Abstract
Opsin proteins are essential molecules in mediating the ability of animals to detect and use light for diverse biological functions. Therefore, understanding the evolutionary history of opsins is key to understanding the evolution of light detection and photoreception in animals. As genomic data have appeared and rapidly expanded in quantity, it has become possible to analyse opsins that functionally and histologically are less well characterized, and thus to examine opsin evolution strictly from a genetic perspective. We have incorporated these new data into a large-scale, genome-based analysis of opsin evolution. We use an extensive phylogeny of currently known opsin sequence diversity as a foundation for examining the evolutionary distributions of key functional features within the opsin clade. This new analysis illustrates the lability of opsin protein-expression patterns, site-specific functionality (i.e. counterion position) and G-protein binding interactions. Further, it demonstrates the limitations of current model organisms, and highlights the need for further characterization of many of the opsin sequence groups with unknown function.
Keywords: opsin, evolution, G-protein, counterion, expression
1. Introduction
The study of visual pigments began in the late nineteenth century, with their discovery by Franz Boll and characterization by Willy Kuhne [1]. George Wald [2] later showed that the visual pigment molecule includes a protein, opsin, covalently bound to a chromophore, typically an aldehyde derived from vitamin A. Today, we know that the opsins are members of the G-protein coupled receptor (GPCR) superfamily—proteins with seven transmembrane helices that are involved in a diverse set of signalling functions. Within the GPCR superfamily, the opsins form a large monophyletic subclass of proteins that are characterized by a lysine in the seventh transmembrane helix that serves as the attachment site for the chromophore. Functional opsin proteins covalently bind a chromophore, gaining photosensitivity. Opsins are essential molecules in mediating the ability of animals to detect and use light for diverse biological functions. Therefore, understanding the evolutionary history of opsins is key to understanding the evolution of light detection and photoreception in animals.
Nathans & Hogness [3] first cloned and sequenced an opsin gene, that of bovine rhodopsin, in 1983. Bovine rhodopsin has since served as a model system for investigating the function of GPCRs owing to its availability in large quantities, and is probably the best-characterized GPCR based on a plethora of biochemical, structural and functional studies. Likewise, Drosophila melanogaster rhodopsin Rh1, first sequenced in 1985 by Zuker et al. [4], has become a model for studies of arthropod visual pigments and phototransduction owing to the power of genetic manipulation in this system. However, as investigations of photopigments have expanded, opsins have been discovered in a wide variety of tissue and cell types, signalling through multiple pathways, and carrying out functions beyond image formation. With the rate of new data acquisition in the post-genomic era, our knowledge of sequence diversity has far outpaced our understanding of the evolution and biological roles of opsin-based photopigments. For example, opsins that exist outside of visual photoreceptors (i.e. those in image-forming organs) now comprise a large diversity of proteins about which very little is known.
The evolutionary history of the opsins has been a topic of much interest since the first sequences became available. Even today, however, with the number of opsin sequences available from public databases and genome projects surpassing 1000 examples, studies of visual pigment function have continued to focus on a limited set of model organisms (e.g. zebrafish, mouse, cow, fruitfly), and most studies have considered the opsins expressed in retinal tissues. Thus, the discovery of consistent differences in photoreceptor structure, primary opsin gene sequence and signal transduction components between vertebrate and protostome visual pigments has led to the conceptual division of opsin diversity into two basic classes: the ‘r-type’ and ‘c-type’ opsins [5,6]. The division reflects the types of photoreceptor cells that house these pigments, described in more detail later. Those placed in the ‘r-type’ group are found in rhabdomeric photoreceptors (as in the eyes of arthropods and cephalopods) that depolarize using a Gq-type signal transduction pathway. ‘R-type’ photopigments are typically bistable, retaining the chromophore in both the native (dark) and photoactivated (light) states [7]. Conversely, the ‘c-type’ opsins are found in ciliary photoreceptors (e.g. vertebrate rods and cones) that respond to light by hyperpolarizing, initiated by a Gt (transducin)-mediated signalling cascade. ‘C-type’ photopigments bleach after photoactivation, losing the bound chromophore. This classification system has dominated the literature since Eakin [8] established the division of photoreceptors into ciliary and rhabdomeric types. Current work, however, reveals that sequence diversity is far greater and more complex than described by these two opsin ‘types’, with additional opsin classes being identified based on unique functions (e.g. retinochromes) and from under-studied taxonomic groups (e.g. the Cnidaria) [9,10].
With the molecular characterization of additional opsins and the development of new tools for investigating molecular phylogenies, studies of opsin evolution based on sequence analyses have appeared [10–13]. This work confirms the existence of new and unsuspected opsin groups. However, it generally has been limited to particular opsin subgroups (for instance, from vertebrates or arthropods) and has typically included sequences derived from expressed opsin genes in defined tissues (e.g. the retina, the pineal gland or other recognized photoreceptive structures). Surprisingly, as of yet, there has been no comprehensive evolutionary look at the currently described opsin sequence diversity. While many studies attempt either to infer the ancestral state of a particular character within the opsin subclass or to make generalizations about the function of a particular group of opsins based on evolutionary relationships, only tentative conclusions are possible without an understanding of the evolution of the opsin subclass as a whole. As genomic data have accumulated, it has become possible to include opsins that are functionally and histologically less well-characterized into a large-scale view of opsin evolution from a genetic perspective. Consequently, we have been inspired to incorporate these new data into a large-scale, genome-based, analysis of opsin evolution. Here, we provide a phylogenetic analysis of the currently known opsin sequence diversity to serve as a foundation for understanding opsin evolution, highlighting where our knowledge of opsin function and diversity is on firm ground and where it is still rudimentary.
To understand opsin evolutionary history, we have reconstructed the phylogenetic relationships among 889 available opsin sequences. About half of these sequences (436) come from analysis of all metazoan genome projects released to GenBank as of December 2010, using bioinformatic techniques to determine full opsin complements from a phylogenetically representative set of metazoan species. The remaining sequences, also from GenBank, mainly represent expressed transcripts, often from taxonomic groups or opsin families where genomic data are lacking (for methods see electronic supplementary material). This new analysis reveals novel relationships and identifies previously unrecognized opsin groups. Establishing the evolutionary pattern allows us to map important protein traits onto the phylogenetic gene tree to examine the distribution of key functional features—such as expression patterns, G-protein-binding partners and counterion placement—within the opsin clade. Further, our extensive analysis allows us to evaluate previous opsin subclass classifications and characterizations based on these features but using more limited datasets. Finally, it demonstrates the limitations of current model organisms, and highlights the need for further experimental characterization of many of the opsin sequence groups with unknown function.
2. Evolution of the opsin family of proteins
Four major monophyletic groups of opsins have been previously defined: those found in ciliary photoreceptors (‘c-opsins’), those from rhabdomeric photoreceptors (‘r-opsins’), the cnidarian opsins (‘Cnidops’) and a mixed group consisting of ‘retinal G-protein coupled receptors’ (RGR), peropsins and neuropsins [6,10,12]. Knowing the relationships among these groups is critical to understanding the key duplication and diversification events that have produced the opsin sequence diversity, expression patterns and functions observed today. Opsin diversity has been further subdivided to form up to 22 subgroups found in the vertebrates alone [11,13,14], mostly defined by their similarity to the first sequence of the group to be characterized. Yet, under-sampling and under-representation of many taxonomic groups (e.g. Cndiarians) and functional groups have concealed the true extent of opsin diversity. As more opsins become characterized, they tend to be placed into existing groups, implying functional similarities that might be specious. Today, however, a long history of opsin research together with the rapidly increasing availability of entire genomes provides a large number of sequences for investigating the evolution and functional diversity of the opsin family at levels deeper than previously seen.
