Abstract
Traditional concepts of the Bering Land Bridge as a zone of predominantly eastward expansion from Eurasia and a staging area for subsequent colonization of lower latitudes in North America led to early inferences regarding biogeographic histories of North American faunas, many of which remain untested. Here we apply a host–parasite comparative phylogeographical (HPCP) approach to evaluate one such history, by testing competing biogeographic hypotheses for five lineages of host-specific parasites shared by the collared pika (Ochotona collaris) and American pika (Ochotona princeps) of North America. We determine whether the southern host species (O. princeps) was descended from a northern ancestor or vice versa. Three parasite phylogenies revealed patterns consistent with the hypothesis of a southern origin, which is corroborated by four additional parasite lineages restricted to O. princeps. This finding reverses the traditional narrative for the origins of North American pikas and highlights the role of dispersal from temperate North America into Beringia in structuring northern diversity considerably prior to the Holocene. By evaluating multiple parasite lineages simultaneously, the study demonstrates the power of HPCP for resolving complex biogeographic histories that are not revealed by characteristics of the host alone.
Keywords: Beringia, coevolution, helminth, host–parasite comparative phylogeography, Ochotona
1. Introduction
Beringia, the region spanning eastern Siberia and northwestern North America, played a central role in structuring Holarctic biotas, and the biogeographic history across this province has consequently been the subject of intense scrutiny [1–4]. The Bering Land Bridge was an intermittent terrestrial corridor that facilitated bi-directional dispersal between the northern continents through much of the Tertiary. With the onset of episodic glaciations during the Pliocene and Pleistocene, faunal expansion across the region became increasingly asymmetrical as greater numbers of Eurasian species colonized eastern Beringia and the Nearctic [5,6]. As a result, during Quaternary interglacials, temperate latitudes of North America were frequently colonized from source populations of species that were first established in Beringia. However, a reciprocal influence for low-latitude faunas relative to Beringian diversity is not well documented.
There are relatively few clear cases of deep (Pliocene or Pleistocene) expansion into Beringia by temperate North American species that then survived in the north through subsequent glacial stages. The colonization of Eurasia by North American equids and camelids provides classic examples [6], and more recent phylogenetic work has revealed evidence for similar histories in some small mammals [7,8]. Although numerous instances of post-glacial (Holocene) range expansion from southern refugia have been described, some of which involve species that successfully invaded Beringia [4,9], the role of such events (or analogous events during previous interglacials) in establishing new and persistent northern lineages has not been fully demonstrated.
Early ideas about North American faunal history were strongly influenced by the observation that colonization across Beringia over the past few million years was dominated by Asian immigrants entering North America [10]. Many hypotheses viewed Beringia as an engine of diversity for descendant components of North American faunas. Critical evaluation of these concepts is increasingly warranted in light of more contemporary perspectives on the complexities of Beringian biogeography [4,9]. Pikas, small lagomorphs of the genus Ochotona, provide a case in point. Pikas originated in the Palearctic and colonized the Nearctic via Beringia. The two extant North American species are sister [11,12], with the collared pika (Ochotona collaris) occurring in Alaska and adjacent Canadian provinces and the American pika (Ochotona princeps) distributed across North America's Intermountain West (figure 1). Based on their Old World origin and geographical distribution in North America, it was suggested that the northern O. collaris evolved directly from the ancestral Beringian colonizers, while O. princeps originated from individuals that expanded southward along the Coastal and Rocky Mountains of western North America [13]. However, evidence for this history is lacking. Indeed, pikas that are morphologically consistent with the extant species appear earlier in the fossil record at low latitudes (approx. 850 000 years before present; kyBP) than at high latitudes (<300 kyBP) [14], raising the alternative hypothesis that the ancestral northern population did not persist, and that O. collaris is descended from a southern ancestor. Under either scenario, speciation probably occurred when continental ice sheets isolated northern and southern populations [13].
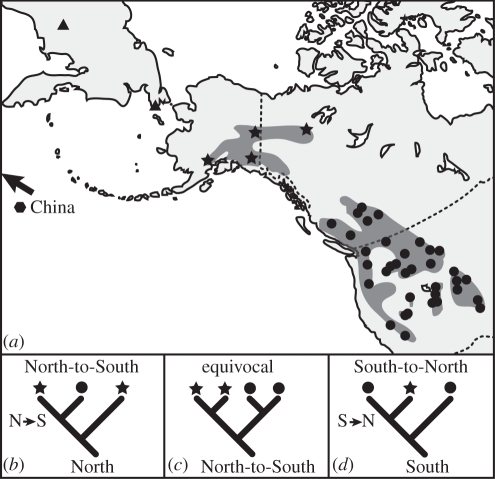
Figure 1.
Species distributions, sampling localities and phylogenetic predictions of biogeographic hypotheses for the history of North American pikas. On the map (a), dark grey patches indicate approximate species distributions for Ochotona princeps and O. collaris, which are distinguished by symbols used to identify sampling localities. Trees (b) and (d) show different patterns of phylogenetic relationships between northern and southern populations predicted by different biogeographic hypotheses. Reciprocal monophyly (c) between populations does not discriminate between hypotheses. Filled circles, O. princeps; filled asterisks, O. collaris; filled triangles, O. hyperborea; filled hexagon, O. cansus.
