the other areas probably owing to constraints on exchanges exerted by the narrow Strait of Sicily and more crucially by the Strait of Otranto. Although it is possible that A. minutum may eventually enter the Adriatic Sea from Siracusa, dispersal in this direction is low as the main flow in the area is towards the eastern Mediterranean and it only enters the Adriatic after cyclonically circulating through the entire eastern Mediterranean Sea [65].
This connectivity model allows us to advance some hypotheses regarding the presence in each of the four populations identified of isolates with genotypes assignable to a different geographical cluster or with mixed genotypic components. Here, it is important to recall that mixed genotypes can arise given that sexual reproduction, being one of the dinoflagellate A. minutum's life stages, permits recombination between two different genotypes of diploid cells and therefore confers greater genetic variability than that displayed by haploid organisms during the late phase of bloom events.
In the northern Adriatic area, allochthonous genotypes with a main Tyrrhenian component (16%) were found (electronic supplementary material, table S7). Furthermore, genotypes with Catalan and Ionian components were retrieved in the Olbia site. While the connectivity model shows that some kind of contact between the Tyrrhenian and Catalan Seas cannot be ruled out, contact between the Tyrrhenian and Adriatic Seas and the Tyrrhenian and Ionian Seas appears very unlikely. Moreover, the northern Adriatic and Tyrrhenian areas are clearly separated by the large- and meso-scale circulation systems and the geographical barrier of the Italian peninsula.
Furthermore, the worth of indirect connectivity as a predictor of the genetic structure within the Mediterranean basin was confirmed by a Mantel test that showed a highly significant correlation between matrices of Fst values among the four populations and the correspondent values of Ci (indirect connectivity, p < 0.001; data not shown).
Anyway, we cannot dismiss that alternative ways of planktonic cell-dispersal exist, and these could include human-mediated transportation. Ballast water discharges and shellfish stock translocation have recently been proposed as a human-aided vector of planktonic microbial species propagation [66], even though the relative importance of this propagation pathway is highly debated [67]. The northern Adriatic and Tyrrhenian sites are characterized by intense commercial shipping, as well as widespread aquaculture with potential mussel translocations; therefore human-mediated transport cannot be excluded.
Application of high-resolution molecular markers, such as microsatellite loci, allowed us to identify a robust population structure within the harmful phytoplankton species A. minutum based on connectivity between distinct areas of the Mediterranean basin. To the best of our knowledge, this study marks the first attempt to correlate genetic differentiation of A. minutum isolates with a model of Mediterranean water circulation. Our model of indirect connectivity was consistent with the results of A. minutum population genetic structure analyses on a regional scale. The low number of sampling sites is certainly a limit for a more comprehensive description of A. minutum population structure in the Mediterranean Sea considering the varied coastal systems and hydrographic regimes in the different Mediterranean regions. Despite the admittedly low number of sampling sites across the Mediterranean region, we obtained a strong signal of differentiation. It seems highly likely that other coastal sites, especially those confined or semi-confined, would host well-differentiated populations and therefore, our data can provide a basis for planning the control and management of harmful algal blooms in Mediterranean waters.
Finally, our results served as validation for estimating population connectivity using an oceanographic approach of Ci for simulating phytoplankton dispersal in the marine environment.
Acknowledgements
We thank Santiago Fraga and Isabel Bravo for providing us with strains; Luglié Antonella and Carmela Caroppo for field samples. Financial support was provided by the EC-funded Research Project SEED (Life cycle transformations among HAB species, and the environmental and physiological factors that regulate them), GOCE-CT-2005-003875; PRIN 2007 Italian Ministry of University and Research, prot. 2007 FXSCL2 Grant and prot. 2007 R8AWYY_003 Grant.