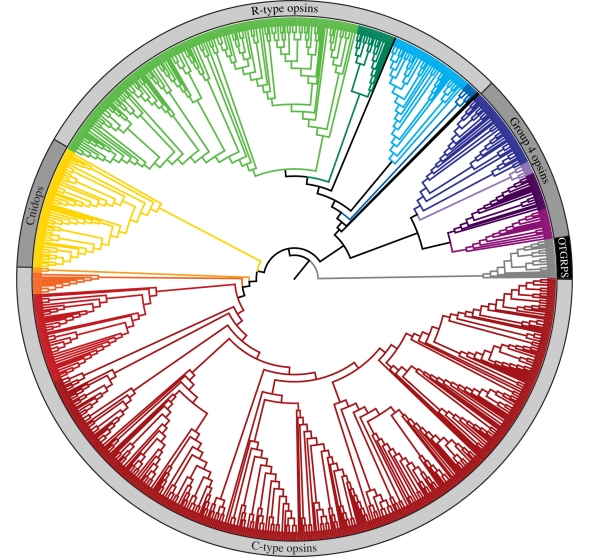
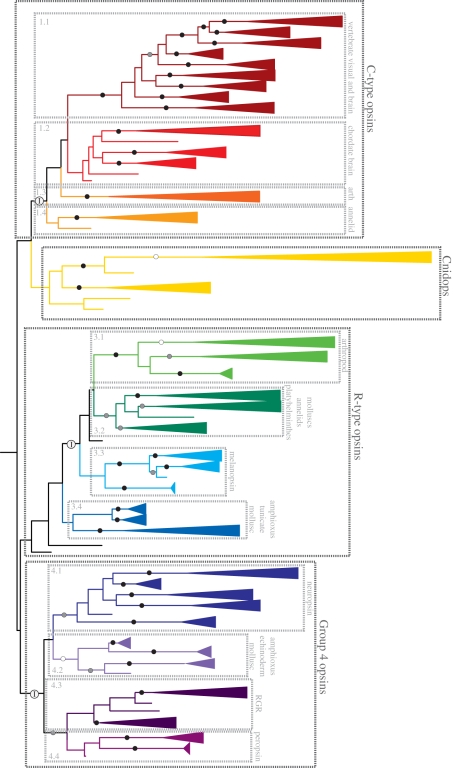
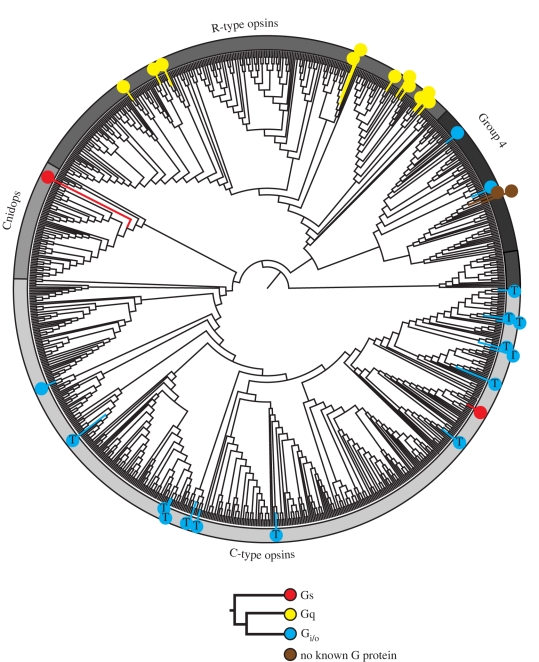
Our phylogenetic analysis of this large and diverse set of opsin sequences recovers the four major lineages described in earlier papers [10,12,15] (figure 1). Previous naming schemes fail to encompass the diversity within these large groups, particularly with regards to the cell type, G-protein-binding partner and counterion diversity (see below for details): we here designate the four major lineages as: (i) the ‘C-type opsins’, (ii) the ‘Cnidops’, (iii) the ‘R-type opsins’, and (iv) Group 4 Opsins (Gr4), which contains an assortment of relatively poorly characterized opsin types. The distribution of opsins into these four major groups is supported by analyses of insertion/deletion events (indels) and intron arrangement, representing rare and thus informative genetic events [16,17]. In particular, intron arrangement supports the C-type and Group 4 opsins, as well as the clustering of the arthropod (group 3.1) and lophotrochozoan (group 3.2) visual opsins with the chordate melanopsins (group 3.3) (figures 1 and 2), while insertion/deletion events support the smaller sequence groups (figure 2). A tantalizing line of evidence functionally uniting the R-type with the Group 4 opsins is the presence of long-term thermal bistability, which has been demonstrated in arthropod and cephalopod visual pigments (R-type) and in chicken neuropsin (Gr4), and suggested for vertebrate melanopsins, also ‘R-type’ [7,18–21].
Figure 1.
Maximum-likelihood tree of 889 genomic and expressed opsin sequences (for reconstruction methods, see electronic supplementary material). The branches of major phylogenetic groups and subgroups have been coloured (see figure 2 for more detail), and the four major opsin clades have been labelled. The closely related, non-opsin GPCRs used to root the tree have branches that have been coloured grey, and the portion of the outer circle denoting outgroups has been coloured black.
Figure 2.
The maximum-likelihood tree from figure 1 (outgroups not shown), with the branches collapsed into well-supported clades where possible. Per cent bootstrap support exceeding 70% is indicated for the major lineages: white circles, 70–79% support; grey circles 80–89%; black circles, 90–100%. Well-supported clades are coloured by hierarchal classifications of related sequences. C-type opsins–group 1.1: vertebrate visual pigments (Rh1, Rh2, SWS1, SWS2, M/LWS), pinopsins, parapinopsins, vertebrate ancient and parietal opsins; group 1.2: teleost multiple tissue opsins (TMTs), encephalopsins and uncharacterized amphioxus and urchin opsins; group 1.3: honeybee ptersopsin, and uncharacterized insect and Daphnia pulex opsins; group 1.4: uncharacterized Platynereis brain and urchin opsins. Cnidops—Ctenophore and cndiarian opsins, including representatives from hydrozoans, anthozoans and cubozoans. R-type opsins–group 3.1: arthropod visual pigments (M/LWS, SWS); group 3.2: annelid, platyhelminthes and mollusc visual pigments; group 3.3: vertebrate melanopsins 1 and 2, and amphioxus sequences; group 3.4: uncharacterized tunicate, amphioxus and mollusc opsins. Group 4 Opsins IT–group 4.1: four separate clades of neuropsins, and amphioxus and urchin opsins; group 4.2: amphioxus, echinoderm and scallop opsins; group 4.3: RGR and uncharacterized mollusc opsins; group 4.4: peropsins, amphioxus and hemichordate opsins. For more detailed information on the members of each group, please see the electronic supplementary material. Major groups also supported by intron analyses have been indicated by white circles containing an ‘I’.
