Abstract
Major histocompatibility complex (MHC)-dependent mating preferences have been observed across vertebrate taxa and these preferences are expected to promote offspring disease resistance and ultimately, viability. However, little empirical evidence linking MHC-dependent mate choice and fitness is available, particularly in wild populations. Here, we explore the adaptive potential of previously observed patterns of MHC-dependent mate choice in a wild population of Atlantic salmon (Salmo salar) in Québec, Canada, by examining the relationship between MHC genetic variation and adult reproductive success and offspring survival over 3 years of study. While Atlantic salmon choose their mates in order to increase MHC diversity in offspring, adult reproductive success was in fact maximized between pairs exhibiting an intermediate level of MHC dissimilarity. Moreover, patterns of offspring survival between years 0+ and 1+, and 1+ and 2+ and population genetic structure at the MHC locus relative to microsatellite loci indicate that strong temporal variation in selection is likely to be operating on the MHC. We interpret MHC-dependent mate choice for diversity as a likely bet-hedging strategy that maximizes parental fitness in the face of temporally variable and unpredictable natural selection pressures.
Keywords: major histocompatibility complex, Salmo salar, genetic quality, mate choice, fitness
1. Introduction
Sexual selection, including intrasexual competition and mate choice, is an important mechanism influencing individual lifetime reproductive success. Because of its potentially important influence on fitness, sexual selection is generally considered an adaptive evolutionary mechanism [1], yet few studies have examined the role of sexual selection as a driver of population viability or adaptation to environmental variation [2]. In particular, the indirect (genetic) benefits obtained via sexual selection may have important fitness effects if the genetic ‘quality’ inherited by offspring confers an advantage under a natural selection scenario [3,4]. The vertebrate major histocompatibility complex (MHC) has been implicated in the evolution of mating preferences owing to the potential genetic benefits that may be conferred to offspring through MHC-dependent mate choice [3,5,6]. MHC molecules are involved in the presentation and recognition of self and non-self peptides to T-cells, and thus play an integral role in the initiation of an adaptive immune response to pathogens [7]. Because of the MHC's integral function in the immune system, the genetic regions that encode for these molecules are thought to be targets of strong parasite-driven natural selection pressures. Indeed, the genetic regions that code for MHC molecules are among the most polymorphic ever described. Genetic diversity at the MHC is thought to be selectively favoured as individuals bearing a diverse MHC repertoire should be able to recognize and mount an immune response against a broad array of pathogens [8–10].
MHC-dependent mate choice has been observed across a variety of vertebrate taxa (e.g. [4,11–13]). Mate choice for enhanced diversity at the MHC may evolve if more diverse offspring exhibit enhanced immunocompetence, particularly when faced with multiple pathogens ([6], e.g. [14,15]). A growing number of studies have also shown evidence of mate choice for intermediate MHC genetic variation potentially owing to the immune system-related trade-offs associated with being too diverse (e.g. [16–19]; see [20] for discussion). Selection may also favour intermediate MHC diversity if highly diverse individuals face outbreeding depression or the disruption of locally adapted gene complexes [3,21,22]. MHC-dependent mating preferences may also be for specific alleles (e.g. [16,23]) and many studies have shown associations between specific MHC alleles and resistance against infections (e.g. [24–28]). Thus, ample evidence exists for MHC-dependent mating preferences and the potential for fitness benefits associated with MHC-dependent mate choice across a variety of organisms. Interestingly, these studies have also shown a large degree of variation in the specific targets of mate choice.
If MHC-dependent mating preferences have evolved in response to the pathogen community, variation in mating preferences observed across studies may be explained, in part, by spatial or temporal variation in pathogen-driven selection [29]. At present, only a few empirical studies conducted on model systems have been able to examine the potential role of temporally or spatially variable natural selection pressures on adaptive mate choice (see [30] for a review). Nevertheless, a growing number of population genetics studies point to the likely importance of temporal variation in selection on MHC evolution (e.g. [31–33]). Thus, mating preferences may also evolve as a type of risk-spreading (bet-hedging) strategy to ensure that at least some offspring are adapted to an uncertain future environment ([34,35]; e.g. [36]). Multiple mating may also be a strategy used to ensure that a diverse complement of genotypes is present within offspring [37]. However, despite the potentially critical role that variable natural selection pressures may have on the evolution of MHC-dependent mating patterns across populations, to our knowledge, no research has yet examined this topic in any organism.