The two mutually exclusive hypotheses make different phylogenetic predictions. The North-to-South (N–S) hypothesis assumes that North American colonization proceeded from Siberia to Alaska and eventually to lower latitudes, resulting in southern diversity nested within the northern clade (figure 1). Conversely, if O. collaris is derived from a southern ancestor (South-to-North; S–N), we would predict southern paraphyly. Under either scenario, northern and southern populations could be reciprocally monophyletic given sufficient time for lineage sorting to occur. Evidence from mitochondrial [12] and allozymic [15] data suggest that collared and American pikas are reciprocally monophyletic, providing no insight into the question of colonization history.
In the absence of informative data from pikas themselves, we can test the alternative histories for North American Ochotona by applying a host–parasite comparative phylogeographical (HPCP) approach. Parasites can be excellent indicators of host biogeographic and ecological histories [16], which can be illuminated using co-phylogenetic methods [17]. Similarly, comparative phylogeography offers a powerful approach for reconstructing regional biogeographic histories from comparisons of co-distributed taxa [18]. A natural union of these related fields has resulted in the development of the HPCP approach [19,20]. Many HPCP studies focus on single parasite lineages, making it impossible to distinguish between patterns that reflect the history of the full assemblage versus taxon-specific processes. The solution is to incorporate at least one additional taxon, representing an independent perspective on the history in question (the ‘Threes Rule’; [17]). Here, we demonstrate the power of HPCP analysis using multiple parasite lineages to resolve aspects of host history that are not revealed by study of the host alone.
At least 17 genera of helminths (roundworms, tapeworms, flukes) parasitize pikas. Only six of these (Murielus, Graphidiella, Ohbayashinema, Labiostomum, Cephaluris and Schizorchis) have been reported from North America, and of these, one tapeworm (Schizorchis) and two pinworm (Cephaluris and Labiostomum) genera parasitize both pika species. Certain Trichostrongylina nematodes (Murielus, Graphidiella and Ohbayashinema) appear to be restricted to O. princeps, having not been detected from over 200 O. collaris specimens examined as part of biodiversity surveys spanning northern Canada and Alaska [21]. Among oxyurid pinworms, Labiostomum is subdivided into the subgenera L. (Labiostomum) and L. (Eugenuris), which we treat as separate units of analysis in this study. We refer to these subgenera and all other parasite genera as the ‘major parasite lineages’ associated with pikas. Each major lineage has greater species diversity in Eurasian pikas, consistent with a Palearctic origin. No lineage is known to parasitize hosts other than pikas, so host-specificity has probably been maintained in North America since the original colonization. Only Schizorchis requires an intermediate host to complete its life cycle, probably an oribatid mite [22], which is unlikely to play a major role in parasite dispersal. Thus, parasite gene flow is mediated by pikas.
To test the competing hypotheses regarding the origins of O. collaris and O. princeps (N–S versus S–N), we focus phylogeographical analyses on Schizorchis, Cephaluris, L. (Labiostomum) and L. (Eugenuris) because they are shared by both pika species. Molecular phylogenetic methods allow us to (i) determine the minimum number of successful North American colonizations accomplished by parasites (representing the number of independent tests of host history in North America), (ii) identify centres of origin for the living North American pikas, and (iii) test for simultaneous isolation between northern and southern populations of parasites.
2. Material and methods
(a). Data collection
We collected parasites from 147 O. princeps and 13 O. collaris specimens distributed across the two hosts' ranges, representing 35 and 4 localities, respectively (figure 1 and electronic supplemental material, table S1). We also sampled parasites from four O. hyperborea specimens (northern pika; two localities in Siberia) and two O. cansus specimens (Gansu pika; one locality in Sichuan, China), which provide a context for understanding relationships between Old and New World parasite lineages. Species identifications were based on original species descriptions and revisions, as well as comparisons with types and vouchers from the United States National Parasite Collection (USNPC): S. caballeroi, holotype: USNPC no. 39024; S. ochotonae, syntype: 37056; S. ryzhikovi, holotype: 77472, paratype: 77473; S. yamashitai, holotype: 59876, voucher: 77471; L. (L.) coloradensis, 39443; L. (E.) utahensis, syntype: 73259; C. alaskensis, vouchers: 73536, 73538. Voucher and tissue samples for all parasites are archived in the USNPC.
We purified genomic DNA from specimens of Cephaluris (n = 92 individuals), L. (Eugenuris) (n = 55), L. (Labiostomum) (n = 49) and Schizorchis (n = 64) using either phenol–chloroform extractions or Qiagen DNeasy kits. We amplified a portion of the mitochondrial (mtDNA) cytochrome oxidase I (COI; 369 bp) gene and ribosomal subunits (rRNA; approx. 810 bp) from the nematodes and cestodes, respectively, using primers COI-F/COI-R [23] and Hym16sF/Hym12sR [24]. Polymerase chain reaction (PCR) conditions were as in Galbreath et al. [25], except for the annealing temperature (50°C). We sequenced PCR products in both directions and aligned sequences with Clustal W [26] using MEGA v. 3.1 [27]. Alignments were checked by eye and indels removed to meet requirements of various subsequent analyses. Unless otherwise noted, further analyses were based on datasets condensed to unique haplotypes. Sequences are deposited in GenBank (HQ189777–HQ190036).