Determining the relationships among early opsin lineages is a difficult problem, compounded by the lack of closely related GPCR sequences to root the phylogeny [10]. The relationships among the four major lineages in our analyses differ from those proposed in other recent studies of opsin evolution [10,12,15]; similar to the same recent studies, however, the statistical support for these relationships is weak. Because we are examining character distributions, the following discussion of patterns of evolution across these opsin groups is not affected by alternative hypotheses of relationships among these groups.
Our phylogeny finds the cnidarian opsins (Cnidops) to be most closely related to the C-type opsin group containing the vertebrate visual pigments, while the group containing the arthropod and cephalopod visual opsins and melanopsins (R-type) is placed with the cluster containing neuropsins, peropsins and photoisomerases (Gr4). The absence of opsins from eukaryotic lineages earlier than ctenophores (e.g. from the genomes of the sponge Amphimedon queenslandica and the choanoflagellate Monosiga brevicollis [10,15,22]), and the relationship between the C-type and the Cnidops (cnidarian+ctenophore) groups in our phylogeny implies that this supergroup arose early in the metazoan lineage, well before the evolution of the bilaterians. Unfortunately, intron analyses of cnidarian genomes fail to shed any light on early relationships owing to either lack of introns or large sequence divergences. As additional genomes become available, particularly from early metazoan lineages (e.g. Ctenophora and Cnidaria), untangling the pre-Cambrian opsin origins should become possible.
Unravelling early opsin evolutionary history is significant for understanding how opsin photopigments became integral components of vision. Figures 1 and 2 reveal that image-forming visual systems are evolutionary novelties, having independently evolved from pre-existing opsin-based photodetection systems several times, including at least once each in the cndarians, the vertebrates and in the protostomes (e.g. platyhelminths, annelids, arthropods, molluscs). C-type visual opsins are derived from ancestors that also led to various annelid, arthropod or vertebrate brain opsins. Similarly, R-type visual opsins have ancestors that also produced a variety of odd molluscan and early deuterostome opsins, as well as the melanopsins. The use of a Gr4, Go-activating, opsin as a visual pigment in scallop suggests the emergence of yet another image-forming visual system based on a divergent opsin.
Based on the presence of widely divergent animal phyla in three of the four major opsin clades resolved in this study (figures 2 and 3), the bilaterian common ancestor already possessed multiple opsin lineages [6,10,23,24]. The C-type opsin group, originally defined based on expression in vertebrate ciliary photoreceptors [25], also contains examples from arthropods, echinoderms and annelids. The R-type group contains not only arthropod ‘rhabdomeric’ opsins, but also the vertebrate melanopsins as well as numerous, poorly characterized, opsin sequences from arthropods, molluscs, chordates, annelids, echinoderms and platyhelminths. The functionally diverse Group 4 opsins are represented by sequences from chordates, molluscs, echinoderms and hemichordates. The Cnidops group, composed entirely of cnidarian and ctenophore opsins, is the only group that lacks representation of broad taxonomic diversity from the major animal phyla.
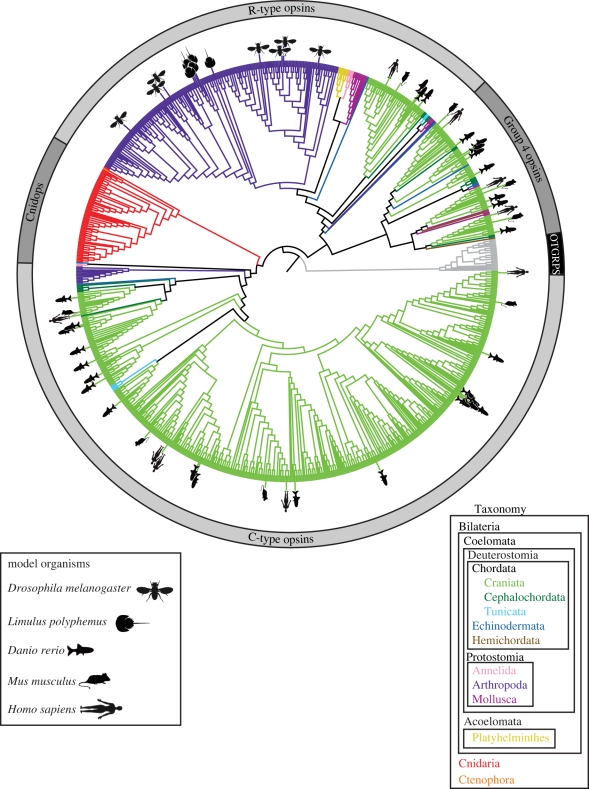
Figure 3.
Maximum-likelihood phylogeny as in figure 1, but with branches coloured by major taxonomic groups as indicated by the key in the lower right. Opsin sequences from the genomes of the model organisms most commonly used for vision studies (e.g. Drosophia melanogaster, Limulus polyphemus, Homo sapiens, Mus musculus, Danio rerio) have been indicated by symbols as indicated in the figure legend, lower left.
The analyses presented in figures 1 and 2 show a more complete picture of the evolutionary relationships among opsins than previously described. Each of the four major lineages contains even more sequence diversity than was previously recognized, represented by large portions of the phylogeny where opsin sequences remain uncharacterized. Each of the four major groups includes sequence clusters from understudied taxonomic groups (e.g. annelids, molluscs, echinoderms, cephalochordates) that contain opsins for which no biological function is yet known. In the C-type opsins, group 1.3 contains opsins from both insects and the Daphnia pulex genome, but of these only honeybee pteropsin has been functionally characterized [26]. Likewise, in the R-type opsins, sequence group 3.4 contains tunicate, cephalochordate and molluscan opsins, none of which has a known function. In Group 4 opsins, the neuropsin group (sequence group 4.1), comprising at least five well-supported and divergent groups, has significant sequence diversity that is not functionally characterized.
3. Functional implications of opsin evolution
Knowing more about the evolutionary history of opsins, we can begin to look at the evolution of opsin functional diversity. In the following sections, we integrate into the phylogeny our knowledge of expression patterns, photoreceptor cell types, signalling transduction pathways and opsin protein function. By combining these data across our large-scale phylogeny, we also highlight where additional research into opsin function should be focused.
(a). Expression patterns and photoreceptor cell types
By definition, photoreceptors are cells specialized for light detection. In photoreceptors devoted to vision, the most obvious morphological modification is the expansion of cellular membrane surface area. In 1972, based on morphological characteristics, Eakin divided photoreceptors into rhabdomeric and ciliary types [7]. In rhabdomeric photoreceptors, membranous, actin-rich, microvilli project from the cell body in densely packed bundles. Ciliary photoreceptors, in contrast, have flattened membranous discs sitting atop one another within an enlarged cellular compartment projecting from a modified cilium. These membrane modifications increase surface area and enable the photoreceptor to house large quantities of visual pigment molecules, thereby increasing the probability of photon capture [27].
