Abstract
The peak sensitivities (λmax) of the short-wavelength-sensitive-1 (SWS1) pigments in mammals range from the ultraviolet (UV) (360–400 nm) to the violet (400–450 nm) regions of the spectrum. In most cases, a UV or violet peak is determined by the residue present at site 86, with Phe conferring UV sensitivity (UVS) and either Ser, Tyr or Val causing a shift to violet wavelengths. In primates, however, the tuning mechanism of violet-sensitive (VS) pigments would appear to differ. In this study, we examine the tuning mechanisms of prosimian SWS1 pigments. One species, the aye-aye, possesses a pigment with Phe86 but in vitro spectral analysis reveals a VS rather than a UVS pigment. Other residues (Cys, Ser and Val) at site 86 in prosimians also gave VS pigments. Substitution at site 86 is not, therefore, the primary mechanism for the tuning of VS pigments in primates, and phylogenetic analysis indicates that substitutions at site 86 have occurred at least five times in primate evolution. The sole potential tuning site that is conserved in all primate VS pigments is Pro93, which when substituted by Thr (as found in mammalian UVS pigments) in the aye-aye pigment shifted the peak absorbance into the UV region with a λmax value at 371 nm. We, therefore, conclude that the tuning of VS pigments in primates depends on Pro93, not Tyr86 as in other mammals. However, it remains uncertain whether the initial event that gave rise to the VS pigment in the ancestral primate was achieved by a Thr93Pro or a Phe86Tyr substitution.
Keywords: visual pigments, primates, colour vision, evolution
1. Introduction
Vertebrate cone visual pigments are classified into four groups according to their evolutionary lineages and spectral properties: short-wavelength-sensitive-1 (SWS1), short-wavelength-sensitive-2 (SWS2), middle-wavelength-sensitive (RH2) and long-wavelength-sensitive (LWS). In mammals, this complement is reduced to just two classes, LWS and SWS, with the latter either from the SWS1 class in marsupial and eutherian mammals, or from the SWS2 class in monotremes [1]. Most mammals are, therefore, dichromats, although monochromacy through the loss of the SWS1 pigment is found in aquatic mammals [2] and in nocturnal species [3–6].
SWS1 pigments are classified as either violet-sensitive (VS) or UV-sensitive (UVS) according to their peak sensitivities (λmax). The ancestral state is UVS and has been retained by many mammalian species that include some rodents [7–10], bats [11,12] and many marsupials [13–15]. The spectral shift to VS is achieved largely by a single amino acid substitution at site 86, with Phe86 in UVS pigments replaced by either Tyr as found in porcine, bovine and marsupial VS pigments [16–18], Val as found in the guinea pig VS pigment [19], or Ser as found in the elephant VS pigment [20].
The λmax values for SWS1 pigments in different primate species are thought to be relatively invariant with peaks in the violet (420–440 nm) region of the spectrum [21]. Microspectrophotometry [22] and in vitro pigment regeneration [23] of the human SWS1 pigment gave peaks at 419 and 414 nm, respectively. Other primates where spectral data are available include eight species of Old World monkeys [21], three species of New World monkeys [24,25] and three species of prosimians [26–28], with their SWS cones exhibiting spectral peaks ranging from 426 to 437 nm. The spectral tuning of these pigments, however, appears to be more complex than in other mammals, as site-directed mutagenesis of the human VS pigment has indicated that substitutions at seven sites that include 86 are required to shift the peak from violet back to UV [29].
Primates are sub-divided into two Suborders, the Haplorrhini comprising the tarsiers, Old and New World monkeys and great apes, and Streptirrhini that include the Lorisiformes, Chiromyiformes and Lemuriformes. The paraphyletic group of prosimian primates combine the latter groups with the Tarsiiformes. Within the Streptirrhini, a functional SWS1 gene is absent from the Lorisiformes and in some species of the Cheirogaleidae family that form part of the Lemuriformes group [6,30]. A disrupted pseudogene is present in these latter species, but an intact SWS1 gene is retained in two close relatives, the grey and Coquerel's mouse lemurs. Members of the Tarsiiformes and the remaining species of Lemuriformes (Lemuridae, Lepilemuridae and Indriidae families) as well as the aye-aye, the sole extant member of the Daubentoniidae family of the Chiromyiformes, all appear to have an intact SWS1 gene [30], although functional information is limited. The residue present at site 86, however, varies substantially across prosimians [30]; Leu86 is found throughout the Haplorrhini but in the Strepsirrhini, either Cys, Ser, Val, Phe, Tyr or Asn is present, although Tyr and Asn are found only in SWS1 pseudogenes of the lesser, small-eared and greater galagos, and the slow loris.