Following survival in wild populations represents a major logistical challenge, and to our knowledge, only a few studies have been able to evaluate the influence of MHC genetic variation on offspring survival in the wild. A recent 10-year study of Seychelles warblers (Acrocephalus sechellensis) showed that juvenile survival was positively associated with MHC diversity and an individual MHC allele [38]. Similarly, in the Soay Sheep (Ovis aries), juvenile survival was associated with specific MHC alleles [39]. In other ‘semi-natural’ experiments, relationships between offspring survival and MHC genetic variation have been somewhat easier to document (e.g. [40–45]). In particular, salmonid fishes represent a particularly promising system in which to evaluate the influence of MHC genetic variation on offspring survival. These species are external fertilizers and thus, family groups of known MHC genotype can be generated in a controlled manner and offspring survival followed in both semi-natural and natural environments. However, long-term studies are still needed to examine the potential fitness benefits of MHC-dependent mate choice on offspring survival and the potential interplay between natural and sexual selection pressures.
In this study, we examined the influence of mate choice for MHC genetic variation on reproductive success and offspring survival across 3 years of study in the Atlantic salmon (Salmo salar) in Québec, Canada. Reproductively mature Atlantic salmon males and females from the Ste-Marguerite River were collected and relocated to an uninhabited stretch of the river, allowed to spawn naturally, and their offspring monitored for the subsequent 3 years. Because all founding parents of this population are known, this system represents an unprecedented opportunity to detail the potential roles of natural and sexual selection on MHC evolution. Previous research has evaluated the mating system of this population [46] and has shown a mating preference for MHC class II diversity [47]. However, the long-term fitness consequences of both multiple mating and MHC-based mate choice have not been examined. Moreover, a recent study showed the potential for temporally variable parasite-imposed natural selection pressures operating on the Atlantic salmon MHC class II in six populations, including the Ste-Marguerite River, in Québec [48]. In this context, our specific objectives were to: (i) examine the roles of MHC class II genetic variation and multiple mating on the reproductive success of adult Atlantic salmon, (ii) examine the potential relationship between MHC class II genetic variation and survival of offspring across years, and (iii) examine the potential for temporally variable natural selection operating on the MHC class II across the 3 years of our study.
2. Methods
(a). Sampling
In July 1995, 41 adult male and 35 adult female Atlantic salmon were collected from the northeast branch of the Ste-Marguerite River in Québec, Canada. The adult spawners were introduced into a previously unoccupied and self-contained (bounded by two impassable waterfalls) 19 km stretch of the river. Sampling methods and study design have been reported elsewhere in detail [46], but briefly, over the following 3 years (late August to early September 1996–1998), offspring belonging to the same cohort were systematically sampled across the entire river area. In total, 650, 352 and 177 offspring were sampled in 1996, 1997 and 1998, respectively. The offspring sampled in 1996 represent the same individuals as were examined in Landry et al. [47]. Adipose fins were collected from each parent and whole offspring were collected and stored in 95 per cent ethanol for genetic analysis. Offspring sampled in 1996, 1997 and 1998 are hereafter referred to as year 0+, year 1+ and year 2+ offspring, respectively.
(b). Genetic analysis
Parents and offspring were genotyped at five microsatellite loci (SSOSL85, Ssa171, Ssa197, Ssa202 and MST-3) and at the MHC class IIβ exon 2 locus using the denaturing gradient gel electrophoresis method. The methods for the microsatellite and MHC genotyping are reported in detail in Garant et al. [46] and Dionne et al. [49].
We assigned parentage to offspring using the microsatellite and MHC markers. Thus, this analysis incorporated an additional marker (MHC) compared to the parentage analysis conducted by Landry et al. [47]. Parentage was assigned using the program CERVUS v. 3.0 [50]. Offspring were identified to a family group when assigned to a parental-pair at 80 per cent or higher confidence.
(c). Statistical analysis
(i). Major histocompatibility complex and non-random mating
Previous studies on Atlantic salmon have shown that individuals select mates so as to increase the MHC diversity found in their offspring [47]. Thus, we calculated amino acid divergence (AAxy) between males and females in a parental-pair by summing the number of amino acid differences (D) between all four parental (two females and two males) MHC alleles (e.g. parental alleles = A, a, B, b; AAxy = DAB + DAb + DaB + Dab; [47]). Amino acid divergence was calculated across the entire 82-codon MHC class IIβ exon 2 sequence (AAxy) and for codons involved directly in pathogen recognition (AAxy-PBR) in MEGA 4 [51]. This peptide-binding region (PBR) was identified by aligning the MHC sequences with the human leucocyte antigen (human MHC), for which the codons involved in pathogen binding are known [52]. The results we obtained for both measures were similar; thus we only report the results based on the AAxy-PBR measurement.