(b). Phylogenetic analyses
We reconstructed phylogenies in separate analyses for each parasite lineage using Bayesian (BA), maximum likelihood (ML) and maximum parsimony (MP) methods. One representative sequence from each of the major pinworm lineages served as outgroups for analyses on the nematodes. Hymenolepis diminuta (GenBank no. AF314223) served as the outgroup for analyses of Schizorchis. Appropriate models of nucleotide evolution were chosen by iterating between ML tree searches in Garli v. 0.96 (D. J. Zwickl, 2006. Unpublished PhD Thesis, University of Texas at Austin, TX, USA) and model searches in DT-Modsel [28] until the model stabilized. All selected models incorporated a gamma distribution to accommodate rate variation among sites. For the COI datasets, we used a simulation procedure to test these null models against alternatives that replaced the gamma distribution with codon site-specific (SS) rates [29]. We (i) estimated the best ML tree and model parameters under the null model using Garli; (ii) simulated 500 datasets on the ML tree under the null model using Mesquite v. 2.5 [30]; (iii) generated ML trees for each simulated dataset under the null model; (iv) calculated the difference δ in the likelihood score for each tree under the null and alternative models using PAUP* v. 4.0b10 [31]; and (v) compared δ for the empirical data with the simulated distribution. In all cases, SS models significantly improved the likelihood score and the null model was rejected.
For ML analyses, we used a version of Garli v. 0.96 (v. r601) that allows SS models, running 10 replicates of each analysis and assessing nodal support with 100 bootstrap replicates. We used MrBayes v. 3.1.2 [32] to conduct BA analyses, running five chains for 10 million steps and discarding one million steps as burn-in. Three independent analyses converged on nearly identical topologies. Stationarity was confirmed by comparing posterior probabilities of splits across all runs using AWTY [33] and ensuring that average standard deviation of split frequencies approached zero (all <0.005). We conducted heuristic MP searches in PAUP with TBR branch swapping and 1000 random addition replicates, summarizing the most parsimonious topologies in strict consensus trees.
(c). Hypothesis testing
To test the robustness of inferred paraphyletic relationships between northern and southern parasite populations, we applied two types of topology tests. First, we used constraint filters in PAUP to calculate the proportion of trees retained by the BA analysis in which paraphyly was not evident [34]. Second, we used parametric bootstrapping (PB; [35]), calculating the difference in likelihood scores between ML phylogenies unconstrained and constrained to match the null hypothesis of monophyly as a test statistic. For each test, we selected a substitution model and parameters as described above based on a ML phylogeny constrained to fit the null hypothesis. To generate null distributions to compare with the empirical result, we used Mesquite to simulate 500 datasets on the best ML tree constrained to fit the null hypothesis and ran constrained and unconstrained ML tree searches on the datasets using Garli. These BA and PB methods were also used to test alternative tree topologies inferred based on different phylogenetic methods.
If the parasite communities of North American pikas were separated in the same event that initiated speciation of O. princeps and O. collaris, we would expect simultaneous divergence for pairs of northern and southern populations of the parasite lineages. We tested relative timing of divergence among nematode lineages using MsBayes v. 20081106 [36]. We excluded Schizorchis because MsBayes assumes equal mutation rates across taxa [37]. Population samples were limited to 50 randomly selected individuals drawn from complete COI sequence datasets and representing the least inclusive, strongly supported clade that included all northern and some southern individuals. We ran one million simulations, generating the posterior distribution from 1000 samples using three summary statistics: mean pairwise differences among sequences between populations, mean pairwise distances among sequences within southern populations and number of segregating sites within southern populations normalized for sample size.
3. Results
(a). Collections and species identifications
Morphological examination of helminths revealed 15 morphospecies and demonstrated that all parasite species known to be associated with Nearctic pikas were present in our sample (electronic supplemental material, table S2). Labiostomum (E.) talkeetnaeuris and L. (L.) rauschi had previously been thought to be restricted to O. collaris, but we found both to be widespread throughout the range of O. princeps [38]. We detected several morphologically distinct groups of L. (Labiostomum), L. (Eugenuris) and Schizorchis that represent putative undescribed species.
(b). Phylogenetic analyses
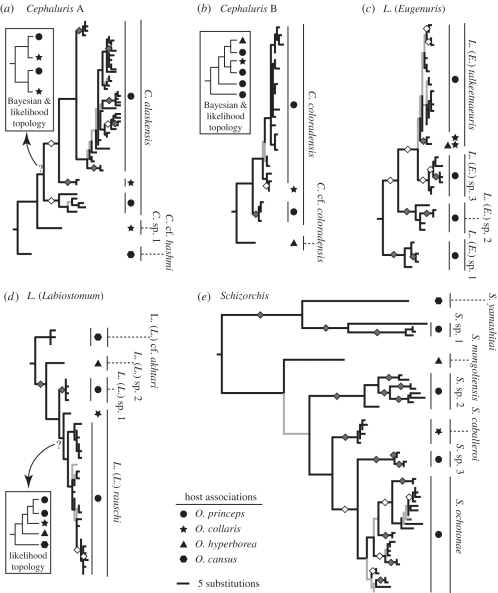
Most species and undescribed morphotypes form monophyletic clades, demonstrating congruence between morphological and molecular structure (figure 2). Cephaluris is divided into two clades that we treated as separate major lineages for analyses (hereafter Cephaluris A and B). The Cephaluris A lineage is unique in that it includes two distinct clades associated with both O. collaris and O. princeps (figure 2). Monophyly of these northern and southern pairs was strongly rejected (table 1). Phylogenies of Cephaluris B, L. (Eugenuris) and Schizorchis revealed that parasite populations associated with American pikas are paraphyletic with respect to those from collared pikas. Southern monophyly was strongly rejected for these lineages (table 1). Tests of L. (Labiostomum) were ambiguous, with contradictory topologies from MP and ML analyses and an unresolved BA topology. Southern monophyly for L. (Labiostomum) was not rejected and tests of the MP topology were equivocal.