For some time, however, it has been known that many photoreceptor cells not involved in vision lack obvious membrane-expanding modifications. For example, rather normal-looking retinal ganglion cells of many (probably all) vertebrates, as well as skin chromatophores in the amphibian Xenopus, contain melanopsin, a visual pigment within the R-type opsin group. The isolation of melanopsin was complemented by the discovery of C-type opsins in isolated groups of cells in the brains of arthropods and annelid worms [6,26]. Clearly, opsins function in a variety of cell types and tissue locations both within and outside of the retina. Here, we use our evolutionary analyses to begin to envision the true diversity of photoreceptor cell types and expression patterns. From the sequences included in our phylogeny, we have mapped 120 with associated data on either the photoreceptor cell type or the tissue of expression (figure 4). This set of sequences, limited as it is, exhibits a plethora of cell and tissue-expression patterns.
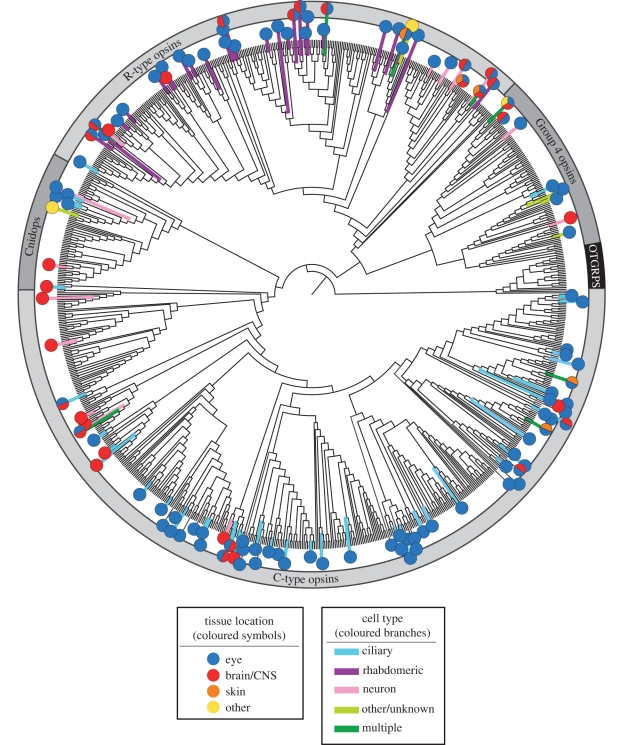
Figure 4.
Maximum-likelihood phylogeny as in figure 1. Based on a literature review, with emphasis on poorly understood or more recently discovered opsins, the branches have been coloured to indicate the photoreceptor cell type, and symbols have been used to illustrate the tissues where each opsin is known to be expressed (see figure key). Acceptable data used for determining cell type or tissue location include in situ hybridization, immunohistochemistry and single cell or tissue-specific reverse transcription-polymerase chain reaction studies. Some opsin sequences had information for tissue location, but not cell type. Opsin sequences that have been found in more than one tissue type have been represented by pie charts. In addition to ‘ciliary’ and ‘rhabdomeric’ cell types, cells are also classed as ‘neurons’ (including retinal and cerebral ganglion and interneuronal cells, as well as cnidarian neural net neurons) and ‘others’ (including epithelial cells in the retina and skin). Tissue location characters include ‘eyes’ (lateral or primary eyes), ‘CNS’ (brains or nerve nets, including specific photosensitive regions within the CNS such as pineal organs, parietal eyes and dorsal ocelli), ‘skin’ (epidermis, chromatophores, melanophores and iridophores) and ‘others’ (gonads and bioluminescent organs). For data related to specific sequences, and their related references, refer to the electronic supplementary material.
All four major opsin groups contain transcripts found in multiple types of photoreceptors and/or multiple tissue locations. Besides vertebrate retinal ciliary photoreceptors, C-type opsins are expressed in neuronal cells in the eye and central nervous system (CNS) and in skin cells lacking membrane specializations (chromatophores and iridophores) [28–30]. The Cnidops are expressed in retinal ciliary photoreceptors, CNS neurons and gonad cells of various cnidarians [10,15,31,32]. R-type opsins are expressed not only in arthropod and molluscan retinal rhabdomeric photoreceptors, but also in neurons, gonads and chromatophores that lack obvious membrane modifications [33–36]. Finally, Group 4 includes opsins expressed in ciliary photoreceptors as well as neuronal, gonad, retinal epithelial and skin cells. Note that related opsins in all four groups can be found in very different, or even multiple, cell types and tissue locations. Furthermore, opsin expression within a given cell type does not strongly correlate with expression in a specific tissue location. Both C-type and R-type opsins exist in specialized photoreceptor cells associated with image-forming vision, as well as non-specialized cells in a variety of tissue locations (e.g. C-type Platynereis opsins and pinopsins in the brain [6,37]; R-type melanopsin in neuronal ganglion cells within the retina [38]).
In summary, our character mapping illustrates that neither cell type nor tissue location is strictly associated with any major evolutionary group of opsin sequences (figure 4). Many opsins expressed in either rhabdomeric or ciliary photoreceptor cells are closely related to others expressed in cells lacking clear secondary membrane modifications. While some animal brain photoreceptors resemble retinal photoreceptors, many others appear to be new types of photosensitive neurons [26,39,40]. Additionally, opsins are expressed in association with dermal-pigmentation systems, including in amphibian, teleost and cephalopod skin, among others [29,41–43]. The diversity of types of opsin-driven photosensitive mechanisms clearly arose through the action of many independent events.
(b). Signal transduction and G-protein interactions
Once activated by light, opsins—like other GPCRs—bind specific G-proteins to activate intracellular signalling pathways. In general, the Gα subunits of such proteins act as the primary effectors for these responses. The activation of multiple molecules of the same G-protein by a single receptor amplifies the response, thereby increasing the dynamic range over which stimuli can induce a physiological change within a cell [44–46].
Approximately 20 Gα subunits have been identified across Metazoa, grouped into four functionally distinct subclasses: Gαs, Gαi/o, Gαq and Gα12/13 [44,47,48]. Opsins activate or couple to at least three of these subclasses (figure 5). Gαi/o proteins, specifically transducin (Gατ), are the major type known to be activated by C-type opsins. The transducin pathway produces a transient decrease in cyclic nucleotide levels, causing the closing of cyclic nucleotide gated channels which leads to photoreceptor hyperpolarization [49,50]. Besides the C-type opsins, only one other example is currently known of an opsin interacting with Gαi/o: a Group 4 scallop opsin from retinal ciliary photoreceptor cells [51]. Of the C-type opsins, where G-protein activation has been investigated, only a single representative has been found that apparently binds to another G-protein (figure 5). This opsin is expressed in tilapia erythrophores [27], and suggests that as a larger diversity of extra-retinal opsin G-protein binding is investigated, more diversity in signal transduction will be found. In contrast, R-type opsins, including arthropod and cephalopod visual pigments as well as the melanopsins, have only been found to use Gαq-mediated pathways [52–54]. These pathways activate phosopholipase C (PLC), which ultimately results in the transient opening of light-sensitive cation channels, generating a depolarization [55–58]. Until recently, only Gαi/o- and Gαq-mediated pathways were known to drive phototransduction. In addition to the tilapia erythrophore opsin, one of the Cnidops opsins is also believed to couple with a Gαs protein, which activates adenylate cyclase [59–61], suggesting that a third phototransduction pathway may be activated by this group (figure 5). Furthermore, it now seems that some opsins do not initiate cell signalling at all. For instance, RGR opsins (in Group 4) play a role in 11-cis-retinal synthesis, either as photoisomerases or as an RPE cofactor, and apparently do not interact with any G-protein [62–64].