The objective of this study was to determine the underlying tuning mechanisms in the evolution of primate SWS1 pigments, by focusing on three prosimian species, each with a different residue at site 86.
2. Material and methods
(a). DNA samples
Genomic DNA (gDNA) was extracted from brown lemur (Eulemur fulvus) liver tissue that was kindly donated by Prof. John Mollon (University of Cambridge, UK). Blood samples from Coquerel's mouse lemur (Mirza coquereli) and the aye-aye (Daubentonia madagascariensis) were purchased from Duke University Primate Centre (USA) and gDNA was extracted using the Nucleon BACC Genomic DNA Extraction Kit (GE Healthcare, UK).
(b). Generation of expression vectors
The human SWS1 construct for in vitro expression was amplified from retinal cDNA using primers complementary to the 5′- and 3′-ends of the coding sequence. The coding sequences for the SWS1 opsin genes of all three prosimian species were generated using the SPLICE technique for joining individual exons amplified from gDNA into a single transcription unit [31]. PCRs were performed using KOD Hot Start polymerase (Novagen, UK) and the primers listed in electronic supplementary material, table S1. The full-length coding sequences were cloned into the pMT4 mammalian expression vector as previously described [16]. Point mutations were introduced using the Quikchange site-directed mutagenesis kit (Stratagene, UK) and the oligonucleotides listed in electronic supplementary material, table S2.
(c). In vitro determination of the λmax of expressed pigments
Opsin expression and pigment production followed the methodology described in Cowing et al. [16]. Twelve 140 cm2 tissue culture plates were used for the brown lemur, Coquerel's mouse lemur and human SWS1 constructs, and 24 140 cm2 tissue culture plates for the aye-aye SWS1 constructs. The absorbance spectra of the purified pigments (dark spectra) were measured at pH 7.2 by UV/visible spectrophotometry using a Spectronic Unicam UV500 dual-beam spectrophotometer, treated with either 1 M HCl to a pH < 2.0 or with hydroxylamine (56 mM) and measured again (treated spectra). For each pigment, the treated spectrum was subtracted from the dark spectrum to generate a difference spectrum. This eliminates protein absorbance scatter that may distort the dark spectrum. The λmax was then determined by fitting a standard visual pigment template [32] to the long-wavelength tail of the difference spectrum using a Solver add-in to Microsoft Excel.
3. Results and discussion
(a). Absorbance peaks of in vitro expressed pigments
Inspection of the SWS1 opsin sequences of 26 primate species listed in GenBank shows that with one exception, Phe86 found in UVS pigments has been replaced by another residue as follows: Leu86 is consistently found in the Haplorrhini group, whereas the Strepsirrhini show much more variation with seven different amino acids at site 86 across the seven families (table 1). The exception is the retention of Phe86 in another member of the Strepsirrhini, the aye-aye [30], suggesting that this pigment may be UVS.
Table 1.
Residues present at seven sites in transmembrane helices II and III of the VS pigments of primates compared with the UVS pigments of two marsupials, the fat-tailed dunnart (GenBank accession number AY7724723) and honey possum (GenBank accession number AY772472), and two rodents, the mouse (GenBank accession number NM_007538) and rat (GenBank accession number NM_031015). The seven sites were identified by Shi et al. [29] from site-directed mutagenesis experiments. Species in italics possess a SWS1 pseudogene. Alignments of the amino acid sequences that form transmembrane helices 2–3 of the different opsins referred to in this table are presented in electronic supplementary material, figure S1, together with the pigments of the other species referred to in the text.