References
- 1.Finlay B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296, 1061–1063 10.1126/science.1070710 (doi:10.1126/science.1070710) [DOI] [PubMed] [Google Scholar]
- 2.Taylor F. J. R., Hoppenrath M., Saldarriaga J. F. 2007. Dinoflagellate diversity and distribution. Biodivers. Conserv. 17, 407–418 10.1007/s10531-007-9258-3 (doi:10.1007/s10531-007-9258-3) [DOI] [Google Scholar]
- 3.Cermeño P., Falkowski P. G. 2009. Controls on diatom biogeography in the ocean. Science 325, 1539–1541 10.1126/science.11741599 (doi:10.1126/science.11741599) [DOI] [PubMed] [Google Scholar]
- 4.Norris R. D. 2000. Pelagic species diversity, biogeography, and evolution. Paleobiology 26, 236–258 10.1666/0094-8373(2000)26[236:PSDBAE]2.0.CO;2 (doi:10.1666/0094-8373(2000)26[236:PSDBAE]2.0.CO;2) [DOI] [Google Scholar]
- 5.Palumbi S. R. 2003. Population genetics, demographic connectivity, and the design of marine reserves. Ecol. Appl. 13, 146–158 10.1890/1051-0761(2003)013 (doi:10.1890/1051-0761(2003)013) [DOI] [Google Scholar]
- 6.De Vargas C. N. R., Zaninetti L., Gibb S. W., Pawlowski J. 1999. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc. Natl Acad. Sci. USA 96, 2864–2868 10.1073/pnas.96.6.2864 (doi:10.1073/pnas.96.6.2864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.John U., Fensome R. A., Medlin L. K. 2003. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense ‘species complex' (Dinophyceae). J. Mol. Evol. 20, 1015–1027 10.1093/molbev/msg105 (doi:10.1093/molbev/msg105) [DOI] [PubMed] [Google Scholar]
- 8.Hughes Martiny J. B., et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. 4, 102–112 10.1038/nrmicro1341 (doi:10.1038/nrmicro1341) [DOI] [PubMed] [Google Scholar]
- 9.Penna A., Fraga S., Battocchi C., Casabianca S., Riobò P., Giacobbe M. G., Vernesi C. 2010. A phylogeography study of the toxic benthic genus Ostreopsis Schmidt. J. Biogeogr. 37, 830–841 10.1111/j.1365-2699.2009.02265.x (doi:10.1111/j.1365-2699.2009.02265.x) [DOI] [Google Scholar]
- 10.Dawson M. N., Sen Gupta A., England M. H. 2005. Coupled biophysical global ocean model and molecular genetic analyses identify multiple introductions of cryptogenic species. Proc. Natl Acad. Sci. USA 102, 11 968–11 973 10.1073/pnas.0503811102 (doi:10.1073/pnas.0503811102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinlan B. P., Gaines S. D. 2003. Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84, 2007–2020 10.1890/01-0622 (doi:10.1890/01-0622) [DOI] [Google Scholar]
- 12.Planes S., Jones G. P., Thorrold S. R. 2009. Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl Acad. Sci. 106, 5693–5697 10.1073/pnas.0808007106 (doi:10.1073/pnas.0808007106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weersing K., Toonen R. J. 2009. Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Progr. Ser. 393, 1–12 10.3354/meps08287 (doi:10.3354/meps08287) [DOI] [Google Scholar]
- 14.Serra I. A., et al. 2010. Genetic structure in the Mediterranean seagrass Posidonia oceanica: disentangling past vicariance events from contemporary patterns of gene flow. Mol. Ecol. 19, 557–568 10.1111/j.1365-294X.2009.04462.x (doi:10.1111/j.1365-294X.2009.04462.x) [DOI] [PubMed] [Google Scholar]