Figure 5.
Maximum-likelihood phylogeny as in figure 1. Coloured symbols have been placed on the tree at locations where the G-protein-binding partner is known for a particular opsin sequence (see legend). Opsins known to couple with transducin are indicated by a Gi/o symbol containing the letter ‘T’. Based on a review of available literature, all the represented G-protein/opsin pairs were determined by either electrophysiological, biochemical or molecular analyses. For specific methods and references, please refer to the electronic supplementary material.
Given these facts, a G-protein-based opsin classification scheme may still hold promise, but the scarcity of opsin/G-protein data available today makes it impossible to confidently resolve any underlying co-evolutionary pattern. This being the case, more biochemical studies are needed to expand our knowledge of opsin/G-protein interactions to truly understand how opsin signalling has evolved.
(c). Counterion diversity
One of opsin's defining characteristics is the presence of a covalently bound chromophore, most commonly 11-cis retinal, which confers light sensitivity to the visual pigment. The chromophore is attached via a Schiff base linkage to a universally conserved lysine in the seventh transmembrane helix. Upon exposure to light, the chromophore undergoes a photoisomerization to all-trans retinal, which drives the activation of the photopigment. Free 11-cis retinal has an absorbance maximum around 380 nm. However, in order to allow visual pigments to function as photodetectors in the visible light range, the Schiff base linkage is protonated, which red-shifts the absorbance of the photopigment to approximately 440 nm. The amino acid side chains lining the binding pocket can further tune the specific absorbance of the pigment to sensitivities ranging from approximately 400–650 nm [65–67].
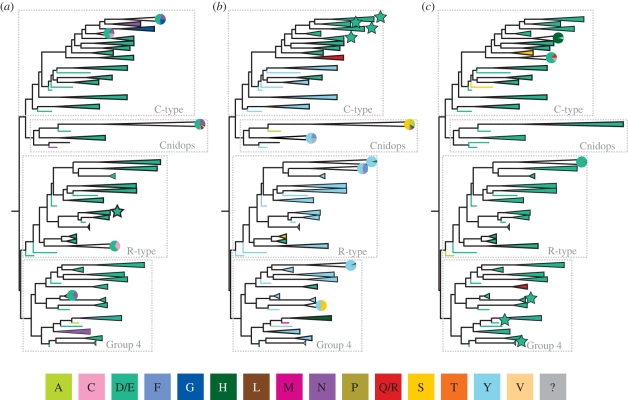
Typically, a negatively charged amino acid, called a counterion, is required to interact with and raise the pKa of the Schiff base, stabilizing the binding of the proton at physiological pH. Loss of the counterion produces a blue shift in the absorbance of the pigment, shifting the peak back to around 380 nm. The counterion also stabilizes the opsin without the chromophore, and loss of the counterion can lead to constitutive activity [67]. In vertebrate visual pigments, glutamic acid 113 (bovine rhodopsin numbering; used throughout the rest of this discussion) located in the third transmembrane helix acts as the counterion. Mutation of this glutamic acid leads both to a blue shift in the peak absorbance of the visual pigment to around 380 nm and to constitutive activity in all vertebrate opsins (C-type sequence group 1.1; figure 2) [66]. However, a negatively charged residue at this position is found only in vertebrate visual pigments (figure 6b). Across most of the rest of known opsin diversity, a conserved tyrosine occurs at this location, which is highly unlikely to be functioning as a counterion since it is uncharged at physiological pH (pKa >10). Therefore, a large percentage of opsin diversity must use one or more other residues as a counterion.
Figure 6.
Maximum-likelihood phylogeny as in figure 2, with the four major lineages indicated. Each well-supported clade has been coloured based on the residue present at three potential counterion sites: (a) bovine rhodopsin site 83, (b) bovine rhodopsin site 113, and (c) bovine rhodopsin site 181. For each group, if more than one amino acid with significantly different properties is found at a particular site, a pie chart representing the proportion of each residue within the group is included. Groups where the counterion position has been demonstrated using biochemical techniques are indicated with a star. The colours used to indicate amino acids are in the figure key; polarity of the reconstructed amino acids is as follows: non polar: A, F, L, M, P, V; no charge: C, G, N, Q, R, S, T, Y; negative: D, E; positive: H, Q, R. Amino acid residues unknown owing to unavailable data are indicated by a ‘?’ and are coloured grey in the figure.
In bovine rhodopsin, a switch occurs during photoactivation, moving the counterion site from E113 to E181, located in extracellular loop two [68]. Experimental evidence suggests that the residue corresponding to E181 also acts as the counterion in the peropsins, the cephalopod photoisomerase retinochrome and amphioxous rhodopsin (all Gr4 opsins) [68,69]. In fact, this glutamic acid is highly conserved throughout the whole opsin family (figure 6c).
Other than vertebrate C-type opsins, it is very difficult to express functional photopigments in tissue culture. As a result, spectral studies investigating counterions in many photopigments are very limited, but recent, mostly unpublished, research is beginning to address this. Our laboratory has found that mutation of the homologue of either E181 or E113 in the R-type opsin melanopsin has no effect on spectral absorbance nor does it create a constitutively active mutant. We have investigated the negative residues conserved across vertebrate melanopsins and found that the aspartic acid corresponding to bovine rhodopsin residue 83 functions as a counterion in mouse melanopsin (P. R. Robinson and B. Nickle, 2011, unpublished data). Thus, there are at least three experimentally demonstrated counterion sites among different groups of opsin proteins. Interestingly, a negative charge at position 83 is conserved among most known opsins (figure 6a). However, work recently carried out in Steven Britt's Laboratory (University of Colorado) has found that neither E181, E113 nor D83 mutants significantly affect the absorption spectrum of Drosophilia melanogaster Rh1. Furthermore, replacement of none of the conserved negatively charged residues found near the chromophore with an uncharged amino acid had any significant effect on the absorbance spectrum (S. G. Britt, 2011, personal communication), leaving the problem unsolved.
Taken together, the evolutionary history and the counterion mutagenic studies paint a more complicated picture than has previously been appreciated: while vertebrate visual pigments (figure 6b, starred groups) use 113 as a counterion, position 181 is used by a diverse group containing photoisomerases and Gi/o-coupled pigments (figure 6c, starred groups), vertebrate melanopsins use position 83 (figure 6a, starred group) and Drosophila uses none of the above nor any other recognized residue near the Schiff base. While 181 clearly is an important and conserved residue, it cannot be unambiguously designated as the ancestral counterion. Based on the limited current information, including counterion determination of only four opsins outside the vertebrate visual pigments, it is impossible to do any meaningful ancestral state reconstruction. Despite this, we would like to point out that the location of the counterion is more flexible than previously thought and that there is surprising consistency in the amino acids present at recognized counterion sites (even when they apparently do not operate functionally as true counterions).