| VS pigments |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| primates |
|||||||||||||||||||||
| UVS pigments |
Simian |
Prosimian |
|||||||||||||||||||
| amino acid sites | dunnart/honey possum | mouse/rat | great apes | Old World monkey | New World monkey | tarsier | aye-aye | brown lemur | ring-tailed lemur | sportive lemur | Coquerel's sifaka | woolly lemur | Coquerel's mouse lemur | grey mouse lemur | fat-tailed dwarf lemur | greater dwarf lemur | lesser galago | small-eared galago | greater galago | pygmy slow loris | slow loris |
| 46 | F | F | T | T | I | V | F | L | L | F | F | F | F | F | F | F | F | F | F | F | F |
| 49 | F | F | L | L | L | S | F | F | F | F | F | F | F | F | F | F | F | F | F | F | F |
| 52 | T | T | F | F | L | I | T | A | A | T | T | T | T | T | T | T | T | T | T | T | T |
| 86 | F | F | L | L | L | L | F | C | C | S | L | V | S | S | S | S | Y | N | N | C | Y |
| 93 | T | T | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 114 | G/Aa | A | G | G/Aa | G/Aa | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G | G |
| 118 | S | S | T | T | T | S | S | S | S | S | S | S | C | C | C | C | S | S | S | S | S |
| Haplorrhini | Strepsirrhini | ||||||||||||||||||||
aAla is present in the honey possum, in three of four species of Old World and one of four species of New World monkeys.
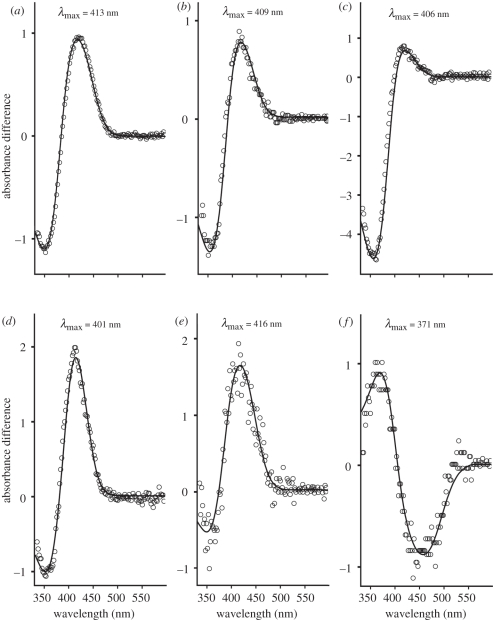
The prosimian SWS1 pigments selected for spectral analysis were derived from the brown lemur (GenBank AB111464), the aye-aye (GenBank DQ1918903-7) and Coquerel's mouse lemur (GenBank DQ191898-902), as each SWS1 pigment possesses a different residue at site 86. The brown lemur pigment (with Cys86) gave a λmax at 413 nm (figure 1a), which is substantially shorter than the spectral peak values obtained by electroretinogram (ERG) flicker photometry for both the brown and ring-tailed lemurs at 437 nm [27]. However, such differences in peak absorbance values are not unexpected since ERG recordings arise from light filtered by the cornea and lens falling on the retina and frequently overestimate the spectral peak for SWS cones. The SWS1 pigment from Coquerel's mouse lemur (with Ser86) gave a λmax of 409 nm (figure 1b). The only other reported pigment with Ser86 is in the elephant with a peak around 422 nm [20], suggesting that other sites may be important for the 13 nm difference between these two mammalian species. Finally, the aye-aye SWS1 pigment (with Phe86) gave a peak at 406 nm within the violet region of the spectrum (figure 1c), which contrasts with all other vertebrate SWS1 pigments with Phe86 that are consistently UVS with λmax values close to 360 nm [33,34].
Figure 1.
Difference spectra for (a) brown lemur wild-type, (b) Coquerel's mouse lemur wild-type, (c) aye-aye wild-type, (d) brown lemur Cys86Val mutant, (e) aye-aye Phe86Ser mutant and (f) aye-aye Pro93Thr mutant pigments. Values shown are the λmax of each pigment obtained by fitting a visual pigment template curve (open circles) [32].