To investigate whether mating between males and females was non-random with respect to MHC amino acid divergence (AAxy-PBR), we used a Monte Carlo simulation. This simulation represents a repetition of that conducted by Landry et al. [47], but uses the reproductive success estimates derived from the new parentage analysis. Each iteration of the Monte Carlo simulation randomly assigned reproductive success to a parental-pair, drawing from the observed reproductive success values across parental-pairs, and then calculated the mean expected AAxy-PBR in all offspring in the population. The simulation was repeated for 5000 iterations to establish the distribution of the mean expected AAxy-PBR values found in offspring under random mating. We then compared the distribution of the mean AAxy-PBR expected in offspring under random mating to that based on the observed mating patterns to determine whether the mating patterns were non-random with respect to MHC AAxy-PBR [53]. We also examined non-random mating with respect to the predicted variance in offspring MHC AAxy-PBR using a Monte Carlo simulation [21]. This type of simulation allows for the investigation of non-random mating for an ‘optimal’ level of MHC AAxy-PBR in offspring.
(ii). Offspring survival
To infer the relative survival of offspring for a given parental-pair across years, we initially calculated the proportion of offspring sampled in each year from each parental-pair as follows:
where Ni is the number of offspring genotyped to the parental-pair i and Nt is the total number of offspring genotyped.
We then examined the change in the proportion of offspring sampled from each parental-pair between years X and Y as an index of relative survival as follows:
This measure was calculated only for parental-pairs with offspring observed in year 0+. The relationships between AAxy-PBR and ΔPi0+–1+ and ΔPi1+–2+ were examined using generalized linear models (GLMs) with a normal distribution, an identity link function, and male and female ID included as factors in the model. AAxy-PBR was included in each model as a potential linear and quadratic predictor (AAxy-PBR × AAxy-PBR).
(iii). Major histocompatibility complex, reproductive success and offspring survival
Relative reproductive success was calculated for each parental-pair in each year as:
where Ni is the number of offspring genotyped to the parental-pair i, and NMi and NFi represent the total number of offspring identified to male and female i, respectively. This measure helped to control for bias associated with the unequal number of offspring sampled among males and females and represents the skew in mating patterns between putative parental-pairs.
The relationship between AAxy-PBR and RS was examined using a GLM with a normal distribution, an identity link function, and AAxy-PBR and AAxy-PBR × AAxy-PBR and their interactions with year included as predictors in the model. Male and female ID were also included as factors in the model.
(iv). Multiple mating, major histocompatibility complex diversity and reproductive success
We examined multiple mating by males and females as a potential strategy to improve offspring survival or expected MHC diversity in offspring [38]. Linear regression was used to examine the relationship between the number of mates for a given male or female (identified via parentage analysis) and ΔPi0+–1+ and ΔPi1+–2+, where i here refers to an individual male or female. We also used linear regression to examine the relationship between number of mates and mean AAxy-PBR expected in a given male or female's offspring.
(v). Temporal variation in major histocompatibility complex and neutral genetic variation
To examine the potential role of temporal variation in selection across years on the MHC, we compared population genetic differentiation between years at the MHC locus with that exhibited by the microsatellite loci. Microsatellites are non-coding regions of the genome and thus, patterns of genetic variation at these markers should generally reflect the influence of neutral evolutionary processes. Thus, any observed differences in population genetic differentiation between years at the MHC relative to the microsatellite markers should result from selection pressures operating on the MHC locus. To estimate pairwise genetic differentiation between years 0+ and 1+, 1+ and 2+, and 0+ and 2+, we initially calculated FST estimates at the MHC locus and FST ± 95% confidence interval (CI) estimates for the microsatellite loci in FSTAT v. 2.9.3 [54]. Because the magnitude of FST is dependent upon the maximum heterozygosity observed at the locus under examination, we corrected our FST estimates as outlined by Hedrick [55]. Thus, we compared genetic differentiation among years using 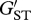 , which is defined as FST/FST(max), where FST(max) = 1 − HS (see eqn. 3 in [55]), and HS is the mean heterozygosity across all subpopulations examined. χ2-tests were used to examine variation in MHC allele frequencies among years in the offspring.
, which is defined as FST/FST(max), where FST(max) = 1 − HS (see eqn. 3 in [55]), and HS is the mean heterozygosity across all subpopulations examined. χ2-tests were used to examine variation in MHC allele frequencies among years in the offspring.