- 15.Darling K. F., Kucera M., Pudsey C. J., Wade C. M. 2004. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc. Natl Acad. Sci. USA 101, 7657–7662 10.1073/pnas.0402401101 (doi:10.1073/pnas.0402401101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casteleyn G., Leliaert F., Backeljau T., Debeer A. E., Kotaki Y., Rhodes L., Lundholm N., Sabbe K., Vyverman W. 2010. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc. Natl Acad. Sci. USA 107, 12 952–12 957 10.1073/pnas.1001380107 (doi:10.1073/pnas.1001380107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iglesias-Rodrıguez M. D., Schofield O. M., Batley J., Medlin L. K., Hayes P. K. 2006. Intraspecific genetic diversity in the marine coccolithophore Emiliania huxleyi (Prymnesiophyceae): the use of microsatellite analysis in marine phytoplankton population studies. J. Phycol. 42, 526–536 10.1111/j.1529-8817.2006.00231.x (doi:10.1111/j.1529-8817.2006.00231.x) [DOI] [Google Scholar]
- 18.Kooistra W. H. C. F., Sarno D., Balzano S., Gu H., Andersen R. A., Zingone A. 2008. Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist 159, 177–193 10.1016/j.protis.2007.09.004 (doi:10.1016/j.protis.2007.09.004) [DOI] [PubMed] [Google Scholar]
- 19.Nagai S., Nishitani G., Sakamoto S., Sugaya T., Lee C. K., Kim C. H., Itakura S., Yamaguchi M. 2009. Genetic structuring and transfer of marine dinoflagellate Cochlodinium polykrikoides in Japanese and Korean coastal waters revealed by microsatellites. Mol. Ecol. 18, 2337–2352 10.1111/j.1365-294X.2009.04193.x (doi:10.1111/j.1365-294X.2009.04193.x) [DOI] [PubMed] [Google Scholar]
- 20.Garcés E., Bravo I., Vila M., Figueroa R. I., Masó M., Sampedro N. 2004. Relationship between vegetative cells and cyst production during Alexandrium minutum bloom in Arenys de Mar harbour (NW Mediterranean). J. Plankton Res. 26, 637–645 10.1093/plankt/fbh065 (doi:10.1093/plankt/fbh065) [DOI] [Google Scholar]
- 21.Anderson D. M., Stock C. A., Keafer B. A., Nelson A. B., Thompson B., McGillicuddy D. J., Keller M., Matrai P. A., Martin J. 2005. Alexandrium fundyense cyst dynamics in the Gulf of Maine. Deep-Sea Res. II 52, 2522–2542 10.1016/j.dsr2.2005.06.014 (doi:10.1016/j.dsr2.2005.06.014) [DOI] [Google Scholar]
- 22.Alpermann T. J., Beszteri B., John U., Tillmann U., Cembella A. D. 2009. Implications of life-history transitions on the population genetic structure of the toxigenic marine dinoflagellate Alexandrium tamarense. Mol. Ecol. 18, 2122–2133 10.1111/j.1365-294X.2009.04165.x (doi:10.1111/j.1365-294X.2009.04165.x) [DOI] [PubMed] [Google Scholar]
- 23.White C., Selkoe K. A., Watson J., Siegel D. A., Zacherl D. C., Toonen R. J. 2010. Ocean currents help explain population genetic structure. Proc. R. Soc. B 277, 1685–1694 10.1098/rspb.2009.2214 (doi:10.1098/rspb.2009.2214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galindo H. M., Pfeiffer-Herbert A. S., McManus M. A., Chao Y. I., Chai F. E. I., Palumbi S. R. 2010. Seascape genetics along a steep cline: using genetic patterns to test predictions of marine larval dispersal. Mol. Ecol. 19, 3692–3707 10.1111/j.1365-294X.2010.04694.x (doi:10.1111/j.1365-294X.2010.04694.x) [DOI] [PubMed] [Google Scholar]
- 25.Unal E., Buckling A. 2010. Basin-scale population genetic structure of the planktonic copepod Calanus finmarchicus in the North Atlantic Ocean. Progr. Oceanogr. 87, 175–185 10.1016/j.pocean.2010.09.017 (doi:10.1016/j.pocean.2010.09.017) [DOI] [Google Scholar]
- 26.Bradbury I. R., Bentzen P. 2007. Non-linear genetic isolation by distance: implications for dispersal estimation in anadromous and marine fish populations. Mar. Ecol. Progr. Ser. 340, 245–257 10.3354/meps340245 (doi:10.3354/meps340245) [DOI] [Google Scholar]