The other residues encoded at site 86 in prosimians with a functional SWS1 gene are Val86 (e.g. as found in the woolly lemur) and Leu86 (e.g. as found in tarsiers and Coquerel's sifaka). Since it has already been established in Old and New World primates that SWS1 pigments with Leu86 peak in the violet, the spectral tuning effect of this residue was not examined further. To determine the effect of Val86, the brown lemur SWS1 coding sequence was mutated to encode a Cys86Val substitution. The resulting mutant pigment gave a λmax at 401 nm (figure 1d). This is 12 nm shorter than the absorbance peak value for the brown lemur wild-type pigment but still spectrally tuned to violet wavelengths. The only other mammalian SWS1 pigment with Val86 is in the guinea pig [19] but this has a λmax some 19 nm more long wavelength-shifted; differences at other tuning sites identified from previous studies [29] may be responsible for this shift. Finally, the effect of Phe86 on the λmax of the aye-aye SWS1 pigment was assessed by substitution with Ser. As shown in figure 1e, the resulting pigment was long wavelength-shifted by 10 nm; Phe86 is able, therefore, to shift the short-wavelength λmax of the aye-aye pigment but not into the UV.
(b). Molecular basis for primate violet sensitivity
Previous studies have indicated that the simultaneous replacement of the residues present at seven sites (46, 49, 52, 86, 93, 114 and 118) in the human VS pigment with those found in the mouse UVS pigment is required to shift the λmax of the human pigment into the UV [29]. However, an examination of these sites in the VS pigments of different primate species shows that only Pro93 is fully conserved across all primate groups (table 1). The other sites show variation in the amino acid present, and there are several instances where the residue present in the UVS pigments of certain rodents [35] and marsupials [14] is also found in primate VS pigments.
If, as seems likely, a single tuning mechanism is responsible for the VS of all primate SWS1 pigments, then the only consistent feature to account for this is Pro93. Substitution at this site may be responsible, therefore, for the long-wavelength shift, regardless of the residue present at site 86. This hypothesis is supported by the observation that in other mammalian pigments, Thr93 is consistently combined with Phe86 in UVS pigments, but it is always replaced in mammalian VS visual pigments by either Val93 as found in the tree shrew, Tupaia belangeri (GenBank accession number: EU487780), and grey squirrel [3], Ile in the cow [16] and elephant [20], Ala in the guinea pig [19] or Ser in the pig [16]. The retention of Thr93 may therefore preclude the generation of a VS pigment.
To investigate this hypothesis, a Pro93Thr mutant was generated in the human pigment. However, although the wild-type pigment gave an in vitro spectral peak at 414 nm as previously reported [29], the Pro93Thr mutant failed to generate a pigment in several repeat experiments (data not shown). As the human SWS1 VS pigment contains Leu86 instead of Phe86, the failure of this mutant pigment to regenerate in vitro is consistent with our hypothesis that UVS pigments require both Phe86 and Thr93. Therefore, we generated a Pro93Thr substitution in the aye-aye SWS1 pigment that naturally exhibits five out of the seven substitutions (Phe46, Phe49, Thr52, Phe86 and Ser118) used previously [29] to shift the human VS pigment into the UV (table 1). Furthermore, the wild-type aye-aye pigment and the UVS pigment of the dunnart possesses common residues at six out of the seven sites (table 1); the aye-aye Pro93Thr mutant will be identical, therefore, over these seven tuning sites to that of the dunnart. As shown in figure 1f, the absorbance of the aye-aye Pro93Thr mutant pigment was significantly shifted by 35 nm from the wild-type VS pigment (λmax value of 406 nm) into the UV region with a peak at 371 nm. Thus, we conclude that a Pro93 is responsible for the VS of primate SWS1 pigments.