Means are reported as ±1 SD. All statistical analyses, excluding the Monte Carlo simulations, were performed in JMP (v. 8, SAS institute). Tests of normality were conducted for all response variables using the Kolmorgorov–Smirnov–Lillefors test. Our measure of RS deviated significantly from a normal distribution, but utilization of square root-transformed RS data yielded analogous results to those using the non-transformed data; thus we report the results using the non-transformed variable. A significance value of 0.05 was used for all tests except for the χ2-tests examining individual allele frequency changes across years. Here we used a Bonferroni-corrected significance threshold of p = 0.0045 (0.05/11) to account for multiple testing.
3. Results
(a). Parentage analysis
We identified a total of 916 offspring to a parental-pair over the 3 years (year 1: n = 512; year 2: n = 267; year 3: n = 137). The parentage analysis revealed that reproductive success was highly variable among males and females. For males, mean proportional reproductive success was 0.02 ± 0.02 (range: 0–0.09) and based on our estimates, two of the 41 (5%) males had no offspring assigned to them through parentage analysis. For females, mean proportional reproductive success was 0.03 ± 0.02 (range: 0–0.07), and similar to what was observed in males, two of the 35 females (5%) had no offspring assigned to them through parentage analysis. Females mated with an average of 6.0 ± 3.7 males (range: 0–13), whereas males mated with an average of 5.5 ± 3.4 females (range: 0–14).
(b). Major histocompatibility complex and reproductive success
The Atlantic salmon adults exhibited a total of 12 MHC class II alleles. Details on the genetic variation observed at the MHC locus in the adults and at the microsatellite loci have previously been reported in Landry et al. [47] and Garant et al. [46]. The Monte Carlo simulation revealed that the mean expected MHC AAxy-PBR in offspring based on the observed matings was significantly higher than expected under random mating, which is indicative of mate choice for MHC diversity (observed MHC AAxy-PBR = 7.54, p = 0.02). However, variance in MHC AAxy-PBR was not significantly different from that expected under random mating (observed variance in MHC AAxy-PBR = 713.45, p = 0.09).
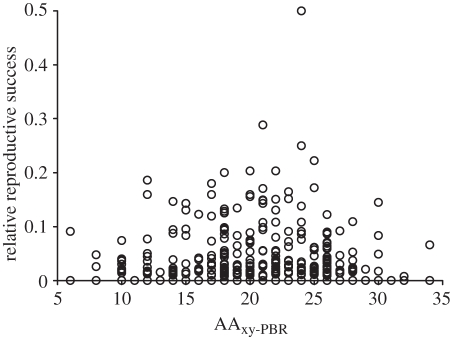
We also found that AAxy-PBR between parents had a significant effect on the RS of a parental-pair; however, the relationship was not linear. Instead, the quadratic amino acid diversity predictor (AAxy-PBR × AAxy-PBR) was significantly associated with RS, indicating that parental-pairs with intermediate MHC amino acid divergence values exhibited the highest RS (table 1 and figure 1). The relationship between the AAxy-PBR × AAxy-PBR predictor and RS held across all years (table 1). The relationship between the AAxy-PBR × AAxy-PBR predictor and RS remained significant even when an extreme RS value observed in year 2+ was excluded from the analysis (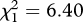 , p = 0.011).
, p = 0.011).
Table 1.
Results of generalized linear models examining the influence of major histocompatibility complex (MHC) amino acid divergence on parental-pair reproductive success and offspring survival in Atlantic salmon (Salmo salar) in the Ste-Marguerite River, Québec, Canada. Model predictors included linear (AAxy-PBR) and quadratic (AAxy-PBR × AAxy-PBR) terms examining amino acid divergence effects on parental-pair relative reproductive success (RS) and offspring survival from year 0+ to 1+ (ΔPi0+–1+ and from year 1+ to year 2+ (ΔPi1+–2+). Male and female IDs were included as factors in each model. Estimates of the effect of each predictor (B), χ2-tests of significance, and the degrees of freedom (d.f.) are indicated. p-values falling below the set critical alpha = 0.05 are indicated in bold.