- 27.Nagai S., et al. 2007. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. J. Phycol. 43, 43–54 10.1111/j.1529-8817.2006.00304.x (doi:10.1111/j.1529-8817.2006.00304.x) [DOI] [Google Scholar]
- 28.Godley B. J., Barbosa C., Bruford M., Broderick A. C., Catry P., Coyne M. S., Formia A., Hays G. C., Witt M. J. 2010. Unravelling migratory connectivity in marine turtles using multiple methods. J. Appl. Ecol. 47, 769–778 10.1111/j.1365-2664.2010.01817 (doi:10.1111/j.1365-2664.2010.01817) [DOI] [Google Scholar]
- 29.Van Lenning K., Masó M., Garcés E., Anglès S., Sampedro N., Morales-Blake A., Camp J. 2007. Short-term variations in development of a recurrent toxic Alexandrium minutum–dominated dinoflagellate bloom induced by meteorological conditions. J. Phycol. 43, 892–907 10.1111/j.1529-8817.2007.00396 (doi:10.1111/j.1529-8817.2007.00396) [DOI] [Google Scholar]
- 30.Heisler J., et al. 2008. Eutrophication and harmful algal blooms: a scientific consensus. Harmful Algae 8, 3–13 10.1016/j.hal.2008.08.006 (doi:10.1016/j.hal.2008.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vila M., Giacobbe M. G., Masò M., Gangemi E., Penna A., Sampedro N., Azzaro F., Camp J., Galluzzi L. 2005. A comparative study on recurrent blooms of Alexandrium minutum in two Mediterranean coastal areas. Harmful Algae 4, 673–695 10.1016/j.hal.2008.08.006 (doi:10.1016/j.hal.2008.08.006) [DOI] [Google Scholar]
- 32.Bravo I., Vila M., Masó M., Ramilo I., Figueroa R. I. 2008. Alexandrium catenella and Alexandrium minutum blooms in the Mediterranean Sea: toward the identification of ecological niches. Harmful Algae 7, 515–522 10.1016/j.hal.2007.11.005 (doi:10.1016/j.hal.2007.11.005) [DOI] [Google Scholar]
- 33.Bravo I., Fraga S., Figueroa R., Pazos Y., Massanet A., Ramilo I. 2010. Bloom dynamics and life cycle strategies of two toxic dinoflagellates in a coastal upwelling system (NW Iberian Peninsula). Deep Sea Res. II 57, 222–234 10.1016/j.dsr2.2009.09.004 (doi:10.1016/j.dsr2.2009.09.004) [DOI] [Google Scholar]
- 34.Penna A., Garcés E., Vila M., Giacobbe M. G., Fraga S., Luglié A., Bravo I., Bertozzini E., Vernesi C. 2005. Alexandrium catenella (Dinophyceae), a toxic ribotype expanding in the NW Mediterranean Sea. Mar. Biol. 148, 13–23 10.1007/s00227-005-0067-5 (doi:10.1007/s00227-005-0067-5) [DOI] [Google Scholar]
- 35.Nagai S., McCauley L., Yasuda N., Erdner D. L., Kulis D. M., Matsuyama Y., Itakura S., Anderson D. M. 2006. Development of microsatellite markers in the toxic dinoflagellate Alexandrium minutum (Dinophyceae). Mol. Ecol. Res. 6, 756–758 10.1111/j.1471-8286.2006.01331.x (doi:10.1111/j.1471-8286.2006.01331.x) [DOI] [Google Scholar]
- 36.Cavalli-Sforza L. L., Edwards A. W. F. 1967. Phylogenetic analysis: models and estimation procedures. Evolution 21, 550–570 10.2307/2406616 (doi:10.2307/2406616) [DOI] [PubMed] [Google Scholar]
- 37.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evanno G., Regnaut S., Goudet J. 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2601–2620 10.1111/j.1365-294X.2005.02553.x (doi:10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 39.Jakobsson M., Rosenberg N. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 10.1093/bioinformatics/btm233 (doi:10.1093/bioinformatics/btm233) [DOI] [PubMed] [Google Scholar]