(c). Evolution of violet-sensitive pigments
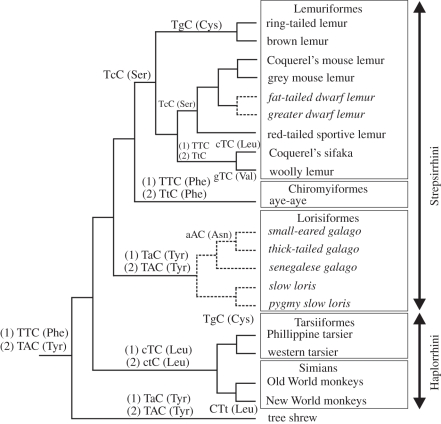
From the above discussion, it would appear that the residue present at site 86 is not responsible for the VS of primate SWS1 pigments. Rather, substitutions at this site may serve to ‘fine-tune’ the pigments in the violet region of the spectrum. However, it does not necessarily follow that substitution at site 86 was not important in the evolution of VS pigments in primates; an alternative explanation is that a Phe86Tyr substitution occurred at the base of the primate lineage but its role in generating a violet-shift was subsequently replaced by Pro93, thereby releasing codon 86 from selective constraint and allowing further changes to occur. To examine this further, the eight different residues found at site 86 in primate SWS1 opsins, together with the corresponding codon sequences, were mapped onto the primate phylogeny reported recently by Perelman et al. [36], together with the tree shrew as the species most closely related to primates for which spectral [37] and sequence data (GenBank accession number EU487780) are available (figure 2). The ancestral Phe residue, as found in non-primate mammalian UVS pigments [14,15,35,38], is encoded by TTC, which is also found in the aye-aye. This codon differs, however, in the SWS1 gene of all other primates. If Phe86 was retained in the ancestral primate and passed unchanged onto the aye-aye, then each residue change at site 86 in the other branches of the tree can be explained by single-step nucleotide changes. The alternative explanation, that a Phe86Tyr substitution occurred at the base of the primate lineage, requires two nucleotide changes in the Tarsiiformes/Simian branch to achieve the Tyr86Leu (TAC to CTC) substitution, and an additional change (TAC to TTC) in the branch to Coquerel's sifaka and the woolly lemur, which makes this the less parsimonious interpretation of the sequence data. It would also mean that Phe86 in the aye-aye is a back mutation. Within the Lemuriformes, Phe (TTC) is replaced by Ser (TCC) in the Coquerel's mouse lemur, the gray mouse lemur, and the fat-tailed and greater dwarf lemurs, Cys (TGC) in the brown and ring-tailed lemurs, Leu (CTC) in the Coquerel's sifaka and Val (GTC) in the woolly lemur. In the Lorisiformes with non-functional SWS1 genes, Phe is replaced initially by Tyr (TAC), followed by Asn (AAC) in the small-eared and thick-tailed galagos. Finally, there is a synonymous transition (CTC to CTT) in New World monkeys. Overall, therefore, if it is assumed that the substitution of Leu and Val in the Lemuriformes were separate events, then there have been either ten or nine amino acid substitutions in the primate lineages depending, respectively, on the presence of either Phe86 or Tyr86 in the ancestral primate. The Phe86Tyr substitution in the tree shrew may also be ancestral to the primate lineage and, as found in many other non-primate species [3,16–20], was almost certainly responsible for the generation of a VS pigment [37,39]. These alternative interpretations are shown in figure 2. Depending, therefore, on the sequence of substitutions at sites 86 and 93, either a Phe86Tyr substitution or a Thr93Pro substitution was the primary change in SWS1 pigments responsible for the evolution of VS in primates.
Figure 2.
Phylogenetic relationships of different primate species showing the different residues found at site 86 in SWS1 opsins, together with the corresponding codon sequences. The effects on nucleotide substitution of the alternative hypotheses of ancestral (1) Phe86 or (2) Tyr86 are shown. Lower case is used to indicate substitutions in codon 86. Solid lines are lineages with functional SWS1 pigments, dashed lines and italics indicate species where SWS1 pseudogenes are present. The tree was modified from Perelman et al. [36]. Branch lengths do not reflect evolutionary distances.
Acknowledgements
This work was supported by grants from the Leverhulme Trust and the BBSRC awarded to D.M.H. and by a National Science Foundation (USA) grant to P.R.R. We are grateful to Dr Rosalie Crouch, Storm Eye Institute, Medical University of South Carolina, USA, for 11-cis-retinal.