4. Conclusions
Our large-scale phylogeny of 889 available opsin sequences, including transcript and genomic sequences, is the first large-scale evolutionary analysis of the opsin subclass of GPCRs. We recover the four previously recognized opsin lineages, but within each clade we find that the opsin subclass contains even more sequence diversity than was previously recognized. Many of the relationships represented in our phylogeny are also supported by intron position within and among groups, which can persist over immense spans of time, with strict conservation often continuing long after the corresponding proteins have become exceedingly diverged [70,71]. As genomes become available for a broader taxonomic set of organisms, our knowledge of opsin groups will certainly continue to expand and opsins with as of yet unknown functions will emerge.
This study emphasizes the large amount of opsin diversity not yet functionally characterized and highlights where future research should be focused. While a large body of literature exists for opsins involved in image formation, many other groups of opsins that have persisted over vast evolutionary timescales are poorly characterized (e.g. neuropsins). We show here that opsin sequences cannot be classified based on the distribution of characters such as G-protein binding, counterion placement, tissue-expression patterns and photoreceptor cell structure. Indeed, the evolutionary history presented here illustrates the lability of the opsin protein in diverse photodetection systems to couple to new G-protein pathways and to be promiscuously expressed in a wide range of cell and tissue types.
Acknowledgements
This study was supported by grants from the National Science Foundation to T.W.C. and P.R.R. (IOS0721608), from the Air Force Office of Scientific Research to T.W.C. (FA9550-09-1-0149) and from the National Eye Institute to P.R.R. (R01EY019053). E.G.C. and J.B. were supported by an NIH training grant (T32 GM066706). We would like to thank B. Nickle and T. Lamb for their valuable insight, and two anonymous reviewers for helpful comments on how to improve this manuscript.
References
- 1.Marmor M. F., Martin L. J. 1978. 100 years of the visual cycle. Survey of Ophthalmology 22, 279–285 10.1016/0039-6257(78)90074-7 (doi:10.1016/0039-6257(78)90074-7) [DOI] [PubMed] [Google Scholar]
- 2.Wald G. 1967. George Wald Nobel Prize Lecture. In Nobel lectures, physiology or medicine 2963-1970. Amsterdam, The Netherlands: Elsevier Publishing Company [Google Scholar]
- 3.Nathans J., Hogness D. S. 1983. Isolation, sequence analysis, and intron–exon arrangement of the gene encoding bovine rhodopsin. Cell 34, 807–814 10.1016/0092-8674(83)90537-8 (doi:10.1016/0092-8674(83)90537-8) [DOI] [PubMed] [Google Scholar]
- 4.Zuker C. S., Cowman A. F., Rubin G. M. 1985. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell 40, 851–858 10.1016/0092-8674(85)90344-7 (doi:10.1016/0092-8674(85)90344-7) [DOI] [PubMed] [Google Scholar]
- 5.Arendt D. 2003. Evolution of eyes and photoreceptor cell types. Int. J. Dev. Biol. 47, 563–571 [PubMed] [Google Scholar]
- 6.Arendt D., Tessmar-Raible K., Snyman H., Dorresteijn A. W., Wittbrodt J. 2004. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871 10.1126/science.1099955 (doi:10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- 7.Tsukamoto H., Terakita A. 2010. Diversity and functional properties of bistable pigments. Photochem. Photobiol. Sci. 9, 1435–1443 10.1039/c0pp00168f (doi:10.1039/c0pp00168f) [DOI] [PubMed] [Google Scholar]
- 8.Eakin R. M. 1972. Structure of invertebrate photoreceptors. In Handbook of sensory physiology: photochemistry of vision (ed. Darnall H. J. A.), pp. 625–684 Berlin/Heidelberg/New York: Springer [Google Scholar]
- 9.Hara-Nishimura I., Matsumoto T., Mori H., Nishimura M., Hara R., Hara T. 1990. Cloning and nucleotide sequence of cDNA for retinochrome, retinal photoisomerase from the squid retina. FEBS Lett. 271, 106–110 10.1016/0014-5793(90)80383-T (doi:10.1016/0014-5793(90)80383-T) [DOI] [PubMed] [Google Scholar]
- 10.Plachetzki D. C., Degnan B. M., Oakley T. H. 2007. The origins of novel protein interactions during animal opsin evolution. PLoS ONE 2, e1054. 10.1371/journal.pone.0001054 (doi:10.1371/journal.pone.0001054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies W. L., Hankins M. W., Foster R. G. 2010. Vertebrate ancient opsin and melanopsin: divergent irradiance detectors. Photochem. Photobiol. Sci. 9, 1444–1457 10.1039/c0pp00203h (doi:10.1039/c0pp00203h) [DOI] [PubMed] [Google Scholar]
- 12.Plachetzki D. C., Fong C. R., Oakley T. H. 2010. The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc. R. Soc. B 277, 1963–1969 10.1098/rspb.2009.1797 (doi:10.1098/rspb.2009.1797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terakita A. 2005. The opsins. Genome Biol. 6, 213. 10.1186/gb-2005-6-3-213 (doi:10.1186/gb-2005-6-3-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster R. G., Bellingham J. 2002. Opsins and melanopsins. Curr. Biol. 12, R543–R544 10.1016/S0960-9822(02)01047-3 (doi:10.1016/S0960-9822(02)01047-3) [DOI] [PubMed] [Google Scholar]
- 15.Suga H., Schmid V., Gehring W. J. 2008. Evolution and functional diversity of jellyfish opsins. Curr. Biol. 18, 51–55 10.1016/j.cub.2007.11.059 (doi:10.1016/j.cub.2007.11.059) [DOI] [PubMed] [Google Scholar]
- 16.Deville J., Rey J., Chabbert M. 2009. An indel in transmembrane helix 2 helps to trace the molecular evolution of class A G-protein-coupled receptors. J. Mol. Evol. 68, 475–489 10.1007/s00239-009-9214-9 (doi:10.1007/s00239-009-9214-9) [DOI] [PubMed] [Google Scholar]
- 17.Murphy W. J., Pringle T. H., Crider T. A., Springer M. S., Miller W. 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 17, 413–421 10.1101/gr.5918807 (doi:10.1101/gr.5918807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdorf K. 1979. The physiology of invertebrate visual pigments. In Handbook of sensory physiology (ed. Autrum H.), pp. 145–224 Berlin, Heidelberg, New York: Springer [Google Scholar]
- 19.Panda S., Nayak S. K., Campo B., Walker J. R., Hogenesch J. B., Jegla T. 2005. Illumination of the melanopsin signaling pathway. Science 307, 600–604 10.1126/science.1105121 (doi:10.1126/science.1105121) [DOI] [PubMed] [Google Scholar]