Figure 2.
Parasite phylogenies. Trees shown are one of the most parsimonious trees identified for (a) Cephaluris A, (b) Cephaluris B, (c) L. (Eugenuris), (d) L. (Labiostomum) and (e) Schizorchis. Inset diagrams indicate incongruent relationships among major clades retrieved from maximum likelihood (ML) and/or Bayesian (BA) analyses if applicable. Specific disputed nodes are indicated with question marks. On all trees, black branches indicate relationships retained by strict consensus of all most parsimonious trees and grey branches were collapsed in the strict consensus tree. Diamonds on branches indicate BA posterior probabilities >0.95, and grey filling within the diamonds denotes ML bootstrap values ≥ 80. Symbols denote host associations for parasite lineages (see the inset key). Species' names or temporary clade designations are given next to major clades that were distinguishable morphologically. Outgroups have been removed for clarity.
Table 1.
Bayesian (BA) and parametric bootstrapping (PB) results for tests of topological hypotheses. n.s. Indicates non-significance owing to identical constrained and unconstrained likelihood trees.
| hypothesis | pBAa | pPBb |
|---|---|---|
| tests of biogeographic hypotheses | ||
| monophyly of southern Cephaluris A | <0.001 | <0.002 |
| monophyly of northern Cephaluris A | <0.001 | <0.002 |
| monophyly of southern Cephaluris B | <0.001 | <0.002 |
| monophyly of southern L. (Eugenuris) | <0.001 | <0.002 |
| monophyly of southern L. (Labiostomum) | 0.319 | n.s. |
| monophyly of southern Schizorchis | 0.008 | 0.022 |
| tests of alternative topologies | ||
| Cephaluris A: Bayesian/likelihood topology | 0.852 | n.s. |
| Cephaluris A: Parsimony topology | 0.001 | <0.002 |
| Cephaluris B: Bayesian/likelihood topology | 0.957 | n.s. |
| Cephaluris B: Parsimony topology | 0.043 | 0.078 |
| L. (Labiostomum): Parsimony topology | 0.681 | 0.004 |
apBA denotes proportion of retained trees from the Bayesian analysis that match the phylogenetic hypothesis.
bpPB denotes proportion of the simulated test statistic distribution that is greater than or equal to the empirical value of the test statistic.
Instances of incongruence between MP, BA and ML trees raise questions about certain relationships, but do not contradict key results. For Cephaluris A, MP showed that parasites associated with O. collaris are either sister to or nested within the southern clade (figure 2). In contrast, ML and BA trees showed a pair of sister relationships between northern and southern clades. Tests of these alternatives rejected the MP topology (table 1). The MP tree for Cephaluris B showed that Palearctic and Nearctic worms were sisters, but in ML and BA trees, the Palearctic haplotype was nested within the southern parasite clade. Tests of these topologies did not reject the BA/ML tree, but only weakly rejected the MP tree. This result does not affect the conclusion that Cephaluris B has a history consistent with S–N colonization, but the ML and BA trees raise the possibility of a westward dispersal into Siberia across the Bering Land Bridge.
All major parasite lineages are of Old World origin; consequently, at least one trans-Beringian colonization event is assumed for each and nodes leading to Palearctic species are expected to predate splits between Nearctic taxa. Thus, we inferred single colonizations of North America for L. (Eugenuris), L. (Labiostomum), Murielus, Graphidiella and Ohbayashinema. A haplotype of L. (Eugenuris) shared between Siberian and Alaskan populations implies recent westward dispersal across Beringia. Like Cephaluris, Schizorchis is divided into two deeply divergent sub-lineages, though one of these is restricted to O. princeps. Each of the Cephaluris and Schizorchis sub-lineages are anchored by basal splits between Nearctic and Palearctic taxa, suggesting that North America was colonized by two species of each genus. In total, we identified nine North American colonization events involving independent parasite lineages, and one (possibly two) secondary dispersal from North America into Siberia.
The MsBayes analysis incorporated five population pairs, including two pairs derived from the ML and BA analyses of the Cephaluris A lineage. We failed to reject the hypothesis of simultaneous divergence between northern and southern pinworm populations. The posterior estimate of hyper-parameter Ψ (number of possible divergence times) did not differ significantly from 1 (median, 95% quantile; 1.00, 1.00–1.01).
4. Discussion
(a). Historical biogeography of North American pikas
Our HPCP analysis rejects the conventional explanation for the origin of O. princeps and O. collaris. In three of the five parasite lineages that are shared by both Nearctic pika species, phylogenies are unambiguously consistent with the S–N dispersal hypothesis. No trees match expectations for the alternative history. Because the parasites have obligate associations with pikas, dispersal and colonization must be mediated by the hosts. Thus, phylogeographical evidence from the parasites is consistent with the disappearance of trans-Beringian migrants from high latitudes, and subsequent recolonization from a southern source. Such a history may explain the apparent absence of O. collaris from northern Alaska [39]. Northward expansion may have been slowed by barriers separating Alaska's southern and northern mountain systems.