References
- 1.Davies W. L., Carvalho L. S., Cowing J. A., Beazley L. D., Hunt D. M., Arrese C. A. 2007. Visual pigments of the platypus: a novel route to mammalian colour vision. Curr. Biol. 17, R161–R163 10.1016/j.cub.2007.01.037 (doi:10.1016/j.cub.2007.01.037) [DOI] [PubMed] [Google Scholar]
- 2.Newman L. A., Robinson P. R. 2005. Cone visual pigments of aquatic mammals. Vis. Neurosci. 22, 873–879 10.1017/S0952523805226159 (doi:10.1017/S0952523805226159) [DOI] [PubMed] [Google Scholar]
- 3.Carvalho L. d. S., Cowing J. A., Wilkie S. E., Bowmaker J. K., Hunt D. M. 2006. Shortwave visual sensitivity in tree and flying squirrels reflects changes in lifestyle. Curr. Biol. 16, R81–R83 10.1016/j.cub.2006.01.045 (doi:10.1016/j.cub.2006.01.045) [DOI] [PubMed] [Google Scholar]
- 4.Jacobs G. H., Deegan J. F. d., Neitz J., Crognale M. A., Neitz M. 1993. Photopigments and color vision in the nocturnal monkey, Aotus. Visi. Res. 33, 1773–1783 10.1016/0042-6989(93)90168-V (doi:10.1016/0042-6989(93)90168-V) [DOI] [PubMed] [Google Scholar]
- 5.Jacobs G. H., Neitz M., Neitz J. 1996. Mutations in S-cone pigment genes and the absence of colour vision in two species of nocturnal primate. Proc. R. Soc. Lond. B 263, 705–710 10.1098/rspb.1996.0105 (doi:10.1098/rspb.1996.0105) [DOI] [PubMed] [Google Scholar]
- 6.Kawamura S., Kubotera N. 2004. Ancestral loss of short wave-sensitive cone visual pigment in lorisiform prosimians, contrasting with its strict conservation in other prosimians. J. Mol. Evol. 58, 314–321 10.1007/s00239-003-2553-z (doi:10.1007/s00239-003-2553-z) [DOI] [PubMed] [Google Scholar]
- 7.Calderone J. B., Jacobs G. H. 1999. Cone receptor variations and their functional consequences in two species of hamster. Vis. Neurosci. 16, 53–63 10.1017/S0952523899161029 (doi:10.1017/S0952523899161029) [DOI] [PubMed] [Google Scholar]
- 8.Chavez A. E., Bozinovic F., Peichl L., Palacios A. G. 2003. Retinal spectral sensitivity, fur coloration, and urine reflectance in the genus octodon (rodentia): implications for visual ecology. Invest. Ophthalmol. Vis. Sci. 44, 2290–2296 10.1167/iovs.02-0670 (doi:10.1167/iovs.02-0670) [DOI] [PubMed] [Google Scholar]
- 9.Jacobs G. H., Deegan J. F., II 1994. Sensitivity to ultraviolet light in the gerbil (Meriones unguiculatus): characteristics and mechanisms. Vis. Res. 34, 1433–1441 10.1016/0042-6989(94)90144-9 (doi:10.1016/0042-6989(94)90144-9) [DOI] [PubMed] [Google Scholar]
- 10.Williams G. A., Calderone J. B., Jacobs G. H. 2005. Photoreceptors and photopigments in a subterranean rodent, the pocket gopher (Thomomys bottae). J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 191, 125–134 10.1007/s00359-004-0578-4 (doi:10.1007/s00359-004-0578-4) [DOI] [PubMed] [Google Scholar]
- 11.Wang D., Oakley T., Mower J., Shimmin L. C., Yim S., Honeycutt R. L., Tsao H., Wen-Hsiung Li L. 2004. Molecular Evolution of Bat Color Vision Genes. Mol. Biol. Evol. 21, 295–302 10.1093/molbev/msh015 (doi:10.1093/molbev/msh015) [DOI] [PubMed] [Google Scholar]
- 12.Winter Y., Lopez J., Von Helversen O. 2003. Ultraviolet vision in a bat. Nature 425, 612–614 10.1038/nature01971 (doi:10.1038/nature01971) [DOI] [PubMed] [Google Scholar]
- 13.Arrese C. A., Hart N. S., Thomas N., Beazley L. D., Shand J. 2002. Trichromacy in Australian marsupials. Curr. Biol. 12, 657–660 10.1016/S0960-9822(02)00772-8 (doi:10.1016/S0960-9822(02)00772-8) [DOI] [PubMed] [Google Scholar]