| RS |
ΔPi0+–1+ |
ΔPi1+–2+ |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| B | χ2 (d.f.) | p | B | χ2 (d.f.) | p | B | χ2 (d.f.) | p | |
| model | 169.14 (79) | <0.001 | 72.84 (67) | 0.292 | 70.01 (67) | 0.377 | |||
| male ID | 87.25 (40) | <0.001 | 33.15 (34) | 0.509 | 41.86 (34) | 0.167 | |||
| female ID | 83.56 (34) | <0.001 | 39.95 (31) | 0.130 | 27.76 (31) | 0.633 | |||
| AAxy-PBR | −2.8 × 10−4 | 2.99 (1) | 0.083 | −2.7 × 10−4 | 5.70 (1) | 0.017 | 1.8 × 10−4 | 3.56 (1) | 0.059 |
| AAxy-PBR × year | −1.8 × 10−4 | 1.23 (1) | 0.267 | ||||||
| AAxy-PBR × AAxy-PBR | −5.2 × 10−5 | 6.94 (1) | 0.008 | 1.1 × 10−5 | 0.41 (1) | 0.524 | 2.4 × 10−7 | 0.00 (1) | 0.983 |
| AAxy-PBR × AAxy-PBR × year | −7.2 × 10−6 | 0.12 (1) | 0.721 | ||||||
Figure 1.
Relationship between major histocompatibility complex (MHC) class II genetic variation and relative reproductive success of parental-pairs in Atlantic salmon (Salmo salar) in the Ste-Marguerite River, Québec, Canada. MHC genetic variation was estimated using mean amino acid divergence (AAxy-PBR) between parents.
(c). Major histocompatibility complex and offspring survival
In contrast to our results for RS, MHC AAxy-PBR between parents was significantly negatively related to ΔPi0+–1+ (table 1). We did not find a significant relationship between AAxy-PBR and ΔPi1+–2+, although in contrast to the results for ΔPi0+–1+, there was a trend towards a positive relationship (table 1).
(d). Multiple mating
We did not find that multiple mating by males or females was related to ΔPi0+–1+ (males: F1,33 = 0.15, p = 0.70; females: F1,30 = 0.003, p = 0.95) or ΔPi1+–2+ (males: F1,33 = 0.42, p = 0.52; females: F1,30 = 0.08, p = 0.78). However, female multiple mating was significantly positively associated with mean expected AAxy-PBR in offspring (F1,30 = 9.22, p = 0.005), a pattern that was not observed in males (F1,33 = 0.42, p = 0.52).
(e). Temporal variation in offspring genetic variation
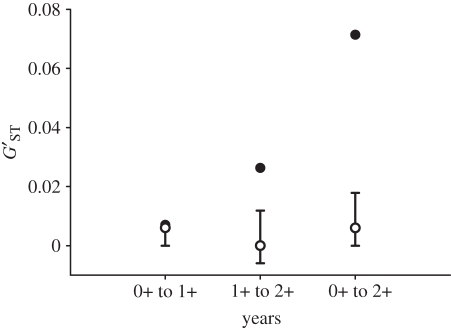
We obtained MHC genotypes for a total of 767 offspring sampled over the 3 years. Eleven MHC class II alleles were identified in the offspring (electronic supplementary material, table S1). The pairwise estimates of genetic differentiation (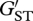 ) between years 0+ and 1+, years 1+ and 2+, and years 0+ and 2+ were significantly higher at the MHC locus than at the microsatellite loci (figure 2). Furthermore, the χ2-test revealed that the allele frequencies at the MHC differed significantly between years 0+ and 1+ (
) between years 0+ and 1+, years 1+ and 2+, and years 0+ and 2+ were significantly higher at the MHC locus than at the microsatellite loci (figure 2). Furthermore, the χ2-test revealed that the allele frequencies at the MHC differed significantly between years 0+ and 1+ (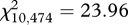 , p = 0.008), and between years 1+ and 2+ (
, p = 0.008), and between years 1+ and 2+ (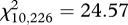 , p = 0.006). Specifically, allele 5 decreased in frequency (
, p = 0.006). Specifically, allele 5 decreased in frequency (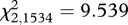 , p = 0.009) and allele 7 (
, p = 0.009) and allele 7 (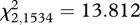 , p = 0.001) increased in frequency among years. The change in the frequency of allele 7 among years was significant, though for allele 5, the change in allele frequency among years was only near-significant when using the conservative Bonferroni-corrected significance threshold of 0.0045.
, p = 0.001) increased in frequency among years. The change in the frequency of allele 7 among years was significant, though for allele 5, the change in allele frequency among years was only near-significant when using the conservative Bonferroni-corrected significance threshold of 0.0045.
Figure 2.