Might hosts and parasites have different biogeographic histories? If the descendants of the original pika colonizers of Beringia persisted in the refugium to the present, it would be necessary to invoke multiple independent events of parasite extinction in the north, followed by secondary contact and parasite exchange between northern and southern host populations. In addition, northern extinctions of one sub-lineage of Schizorchis and three nematodes (Murielus, Graphidiella and Ohbayashinema) also must be assumed to reconcile the N–S hypothesis [21,40]. The S–N hypothesis is more parsimonious, explaining the loss of ancestral parasite populations in the north via host extinction and parasite re-colonization via host re-colonization. A single re-colonization event is also consistent with failure to reject the hypothesis of simultaneous divergence between northern and southern populations. Parasite lineages that are absent from O. collaris probably ‘missed the boat’ during re-colonization [41], which is prone to happen if founder populations are small and isolated, just as founder effect can lead to a loss of genetic diversity at the leading edge of expanding populations [42]. In summary, evidence from seven of nine independent parasite lineages points to a southern origin for the northern parasite assemblage, strongly implying that O. collaris, itself, was derived from a northern isolate of its southern relative.
This is not the first evidence for a deep and complex history for North American pikas that contrasts with the conventional understanding of pika origins. For example, the earliest record of Ochotona in North America is from Oregon (O. spanglei) and dates to the Miocene–Pliocene transition [43], and middle Pleistocene records of pikas that are morphologically similar to both O. collaris and O. princeps [14] have been identified in eastern North America where pikas no longer persist. Though the relationship between these fossils and the modern species is unknown, these early records hint at a complex history of diversification and dispersal at low latitudes. Likewise, molecular clock estimates indicate that pikas occupied North America continuously since at least the Early Pleistocene, with the split between O. collaris and O. princeps possibly dating as far back as the Pliocene [12].
Our findings have implications for understanding the complementary roles of Beringia and temperate North America in structuring biotic diversity across the Nearctic. Beringia was a continental crossroad and a centre of diversification [1], in both capacities serving as a source of new diversity for North America. During the Quaternary, continental ice sheets and a mosaic of Arctic environments helped to block temperate North American species from entering Beringia and crossing into Asia [10]. Northward range expansion from low latitudes is therefore predominantly considered in the context of post-Pleistocene population responses to glacial retreat [9], which generally result in genetically homogeneous northern populations that may retract southward during future cooling events [4]. The original colonization of North America from Asia by ancestral Ochotona fits the traditional Beringian narrative of eastward colonization from Asia. In contrast, the finding that pikas returned to Beringia from temperate latitudes and persisted there, undergoing speciation and contributing to the evolution of a diverse North American parasite fauna (up to 18 species descended from nine ancestral parasites) emphasizes the biogeographic complexity of the region. Further, the indication that at least one parasite lineage recently crossed from eastern to western Beringia raises the possibility of Late Pleistocene contact between collared pikas and Ochotona hyperborea in Asia. Our results suggest a particular role for S–N colonization in cold-adapted species like pikas that may face fewer obstacles to dispersal between low and high latitudes [44]. Histories of dispersal and diversification in other North American species associated with cold environments should be reviewed, with an emphasis on evaluating the potential for HPCP to enrich biogeographic inferences.
(b). Host–parasite comparative phylogeography
As one of the most taxonomically comprehensive phylogeographical studies of a parasite assemblage to date, our work demonstrates the power of HPCP for resolving host biogeographic histories. The strength of this approach is particularly evident in consideration of weaknesses of our study. For example, our phylogenetic reconstructions are based on small fragments of a single DNA locus, which yielded limited support for certain nodes in some trees. From such data, conclusions drawn from a single parasite lineage would be weak. However, support for a given biogeographic hypothesis increases with each additional taxon that meets the predictions of the hypothesis. Though individually weak, concordance among biogeographic perspectives offered by multiple parasites suggests that the overall pattern is robust.
Comprehensive sampling of diversity is important for HPCP studies. Though our survey of parasites from O. princeps was extensive, sampling of O. collaris was less thorough, and sampling of Asian diversity was obviously incomplete. The latter point certainly has implications for our understanding of the number, timing and direction of dispersal events across Beringia, and future work will focus on establishing this Old World context for North American diversity. However, a more comprehensive Palearctic perspective should not substantively influence relationships between the northern and southern Nearctic parasite faunas. Sampling within O. collaris is a much more critical issue, but we are cautiously optimistic that the current dataset is sufficient to support our conclusions. First, collection localities were widely dispersed across the full host range. Second, most of the current range of O. collaris was colonized post-glacially from a restricted Beringian population. Range fluctuation around a single glacial-age refugium could have helped homogenize the parasite assemblage, breaking up pockets of locally endemic faunas and spreading species among host populations [45]. Third, our sample of O. collaris parasites includes all species previously reported from extensive biotic surveys conducted over the past century, so at minimum it represents a comprehensive sample of known diversity.
If we did fail to detect some parasite diversity from O. collaris, our major conclusions could be weakened but probably would not be overturned. An undetected parasite haplotype could fall (i) within or sister to an existing clade of O. collaris parasites, (ii) not sister to other O. collaris parasites, but still nested within the phylogeny of parasites from O. princeps, or (iii) sister to all the O. princeps parasites. The first two scenarios would not affect our conclusions. The third could lead to an equivocal biogeographic inference, but it would not support the competing biogeographic hypothesis unless many deeply divergent lineages were discovered. Also, the fact that we detected congruent histories across multiple parasite lineages reduces the likelihood that the observed pattern is simply a consequence of sampling bias. Although we cannot exclude the possibility that more intensive sampling in the north might reveal isolated populations that maintain deeply divergent parasite lineages, the homogenizing influence of glacial–interglacial range fluctuation, which is evident in low levels of genetic structure observed in O. collaris [12], indicates that such ancient and persistent isolates are unlikely.