- 14.Cowing J. A., Arrese C. A., Davies W. L., Beazley L. D., Hunt D. M. 2008. Cone visual pigments in two marsupial species: the fat-tailed dunnart (Sminthopsis crassicaudata) and the honey possum (Tarsipes rostratus). Proc. R. Soc. B 275, 1491–1499 10.1098/rspb.2008.0248 (doi:10.1098/rspb.2008.0248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt D. M., Chan J., Carvalho L. S., Hokoc J. N., Ferguson M. C., Arrese C. A., Beazley L. D. 2009. Cone visual pigments in two species of South American marsupials. Gene 433, 50–55 10.1016/j.gene.2008.12.006 (doi:10.1016/j.gene.2008.12.006) [DOI] [PubMed] [Google Scholar]
- 16.Cowing J. A., Poopalasundaram S., Wilkie S. E., Robinson P. R., Bowmaker J. K., Hunt D. M. 2002. The molecular mechanism for the spectral shifts between vertebrate ultraviolet- and violet-sensitive cone visual pigments. Biochem. J. 367, 129–135 10.1042/BJ20020483 (doi:10.1042/BJ20020483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deeb S. S., Wakefield M. J., Tada T., Marotte L., Yokoyama S., Marshall Graves J. A. 2003. The cone visual pigments of an Australian marsupial, the tammar wallaby (Macropus eugenii): sequence, spectral tuning, and evolution. Mol. Biol. Evol. 20, 1642–1649 10.1093/molbev/msg181 (doi:10.1093/molbev/msg181) [DOI] [PubMed] [Google Scholar]
- 18.Fasick J. I., Applebury M. L., Oprian D. D. 2002. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry 41, 6860–6865 10.1021/bi0200413 (doi:10.1021/bi0200413) [DOI] [PubMed] [Google Scholar]
- 19.Parry J. W., Poopalasundaram S., Bowmaker J. K., Hunt D. M. 2004. A novel amino acid substitution is responsible for spectral tuning in a rodent violet-sensitive visual pigment. Biochemistry 43, 8014–8020 10.1021/bi049478w (doi:10.1021/bi049478w) [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama S., Takenaka N., Agnew D. W., Shoshani J. 2005. Elephants and human color-blind deuteranopes have identical sets of visual pigments. Genetics 170, 335–344 10.1534/genetics.104.039511 (doi:10.1534/genetics.104.039511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowmaker J. K., Astell S., Hunt D. M., Mollon J. D. 1991. Photosensitive and photostable pigments in the retinae of Old World monkeys. J. Exp. Biol. 156, 1–19 [DOI] [PubMed] [Google Scholar]
- 22.Dartnall H. J., Bowmaker J. K., Mollon J. D. 1983. Human visual pigments: microspectrophotometric results from the eyes of seven persons. Proc. R. Soc. Lond. B 220, 115–130 10.1098/rspb.1983.0091 (doi:10.1098/rspb.1983.0091) [DOI] [PubMed] [Google Scholar]
- 23.Fasick J. I., Lee N., Oprian D. D. 1999. Spectral tuning in the human blue cone pigment. Biochemistry 38, 11 593–11 596 10.1021/bi991600h (doi:10.1021/bi991600h) [DOI] [PubMed] [Google Scholar]
- 24.Jacobs G. H., Deegan J. F., 2nd 2003. Cone pigment variations in four genera of new world monkeys. Vis. Res. 43, 227–236 10.1016/S0042-6989(02)00565-5 (doi:10.1016/S0042-6989(02)00565-5) [DOI] [PubMed] [Google Scholar]
- 25.Mollon J. D., Bowmaker J. K., Jacobs G. H. 1984. Variations of colour vision in a New World primate can be explained by polymorphism of retinal photopigments. Proc. R. Soc. Lond. B 222, 373–399 10.1098/rspb.1984.0071 (doi:10.1098/rspb.1984.0071) [DOI] [PubMed] [Google Scholar]