- 20.Stavenga D. G., Schwemer J. 1984. Visual pigments of invertebrates. In Photoreception and vision in invertebrates (ed. Ali M. A.), pp. 11–61 New York, London: Plenum Press [Google Scholar]
- 21.Yamashita T., Ohuchi H., Tomonari S., Ikeda K., Sakai K., Shichida Y. 2010. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc. Natl Acad. Sci. USA 107, 22 084–22 089 10.1073/pnas.1012498107 (doi:10.1073/pnas.1012498107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava M., et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726 10.1038/nature09201 (doi:10.1038/nature09201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson E. 2005. Photoreceptor evolution: ancient siblings serve different tasks. Curr. Biol. 15, R94–R96 10.1016/j.cub.2005.01.027 (doi:10.1016/j.cub.2005.01.027) [DOI] [PubMed] [Google Scholar]
- 24.Vopalensky P., Kozmik Z. 2009. Eye evolution: common use and independent recruitment of genetic components. Phil. Trans. R. Soc. B 364, 2819–2832 10.1098/rstb.2009.0079 (doi:10.1098/rstb.2009.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arendt D., Wittbrodt J. 2001. Reconstructing the eyes of Urbilateria. Phil. Trans. R. Soc. Lond. B 356, 1545–1563 10.1098/rstb.2001.0971 (doi:10.1098/rstb.2001.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velarde R. A., Sauer C. D., Walden K. K., Fahrbach S. E., Robertson H. M. 2005. Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 35, 1367–1377 10.1016/j.ibmb.2005.09.001 (doi:10.1016/j.ibmb.2005.09.001) [DOI] [PubMed] [Google Scholar]
- 27.Witkovsky P., Gallin E., Hollyfield J. G., Ripps H., Bridges C. D. 1976. Photoreceptor thresholds and visual pigment levels in normal and vitamin A-deprived Xenopus tadpoles. J. Neurophysiol. 39, 1272–1287 [DOI] [PubMed] [Google Scholar]
- 28.Ban E., Kasai A., Sato M., Yokozeki A., Hisatomi O., Oshima N. 2005. The signaling pathway in photoresponses that may be mediated by visual pigments in erythrophores of Nile tilapia. Pigment Cell Res. 18, 360–369 10.1111/j.1600-0749.2005.00267.x (doi:10.1111/j.1600-0749.2005.00267.x) [DOI] [PubMed] [Google Scholar]
- 29.Kasai A., Oshima N. 2006. Light-sensitive motile iridophores and visual pigments in the neon tetra, Paracheirodon innesi. Zool. Sci. 23, 815–819 10.2108/zsj.23.815 (doi:10.2108/zsj.23.815) [DOI] [PubMed] [Google Scholar]
- 30.Kawano-Yamashita E., Terakita A., Koyanagi M., Shichida Y., Oishi T., Tamotsu S. 2007. Immunohistochemical characterization of a parapinopsin-containing photoreceptor cell involved in the ultraviolet/green discrimination in the pineal organ of the river lamprey Lethenteron japonicum. J. Exp. Biol. 210, 3821–3829 10.1242/jeb.007161 (doi:10.1242/jeb.007161) [DOI] [PubMed] [Google Scholar]
- 31.Eakin R. M., Westfall J. A. 1962. Fine structure of photoreceptors in the hydromedusan, Polyorchis penicillatus. Proc. Natl Acad. Sci. USA 48, 826–833 10.1073/pnas.48.5.826 (doi:10.1073/pnas.48.5.826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin V. J. 2002. Photoreceptors of cnidarians. Can. J. Zool. 80, 1703–1722 10.1139/z02-136 (doi:10.1139/z02-136) [DOI] [Google Scholar]
- 33.Lampel J., Briscoe A. D., Wasserthal L. T. 2005. Expression of UV-, blue-, long-wavelength-sensitive opsins and melatonin in extraretinal photoreceptors of the optic lobes of hawk moths. Cell Tissue Res. 321, 443–458 10.1007/s00441-004-1069-1 (doi:10.1007/s00441-004-1069-1) [DOI] [PubMed] [Google Scholar]
- 34.Shimizu I., Yamakawa Y., Shimazaki Y., Iwasa T. 2001. Molecular cloning of Bombyx cerebral opsin (Boceropsin) and cellular localization of its expression in the silkworm brain. Biochem. Biophys. Res. Commun. 287, 27–34 10.1006/bbrc.2001.5540 (doi:10.1006/bbrc.2001.5540) [DOI] [PubMed] [Google Scholar]
- 35.Spaethe J., Briscoe A. D. 2005. Molecular characterization and expression of the UV opsin in bumblebees: three ommatidial subtypes in the retina and a new photoreceptor organ in the lamina. J. Exp. Biol. 208, 2347–2361 10.1242/jeb.01634 (doi:10.1242/jeb.01634) [DOI] [PubMed] [Google Scholar]
- 36.Tong D., Rozas N. S., Oakley T. H., Mitchell J., Colley N. J., McFall-Ngai M. J. 2009. Evidence for light perception in a bioluminescent organ. Proc. Natl Acad. Sci. USA 106, 9836–9841 10.1007/s00114-006-0119-9 (doi:10.1007/s00114-006-0119-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frigato E., Vallone D., Bertolucci C., Foulkes N. S. 2006. Isolation and characterization of melanopsin and pinopsin expression within photoreceptive sites of reptiles. Naturwissenschaften 93, 379–385 10.1007/s00114-006-0119-9 (doi:10.1007/s00114-006-0119-9) [DOI] [PubMed] [Google Scholar]
- 38.Provencio I., Rodriguez I. R., Jiang G., Hayes W. P., Moreira E. F., Rollag M. D. 2000. A novel human opsin in the inner retina. J. Neurosci. 20, 600–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blackshaw S., Snyder S. H. 1999. Encephalopsin: a novel mammalian extraretinal opsin discretely localized in the brain. J. Neurosci. 19, 3681–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikawa T., Okano T., Oishi T., Fukada Y. 1998. A deep brain photoreceptive molecule in the toad hypothalamus. FEBS Lett. 424, 69–72 10.1016/S0014-5793(98)00139-2 (doi:10.1016/S0014-5793(98)00139-2) [DOI] [PubMed] [Google Scholar]
- 41.Mathger L. M., Roberts S. B., Hanlon R. T. 2010. Evidence for distributed light sensing in the skin of cuttlefish, Sepia officinalis. Biol. Lett. 6, 600–603 10.1098/rsbl.2010.0223 (doi:10.1098/rsbl.2010.0223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provencio I., Jiang G., De Grip W. J., Hayes W. P., Rollag M. D. 1998. Melanopsin: an opsin in melanophores, brain, and eye. Proc. Natl Acad. Sci. USA 95, 340–345 10.1073/pnas.95.1.340 (doi:10.1073/pnas.95.1.340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez M. D., Speiser D. I., Pankey M. S., Oakley T. H. 2011. Understanding dermal light sense in the context of integrative photoreceptor cell biology. Vis. Neurosci. 28, 265–279 10.1017/S0952523811000150 (doi:10.1017/S0952523811000150) [DOI] [PubMed] [Google Scholar]
- 44.Neves S. R., Ram P. T., Iyengar R. 2002. G protein pathways. Science 296, 1636–1639 10.1126/science.1071550 (doi:10.1126/science.1071550) [DOI] [PubMed] [Google Scholar]