Advocates of HPCP have suggested that genealogical congruence between hosts and parasites is important for parasites to be useful as proxies for host history [19,20,46]. Concordant phylogeographical patterns between host and parasites would then presumably reflect co-differentiation owing to extrinsic barriers to gene flow, and additional substructure within parasites could indicate cryptic host isolation events (e.g. [47]). This emphasis has led some to conclude that HPCP studies should focus on parasites with vertical (parent-to-offspring) transmission, small effective population sizes, high mutation rates and short generation times, thereby maximizing the likelihood of concordant parasite and host genealogies [20,46]. We suggest that a focus on genealogical congruence and vicariance restricts the types of questions that can be asked, and can miss complex patterns of colonization (geographical and host) and biotic expansion, which are often dominant processes in host–parasite histories [48,49]. Here, incomplete sorting of parasite lineages provided the key to testing hypotheses. Had the parasites in this study conformed to the criteria outlined above, they would have been more likely to yield uninformative phylogenies (figure 1c).
As in any comparative phylogeographical analysis, investigators of HPCP should define the specific hypotheses to test and consider the phylogenetic, biogeographic, demographic or population genetic predictions that follow. The characteristics of the ideal parasites for study may vary with the question being addressed. Further, using HPCP to test an explicit a priori hypothesis for historical biogeography should not be confused with approaches in cladistic biogeography that seek a maximum fit of data to a particular model of distribution and diversification [50]. Both HPCP as invoked here, and a posteriori methods for exploring diverse assemblages of parasites with varying life-history characteristics, can reveal complex species histories in the absence of constraints and formalized assumptions [17].
Acknowledgements
This work was supported by grants from the National Science Foundation (DEB 0506042 to K.E.G., DEB 0196095 and DEB 0415668 to E.P.H., DEB 0343526 to Kelly Zamudio), American Society of Mammalogists, Sigma Xi, Andrew Mellon Foundation (Cornell), American Museum of Natural History and Cornell Department of Ecology and Evolutionary Biology. We used the resources of the Computational Biology Service Unit at Cornell University, which is partially funded by Microsoft Corporation. J. Cook, C. Dardia, M. Matocq, A. Barroll, S. Kiphart, K. Kiphart and K. Fryzel supported fieldwork. P. Allen and S. Stockwell provided computing assistance. Specimens were contributed by A. Veitch and R. Popko, the Beringian Coevolution Project and P Giraudoux, F. Raoul and J. P. Quéré of the University of Franche-Comté who are supported by the US National Institutes of Health through the Fogarty International Centre (EID TW001565-05). K. Zamudio, R. Harrison, I. Lovette and two anonymous reviewers offered insightful comments on the manuscript.
References
- 1.Sher A. 1999. Traffic lights at the Beringian crossroads. Nature 397, 103–104 10.1038/16341 (doi:10.1038/16341) [DOI] [Google Scholar]
- 2.Hultén E. 1937. Outline of the History of Arctic and boreal biota during the Quaternary Period. New York, NY: Lehre J. Cramer [Google Scholar]
- 3.Hopkins D. M. 1959. Cenozoic history of the Bering Land Bridge. Science 129, 1519–1528 10.1126/science.129.3362.1519 (doi:10.1126/science.129.3362.1519) [DOI] [PubMed] [Google Scholar]
- 4.Hewitt G. 2004. The structure of biodiversity: insights from molecular phylogeography. Front. Zool. 1, 4. 10.1186/1742-9994-1-4 (doi:10.1186/1742-9994-1-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waltari E., Hoberg E. P., Lessa E. P., Cook J. A. 2007. Eastward Ho: phylogeographical perspectives on colonization of hosts and parasites across the Beringian nexus. J. Biogeogr. 34, 561–574 10.1111/j.1365-2699.2007.01705.x (doi:10.1111/j.1365-2699.2007.01705.x) [DOI] [Google Scholar]
- 6.Kurtén B. 1971. The age of mammals. London, UK: Weidenfeld and Nicolson [Google Scholar]
- 7.Demboski J. R., Cook J. A. 2003. Phylogenetic diversification within the Sorex cinereus group (Soricidae). J. Mammal. 84, 144–158 (doi:10.1644/1545-1542(2003)084<0144:PDWTSC>2.0.CO;2) [DOI] [Google Scholar]
- 8.Galbreath K. E., Cook J. A., Eddingsaas A. A., DeChaine E. G. In press Diversity and demography in Beringia: multi-locus tests of paleodistribution models reveal complex histories for arctic ground squirrels. Evolution 65 10.1111/j.1558-5646.2011.01287.x (doi:10.1111/j.1558-5646.2011.01287.x) [DOI] [PubMed] [Google Scholar]