- 40.Nordborg M., et al. 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3, 1289–1299 10.1371/journal.pbio.0030196 (doi:10.1371/journal.pbio.0030196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordi A., Wang D. P. 2009. Mean dynamic topography and eddy kinetic energy in the Mediterranean Sea: comparison between altimetry and a 1/16 degree ocean circulation model. Ocean Model 29, 137–146 10.1016/j.ocemod.2009.04.001 (doi:10.1016/j.ocemod.2009.04.001) [DOI] [Google Scholar]
- 42.Godsil C., Gordon R. 2001. Algebraic graph theory, pp. 207 New York, NY: Springer [Google Scholar]
- 43.Penna A., et al. 2008. Phylogenetic relationships among the Mediterranean Alexandrium (Dinophyceae) species based on sequences of 5.8S gene and internal transcribed spacers of the rRNA operon. Eur. J. Phycol. 43, 163–178 10.1080/09670260701783730 (doi:10.1080/09670260701783730) [DOI] [Google Scholar]
- 44.McCauley L., Erdner D., Nagai S., Richlen M., Anderson D. 2009. Biogeographic analysis of the globally distributed harmful algal bloom species Alexandrium minutum (Dinophyceae) based on rRNA gene sequences and microsatellite markers. J. Phycol. 45, 454–463 10.1111/j.1529-8817.2009.00650.x (doi:10.1111/j.1529-8817.2009.00650.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godhe A., Härnström K. 2010. Linking the planktonic and benthic habitat: genetic structure of the marine diatom Skeletonema marinoi. Mol. Ecol. 19, 4478–4490 10.1111/j.1365-294X.2010.04841.x (doi:10.1111/j.1365-294X.2010.04841.x) [DOI] [PubMed] [Google Scholar]
- 46.Halkett F., Simon J. C., Balloux F. 2005. Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol. Evol. 20, 194–201 10.1016/j.tree.2005.01.001 (doi:10.1016/j.tree.2005.01.001) [DOI] [PubMed] [Google Scholar]
- 47.Estrada M., Solè J., Anglès S., Garcés E. 2010. The role of resting cysts in Alexandrium minutum population dynamics. Deep Sea Res. II 57, 308–321 10.1016/j.dsr2.2009.09.007 (doi:10.1016/j.dsr2.2009.09.007) [DOI] [Google Scholar]
- 48.Penna A., et al. 2010. Detection of microalgal resting cysts in European coastal sediments using a PCR-based assay. Deep Sea Res. II 57, 288–300 10.1016/j.dsr2.2009.09.010 (doi:10.1016/j.dsr2.2009.09.010) [DOI] [Google Scholar]
- 49.Mitarai S., Siegel D. A., Watson J. R., Dong C., McWilliams J. C. 2009. Quantifying connectivity in the coastal ocean with application to the Southern California Bight. J. Geophys. Res. 114, C10026. 10.1029/2008JC005166 (doi:10.1029/2008JC005166) [DOI] [Google Scholar]
- 50.Yasuda N., Nagai S., Masami H., Ken O., Karin G., Nadaoka A. K. 2009. Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Mol. Ecol. 18, 1574–1590 10.1111/j.1365-294X.2009.04133.x (doi:10.1111/j.1365-294X.2009.04133.x) [DOI] [PubMed] [Google Scholar]
- 51.Fraser C. I., Nikula R., Waters J. M. 2010. Oceanic rafting by a coastal community. Proc. R. Soc. B 278, 649–655 10.1098/rspb.2010.1117 (doi:10.1098/rspb.2010.1117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wares J. P., Gaines S. D., Cunningham C. W. 2001. A comparative study of asymmetric migration events across a marine biogeographic boundary. Evolution 55, 295–306 10.1111/j.0014-3820.2001.tb01294.x (doi:10.1111/j.0014-3820.2001.tb01294.x) [DOI] [PubMed] [Google Scholar]
- 53.Matschiner M., Hanel R., Salzburger W. 2009. Gene flow by larval dispersal in the Antarctic notothenioid fish Gobionotothen gibberifrons. Mol. Ecol. 18, 2574–2587 10.1111/j.1365-294X.2009.04220.x (doi:10.1111/j.1365-294X.2009.04220.x) [DOI] [PubMed] [Google Scholar]