- 26.Jacobs G. H. 1996. Primate photopigments and primate color vision. Proc. Natl Acad. Sci. USA 93, 577–581 10.1073/pnas.93.2.577 (doi:10.1073/pnas.93.2.577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs G. H., Deegan J. F. 1993. Photopigments underlying color vision in ringtail lemurs (Lemur catta) and brown lemurs (Eulemur fulvus). Am. J. Primatol. 30, 243–256 10.1002/ajp.1350300307 (doi:10.1002/ajp.1350300307) [DOI] [PubMed] [Google Scholar]
- 28.Jacobs G. H., Deegan J. F., 2nd, Tan Y., Li W. H. 2002. Opsin gene and photopigment polymorphism in a prosimian primate. Vis. Res. 42, 11–18 10.1016/S0042-6989(01)00264-4 (doi:10.1016/S0042-6989(01)00264-4) [DOI] [PubMed] [Google Scholar]
- 29.Shi Y., Radlwimmer F. B., Yokoyama S. 2001. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc. Natl Acad. Sci. USA 98, 11 731–11 736 10.1073/pnas.201257398 (doi:10.1073/pnas.201257398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y., Yoder A. D., Yamashita N., Li W. H. 2005. Evidence from opsin genes rejects nocturnality in ancestral primates. Proc. Natl Acad. Sci. USA 102, 14 712–14 716 10.1073/pnas.0507042102 (doi:10.1073/pnas.0507042102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies W. L., Carvalho L. S., Hunt D. M. 2007. SPLICE: a technique for generating in vitro spliced coding sequences from genomic DNA. Biotechniques 43, 785–789 10.2144/000112588 (doi:10.2144/000112588) [DOI] [PubMed] [Google Scholar]
- 32.Govardovskii V. I., Fyhrquist N., Reuter T., Kuzmin D. G., Donner K. 2000. In search of the visual pigment template. Vis. Neurosci. 17, 509–528 10.1017/S0952523800174036 (doi:10.1017/S0952523800174036) [DOI] [PubMed] [Google Scholar]
- 33.Hunt D. M., Cowing J. A., Wilkie S. E., Parry J., Poopalasundaram S., Bowmaker J. K. 2004. Divergent mechanisms for the tuning of shortwave sensitive visual pigments in vertebrates. Photochem. Photobiol. Sci. 3, 713–720 10.1039/b314693f (doi:10.1039/b314693f) [DOI] [PubMed] [Google Scholar]
- 34.Yokoyama S., Radlwimmer F. B., Kawamura S. 1998. Regeneration of ultraviolet pigments of vertebrates. FEBS Lett. 423, 155–158 10.1016/S0014-5793(98)00086-6 (doi:10.1016/S0014-5793(98)00086-6) [DOI] [PubMed] [Google Scholar]
- 35.Chiu M. I., Zack D. J., Wang Y., Nathans J. 1994. Murine and bovine blue cone pigment genes: cloning and characterization of two new members of the S family of visual pigments. Genomics 21, 440–443 10.1006/geno.1994.1292 (doi:10.1006/geno.1994.1292) [DOI] [PubMed] [Google Scholar]
- 36.Perelman P., et al. 2011. A molecular phylogeny of living primates. PLoS Genet. 7, e1001342. 10.1371/journal.pgen.1001342 (doi:10.1371/journal.pgen.1001342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petry H. M., Harosi F. I. 1990. Visual pigments of the tree shrew (Tupaia belangeri) and greater galago (Galago crassicaudatus): a microspectrophotometric investigation. Vis. Res. 30, 839–851 10.1016/0042-6989(90)90053-N (doi:10.1016/0042-6989(90)90053-N) [DOI] [PubMed] [Google Scholar]
- 38.Strachan J., Chang L. Y., Wakefield M. J., Graves J. A., Deeb S. S. 2004. Cone visual pigments of the Australian marsupials, the stripe-faced and fat-tailed dunnarts: sequence and inferred spectral properties. Vis. Neurosci. 21, 223–229 10.1017/S0952523804213281 (doi:10.1017/S0952523804213281) [DOI] [PubMed] [Google Scholar]
- 39.Jacobs G. H., Neitz J. 1986. Spectral mechanisms and color vision in the tree shrew (Tupaia belangeri). Vis. Res. 26, 291–298 10.1016/0042-6989(86)90026-X (doi:10.1016/0042-6989(86)90026-X) [DOI] [PubMed] [Google Scholar]