- 9.Shafer A. B. A., Cullingham C. I., Côté S. D., Coltman D. W. 2010. Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol. Ecol. 19, 4589–4621 10.1111/j.1365-294X.2010.04828.x (doi:10.1111/j.1365-294X.2010.04828.x) [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann R. S. 1981. Different voles for different holes: environmental restrictions on refugial survival of mammals. In Evolution today. Proc. of the Second International Congress of Systematic and Evolutionary Biology, pp. 25–45 Pittsburgh, PA: Carnegie-Mellon University [Google Scholar]
- 11.Lissovsky A. A., Ivanova N. V., Borisenko A. V. 2007. Molecular phylogenetics and taxonomy of the subgenus Pika (Ochotona, Lagomorpha). J. Mammal. 88, 1195–1204 10.1644/06-MAMM-A-363R.1 (doi:10.1644/06-MAMM-A-363R.1) [DOI] [Google Scholar]
- 12.Lanier H. C., Olson L. E. 2009. Inferring divergence times within pikas (Ochotona spp.) using mtDNA and relaxed molecular dating techniques. Mol. Phylogenet. Evol. 53, 1–12 10.1016/j.ympev.2009.05.035 (doi:10.1016/j.ympev.2009.05.035) [DOI] [PubMed] [Google Scholar]
- 13.Guthrie R. D. 1973. Mummified pika (Ochotona) carcass and dung pellets from Pleistocene deposits in interior Alaska. J. Mammal. 54, 970–971 10.2307/1379093 (doi:10.2307/1379093) [DOI] [Google Scholar]
- 14.Mead J. I., Grady F. 1996. Ochotona (Lagomorpha) from Late Quaternary cave deposits in eastern North America. Quat. Res. 45, 93–101 10.1006/qres.1996.0009 (doi:10.1006/qres.1996.0009) [DOI] [Google Scholar]
- 15.Hafner D. J., Sullivan R. M. 1995. Historical and ecological biogeography of Nearctic pikas (Lagomorpha, Ochotonidae). J. Mammal. 76, 302–321 10.2307/1382343 (doi:10.2307/1382343) [DOI] [Google Scholar]
- 16.Manter H. W. 1966. Parasites of fishes as biological indicators of recent and ancient conditions. In Host–parasite relationships (ed. McCauley J. E.), pp. 59–71 Corvallis, OR: Oregon State University Press [Google Scholar]
- 17.Brooks D. R., McLennan D. A. 2002. The nature of diversity: an evolutionary voyage of discovery. Chicago, IL: University of Chicago Press [Google Scholar]
- 18.Avise J. C. 2000. Phylogeography: the history and formation of species. Cambridge, UK: Harvard University Press [Google Scholar]
- 19.Criscione C. D., Poulin R., Blouin M. S. 2005. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol. Ecol. 14, 2247–2257 10.1111/j.1365-294X.2005.02587.x (doi:10.1111/j.1365-294X.2005.02587.x) [DOI] [PubMed] [Google Scholar]
- 20.Nieberding C. M., Olivieri I. 2007. Parasites: proxies for host genealogy and ecology. Trends Ecol. Evol. 22, 156–165 10.1016/j.tree.2006.11.012 (doi:10.1016/j.tree.2006.11.012) [DOI] [PubMed] [Google Scholar]
- 21.Durette-Desset M. C., Galbreath K. E., Hoberg E. P. 2010. Discovery of new Ohbayashinema spp. (Nematoda: Heligomosomoidea) in Ochotona princeps and Ochotona cansus (Lagomorpha: Ochotonidae) from western North America and Central Asia, with considerations of historical biogeography. J. Parasitol. 96, 569–579 10.1645/GE-2397.1 (doi:10.1645/GE-2397.1) [DOI] [PubMed] [Google Scholar]
- 22.Guan J., Lin Y. 1988. Life cycle of Schizorchis altaica Gvozdev (Cestoda: Anoplocephalidae). J. Xiamen Univ. Nat. Sci. 27, 709–713 [Google Scholar]
- 23.McDonnell A., Love S., Tait A., Lichtenfels J. R., Matthews J. B. 2000. Phylogenetic analysis of partial mitochondrial cytochrome oxidase c subunit I and large ribosomal RNA sequences and nuclear internal transcribed spacer I sequences from species of Cyathostominae and Strongylinae (Nematoda, Order Strongylida), parasites of the horse. Parasitology 121, 649–659 10.1017/S003118200000696X (doi:10.1017/S003118200000696X) [DOI] [PubMed] [Google Scholar]
- 24.von Nickisch-Rosenegk M., Brown W. M., Boore J. L. 2001. Complete sequence of the mitochondrial genome of the tapeworm Hymenolepis diminuta: gene arrangements indicate that platyhelminths are eutrochozoans. Mol. Biol. Evol. 18, 721–730 [DOI] [PubMed] [Google Scholar]
- 25.Galbreath K. E., Hafner D. J., Zamudio K. R. 2009. When cold is better: climate-driven elevation shifts yield complex patterns of diversification and demography in an alpine specialist (American pika, Ochotona princeps). Evolution 63, 2848–2863 10.1111/j.1558-5646.2009.00803.x (doi:10.1111/j.1558-5646.2009.00803.x) [DOI] [PubMed] [Google Scholar]
- 26.Thompson J. D., Higgins D. G., Gibson T. J. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 10.1093/nar/22.22.4673 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S., Tamura K., Nei M. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163 10.1093/bib/5.2.150 (doi:10.1093/bib/5.2.150) [DOI] [PubMed] [Google Scholar]
- 28.Minin V., Abdo Z., Joyce P., Sullivan J. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52, 674–683 10.1080/10635150390235494 (doi:10.1080/10635150390235494) [DOI] [PubMed] [Google Scholar]