- 54.Collins C. J., Fraser C. I., Ashcroft A., Waters J. M. 2010. Asymmetric dispersal of southern bull-kelp (Durvillaea antarctica) adults in coastal New Zealand: Testing an oceanographic hypothesis. Mol. Ecol. 19, 4572–4580 10.1111/j.1365-294X.2010.04842.x (doi:10.1111/j.1365-294X.2010.04842.x) [DOI] [PubMed] [Google Scholar]
- 55.Andreakis N., Kooistra W. H. C. F., Procaccini G. 2009. High genetic diversity and connectivity in the polyploid invasive seaweed Asparagopsis taxiformis (Bonnemaisoniales) in the Mediterranean, explored with microsatellite alleles and multilocus genotypes. Mol. Ecol. 18, 212–226 10.1111/j.1365-294X.2008.04022.x (doi:10.1111/j.1365-294X.2008.04022.x) [DOI] [PubMed] [Google Scholar]
- 56.Vila M., Camp J., Garcés E., Masó M., Delgado M. 2001. High resolution spatio-temporal detection of potentially harmful dinoflagellates in confined waters of the NW Mediterranean. J. Plankton Res. 23, 497–514 10.1093/plankt/23.5.497 (doi:10.1093/plankt/23.5.497) [DOI] [Google Scholar]
- 57.Bethoux J. P., Gentili B., Morin P., Nicolas E., Pierre C., Ruiz-Pino D. 1999. The Mediterranean Sea: a miniature ocean for climatic and environmental studies and a key for the climatic functioning of the North Atlantic. Progr. Oceanogr. 44, 131–146 10.1016/S0079-6611(99)00023-3 (doi:10.1016/S0079-6611(99)00023-3) [DOI] [Google Scholar]
- 58.Spalding M. D., et al. 2007. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57, 573–583 10.1641/B570707 (doi:10.1641/B570707) [DOI] [Google Scholar]
- 59.Demirov E., Pinardi N. 2002. Simulation of the Mediterranean Sea circulation from 1979 to 1993. Part I. The interannual variability. J. Mar. Syst. 33, 23–50 10.1016/S0924-7963(02)00051-9 (doi:10.1016/S0924-7963(02)00051-9) [DOI] [Google Scholar]
- 60.Polimene L., Pinardi N., Zavatarelli M., Colella S. 2006. The Adriatic Sea ecosystem seasonal cycle: validation of a three dimensional numerical model. J. Geophys. Res. 111, C03S19. 10.1029/2005JC003260 (doi:10.1029/2005JC003260) [DOI] [Google Scholar]
- 61.Russo A., Artegiani A. 1996. The Adriatic Sea hydrography. Sci. Mar. 60, 33–43 [Google Scholar]
- 62.Artegiani A., Bregant D., Paschini E., Pinardi N., Raicich F., Russo A. 1997. The Adriatic Sea general circulation. Part II: Baroclinic circulation structure. J. Phys. Oceanogr. 27, 1515–1532 (doi:10.1175/1520-0485(1997)027<1515:TASGCP>2.0.CO;2) [DOI] [Google Scholar]
- 63.Millot C. 1999. Circulation in the Western Mediterranean Sea. J. Mar. Syst. 20, 423–442 10.1016/S0924-7963(98)00078-5 (doi:10.1016/S0924-7963(98)00078-5) [DOI] [Google Scholar]
- 64.Artale V., Astraldi M., Buffoni G., Gasparini G. P. 1994. Seasonal variability of gyre-scale circulation in the northern Tyrrhenian Sea. J. Geophys. Res. 99, 14 127–14 137 10.1029/94JC00284 (doi:10.1029/94JC00284) [DOI] [Google Scholar]
- 65.Robinson A., Leslie W., Theocharis A., Lascaratos A. 2001. Mediterranean Sea circulation. In Encyclopedia of ocean sciences, vol. 3, pp. 1689–1705 New York, NY: Elsevier [Google Scholar]
- 66.Hallegraeff G. M. 1998. Transport of toxic dinoflagellates via ships' ballast water: bioeconomic risk assessment and efficacy of possible ballast water management strategies. Mar. Ecol. Progr. Ser. 168, 297–309 10.3354/meps168297 (doi:10.3354/meps168297) [DOI] [Google Scholar]
- 67.Smayda T. J. 2007. Reflections on the ballast water dispersal—harmful algal bloom paradigm. Harmful Algae 6, 601–622 10.1016/j.hal.2007.02.003 (doi:10.1016/j.hal.2007.02.003) [DOI] [Google Scholar]