- 45.Pedersen S. E., Ross E. M. 1982. Functional reconstitution of beta-adrenergic receptors and the stimulatory GTP-binding protein of adenylate cyclase. Proc. Natl Acad. Sci. USA 79, 7228–7232 10.1073/pnas.79.23.7228 (doi:10.1073/pnas.79.23.7228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stryer L. 1983. Transducin and the cyclic GMP phosphodiesterase: amplifier proteins in vision. Cold Spring Harb. Symp. Quant. Biol. 48 (Pt 2), 841–852 [DOI] [PubMed] [Google Scholar]
- 47.Simon M. I., Strathmann M. P., Gautam N. 1991. Diversity of G proteins in signal transduction. Science 252, 802–808 10.1126/science.1902986 (doi:10.1126/science.1902986) [DOI] [PubMed] [Google Scholar]
- 48.Woehler A., Ponimaskin E. G. 2009. G protein-mediated signaling: same receptor, multiple effectors. Curr. Mol. Pharmacol. 2, 237–248 10.2174/1874467219092939237 (doi:10.2174/1874467219092939237) [DOI] [PubMed] [Google Scholar]
- 49.Baylor D. A., Yau K. W., Lamb T. D., Matthews G. 1978. Properties of the membrane current of rod outer segments. Sens. Process. 2, 300–305 [PubMed] [Google Scholar]
- 50.Hagins W. A., Penn R. D., Yoshikami S. 1970. Dark current and photocurrent in retinal rods. Biophys. J. 10, 380–412 10.1016/S0006-3495(70)86308-1 (doi:10.1016/S0006-3495(70)86308-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kojima D., Terakita A., Ishikawa T., Tsukahara Y., Maeda A., Shichida Y. 1997. A novel Go-mediated phototransduction cascade in scallop visual cells. J. Biol. Chem. 272, 22 979–22 982 10.1074/jbc.272.37.22979 (doi:10.1074/jbc.272.37.22979) [DOI] [PubMed] [Google Scholar]
- 52.Fein A., Cavar S. 2000. Divergent mechanisms for phototransduction of invertebrate microvillar photoreceptors. Vis. Neurosci. 17, 911–917 10.1017/S0952523800176102 (doi:10.1017/S0952523800176102) [DOI] [PubMed] [Google Scholar]
- 53.Garger A. V., Richard E. A., Lisman J. E. 2004. The excitation cascade of Limulus ventral photoreceptors: guanylate cyclase as the link between InsP3-mediated Ca2+ release and the opening of cGMP-gated channels. BMC Neurosci. 5, 7. 10.1186/1471-2202-5-7 (doi:10.1186/1471-2202-5-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardie R. C. 2001. Phototransduction in Drosophila melanogaster. J. Exp. Biol. 204, 3403–3409 [DOI] [PubMed] [Google Scholar]
- 55.Chyb S., Raghu P., Hardie R. C. 1999. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature 397, 255–259 10.1038/16703 (doi:10.1038/16703) [DOI] [PubMed] [Google Scholar]
- 56.Cockcroft S., Gomperts B. D. 1985. Role of guanine nucleotide binding protein in the activation of polyphosphoinositide phosphodiesterase. Nature 314, 534–536 10.1038/314534a0 (doi:10.1038/314534a0) [DOI] [PubMed] [Google Scholar]
- 57.Cook B., Minke B. 1999. TRP and calcium stores in Drosophila phototransduction. Cell Calcium 25, 161–171 10.1054/ceca.1998.0018 (doi:10.1054/ceca.1998.0018) [DOI] [PubMed] [Google Scholar]
- 58.Raghu P., Colley N. J., Webel R., James T., Hasan G., Danin M., Selinger Z., Hardie R. C. 2000. Normal phototransduction in Drosophila photoreceptors lacking an InsP(3) receptor gene. Mol. Cell. Neurosci. 15, 429–445 10.1006/mcne.2000.0846 (doi:10.1006/mcne.2000.0846) [DOI] [PubMed] [Google Scholar]
- 59.Koyanagi M., Takano K., Tsukamoto H., Ohtsu K., Tokunaga F., Terakita A. 2008. Jellyfish vision starts with cAMP signaling mediated by opsin-G(s) cascade. Proc. Natl Acad. Sci. USA 105, 15 576–15 580 10.1073/pnas.0806215105 (doi:10.1073/pnas.0806215105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Northup J. K., Sternweis P. C., Smigel M. D., Schleifer L. S., Ross E. M., Gilman A. G. 1980. Purification of the regulatory component of adenylate cyclase. Proc. Natl Acad. Sci. USA 77, 6516–6520 10.1073/pnas.77.11.6516 (doi:10.1073/pnas.77.11.6516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross E. M., Gilman A. G. 1980. Biochemical properties of hormone-sensitive adenylate cyclase. Annu. Rev. Biochem. 49, 533–564 10.1146/annurev.bi.49.070180.002533 (doi:10.1146/annurev.bi.49.070180.002533) [DOI] [PubMed] [Google Scholar]
- 62.Chen P., et al. 2001. A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet. 28, 256–260 10.1038/90089 (doi:10.1038/90089) [DOI] [PubMed] [Google Scholar]
- 63.Maeda T., Van Hooser J. P., Driessen C. A., Filipek S., Janssen J. J., Palczewski K. 2003. Evaluation of the role of the retinal G protein-coupled receptor (RGR) in the vertebrate retina in vivo. J. Neurochem. 85, 944–956 10.1046/j.1471-4159.2003.01741.x (doi:10.1046/j.1471-4159.2003.01741.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wenzel A., et al. 2005. The retinal G protein-coupled receptor (RGR) enhances isomerohydrolase activity independent of light. J. Biol. Chem. 280, 29 874–29 884 10.1074/jbc.M503603200 (doi:10.1074/jbc.M503603200) [DOI] [PubMed] [Google Scholar]
- 65.Nathans J. 1990. Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry 29, 9746–9752 10.1021/bi00493a034 (doi:10.1021/bi00493a034) [DOI] [PubMed] [Google Scholar]
- 66.Nickle B., Wilkie S. E., Cowing J. A., Hunt D. M., Robinson P. R. 2006. Vertebrate opsins belonging to different classes vary in constitutively active properties resulting from salt-bridge mutations. Biochemistry 45, 7307–7313 10.1021/bi060234g (doi:10.1021/bi060234g) [DOI] [PubMed] [Google Scholar]
- 67.Robinson P. R., Cohen G. B., Zhukovsky E. A., Oprian D. D. 1992. Constitutively active mutants of rhodopsin. Neuron 9, 719–725 10.1016/0896-6273(92)90034-B (doi:10.1016/0896-6273(92)90034-B) [DOI] [PubMed] [Google Scholar]
- 68.Terakita A., Koyanagi M., Tsukamoto H., Yamashita T., Miyata T., Shichida Y. 2004. Counterion displacement in the molecular evolution of the rhodopsin family. Nat. Struct. Mol. Biol. 11, 284–289 10.1038/nsmb731 (doi:10.1038/nsmb731) [DOI] [PubMed] [Google Scholar]
- 69.Terakita A., Yamashita T., Shichida Y. 2000. Highly conserved glutamic acid in the extracellular IV-V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc. Natl Acad. Sci. USA 97, 14 263–14 267 10.1073/pnas.260349597 (doi:10.1073/pnas.260349597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shichida Y., Matsuyama T. 2009. Evolution of opsins and phototransduction. Phil. Trans. R. Soc. B 364, 2881–2895 10.1098/rstb.2009.0051 (doi:10.1098/rstb.2009.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sullivan J. C., Reitzel A. M., Finnerty J. R. 2006. A high percentage of introns in human genes were present early in animal evolution: evidence from the basal metazoan Nematostella vectensis. Genome Inform. 17, 219–229 [PubMed] [Google Scholar]