- 29.Savolainen R., Vepsäläinen K. 2003. Sympatric speciation through intraspecific social parasitism. Proc. Natl Acad. Sci. USA 100, 7169–7174 10.1073/pnas.1036825100 (doi:10.1073/pnas.1036825100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddison W. P., Maddison D. R. 2008. Mesquite: a modular system for evolutionary analysis, Version 2.5. See http://mesquiteproject.org/
- 31.Swofford D. L. 2000. PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4.0 Sunderland, MA: Sinauer Associates [Google Scholar]
- 32.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: a program for the Bayesian inference of phylogeny. See http://mrbayescsitfsuedu//ref://mrbayescsitfsuedu/
- 33.Nylander J. A., Wilgenbusch J. C., Warren D. L., Swofford D. L. 2008. AWTY (Are We There Yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583 10.1093/bioinformatics/btm388 (doi:10.1093/bioinformatics/btm388) [DOI] [PubMed] [Google Scholar]
- 34.Carstens B. C., Brunsfeld S. J., Demboski J. R., Good J. M., Sullivan J. 2005. Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution 59, 1639–1652 10.1554/04-661.1 (doi:10.1554/04-661.1) [DOI] [PubMed] [Google Scholar]
- 35.Goldman N. 1993. Statistical tests of models of DNA substitution. J. Mol. Evol. 36, 182–198 10.1007/BF00166252 (doi:10.1007/BF00166252) [DOI] [PubMed] [Google Scholar]
- 36.Hickerson M. J., Stahl E., Takebayashi N. 2007. MsBayes: pipeline for testing comparative phylogeographic histories using hierarchical approximate Bayesian computation. BMC Bioinform. 8, (doi:10.1186/1471-2105-8-268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickerson M. J., Stahl E. A., Lessios H. A. 2006. Test for simultaneous divergence using approximate Bayesian computation. Evolution 60, 2435–2453 [PubMed] [Google Scholar]
- 38.Hoberg E. P., Pilitt P. A., Galbreath K. E. 2009. Why museums matter: a tale of pinworms (Oxyuroidea: Heteroxynematidae) among pikas (Ochotona princeps and O. collaris) in the American West. J. Parasitol. 95, 490–501 10.1645/GE-1823.1 (doi:10.1645/GE-1823.1) [DOI] [PubMed] [Google Scholar]
- 39.MacDonald S. O., Jones C. 1987. Ochotona collaris Mammal. Spec. 281, 1–4 10.2307/3503971 (doi:10.2307/3503971) [DOI] [Google Scholar]
- 40.Hoberg E. P. 2005. Coevolution and biogeography among Nematodirinae (Nematoda: Trichostrongylina) Lagomorpha and Artiodactyla (Mammalia): exploring determinants of history and structure for the northern fauna across the Holarctic. J. Parasitol. 91, 358–369 10.1645/GE-3466 (doi:10.1645/GE-3466) [DOI] [PubMed] [Google Scholar]
- 41.Paterson A. M., Gray R. D. 1997. Host–parasite cospeciation, host-switching and missing the boat. In Host–parasite evolution: general principles and avian models (eds Clayton D. H., Moore J.), pp. 236–250 Oxford, UK: Oxford University Press [Google Scholar]
- 42.Hewitt G. M. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276 [Google Scholar]
- 43.Shotwell J. A. 1956. Hemphillian mammalian assemblage from northeastern Oregon. Geol. Soc. Am. Bull 67, 717–738 10.1130/0016-7606(1956)67[717:HMAFNO]2.0.CO;2 (doi:10.1130/0016-7606(1956)67[717:HMAFNO]2.0.CO;2) [DOI] [Google Scholar]
- 44.Smith A. T. 2008. The world of pikas. In Lagomorph biology: evolution, ecology, and conservation (eds Alves P. C., Ferrand N., Hackländer K.), pp. 89–102 Berlin, Germany: Springer [Google Scholar]
- 45.Coope G. 2004. Several million years of stability among insect species because of, or in spite of, Ice Age climatic instability? Phil. Trans. R. Soc. Lond. B 359, 209–214 10.1098/rstb.2003.1393 (doi:10.1098/rstb.2003.1393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiteman N. K., Parker P. G. 2005. Using parasites to infer host population history: a new rationale for parasite conservation. Anim. Conserv. 8, 175–181 10.1017/S1367943005001915 (doi:10.1017/S1367943005001915) [DOI] [Google Scholar]
- 47.Nieberding C., Morand S., Libois R., Michaux J. R. 2004. A parasite reveals cryptic phylogeographic history of its host. Proc. R. Soc. Lond. B 271, 2559–2568 10.1098/rspb.2004.2930 (doi:10.1098/rspb.2004.2930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoberg E. P., Brooks D. R. 2008. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems. J. Biogeogr. 35, 1533–1550 10.1111/j.1365-2699.2008.01951.x (doi:10.1111/j.1365-2699.2008.01951.x) [DOI] [Google Scholar]
- 49.Hoberg E. P., Brooks D. R. 2010. Beyond vicariance: integrating taxon pulses, ecological fitting and oscillation in historical biogeography and evolution. In The geography of host–parasite interactions (eds Morand S., Krasnov B.), pp. 7–20 Oxford, UK: Oxford University Press [Google Scholar]
- 50.van Veller M. G. P., Kornet D. J., Zandee M. 2002. A posteriori and a priori methodologies for testing hypotheses of causal processes in vicariance biogeography. Cladistics 18, pp. 207–217 [DOI] [PubMed] [Google Scholar]