MHC and microsatellite population genetic differentiation exhibited across 3 years of study in Atlantic salmon (Salmo salar) offspring in the Ste-Marguerite River in Québec, Canada. Closed circles indicate pairwise estimates of population differentiation (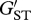 ) between the years for the MHC class II. Population genetic differentiation at microsatellite loci is indicated by open circles and the error bars indicate 95% CI surrounding the estimate of differentiation.
) between the years for the MHC class II. Population genetic differentiation at microsatellite loci is indicated by open circles and the error bars indicate 95% CI surrounding the estimate of differentiation.
4. Discussion
Much attention has been paid to documenting MHC-dependent mating patterns because of the potentially important fitness benefits associated with these behaviours [6]. In this study, we incorporated an additional genetic marker (MHC) in order to refine the parentage assignment conducted by Landry et al. [47] and re-examine mating patterns for evidence of non-random mating for MHC diversity in Atlantic salmon. Our results are concordant with those originally described in Landry et al. [47], revealing a strong pattern of MHC disassortative mating. Several other studies have shown evidence of mate choice for MHC dissimilar mates. For example, Chinook salmon (Oncorhynchus tshawytscha), grey mouse lemurs (Microcebus murinus), savannah sparrows (Passerculus sandwichensis) and Seychelles warblers select mates in order to increase MHC diversity in their offspring [11,12,38,56,57]. Our results also point to female multiple mating as a behavioural mechanism used to achieve higher MHC diversity in offspring in the Atlantic salmon. We are only aware of one other study system that has examined the potential role of multiple mating in obtaining MHC genetic benefits. In Seychelles warblers, females mated to similar MHC males are more likely to seek out extra-pair matings in order to increase the MHC diversity found in their offspring [38,58]. Other studies support a role for genetic benefits obtained through multiple mating behaviours, although the target genes involved in mate choice have not been identified (e.g. [59–61]). Together, our results provide further evidence that mating behaviours, namely MHC-driven disassortative mating and multiple mating, enhance offspring MHC genetic diversity in the Atlantic salmon.
MHC-dependent mate choice is expected to evolve when variation at these genes directly influences fitness [4]. Our results show that Atlantic salmon parental-pairs exhibiting intermediate levels of MHC amino acid diversity achieved the highest reproductive success, despite observing mate choice for maximal MHC diversity. Kalbe et al. [17] also showed that stickleback males and females with an intermediate number of MHC alleles achieved the highest reproductive success. For sticklebacks, it has been suggested that individuals bearing intermediate levels of diversity achieve optimal fitness because they are able to benefit from the advantages of being able to mount an immune response against a diverse pathogen community while also minimizing the costs of potentially self-damaging immune responses associated with extreme MHC diversity across multiple loci [20]. However, most salmonids exhibit one expressed MHC class II locus [62]; thus, it is unlikely that there are significant auto-immune-related costs associated with being diverse at the MHC. Instead, we suggest that our results reflect an evolutionary trade-off between sexual selection for offspring diversity and the potentially unpredictable parasite-imposed natural selection pressures that will be faced by offspring. Under fluctuating environmental selection pressures, parental mating decisions should be to assure diversity within offspring, but will result in some proportion of offspring being maladapted to the future pathogen community (i.e. genetic bet-hedging; [63,64]). At the genomic level, it has been previously suggested that both natural and sexual selection may be driving the evolution of intermediate levels of diversity as a result of trade-offs between inbreeding and outbreeding depression [22,65–67]. Our results demonstrate that intermediate levels of genetic diversity at the MHC are associated with reproductive success in parents and we suggest that mate choice for genetic divergence in offspring is adaptive when offspring are faced with temporally variable parasite-driven selection pressures.
The observed representation of Atlantic salmon family groups in the Ste-Marguerite river over the three years of our study implicated an important role for MHC genetic variation in offspring survival. Our results indicate that offspring derived from matings between MHC similar parents exhibit higher survival than offspring derived from MHC dissimilar parents between years 0+ and 1+, suggesting that directional selection may be operating on an individual MHC allele or a complement of similar MHC alleles over this time period. On the other hand, we observed a near-significant positive relationship between offspring survival and MHC amino acid diversity between years 1+ and 2+, suggesting that MHC diversity may be favoured by natural selection over that time period. The only other multi-year study to examine the relationship between MHC genetic variation and survival reported evidence of an MHC class II diversity advantage on juvenile survival in the Seychelles warbler, but did not find a relationship between survival in later life-history stages and MHC genetic variation [38]. Incongruencies in the relationships observed between MHC diversity and survival between studies and across years could reflect the influence of genotype-by-environment effects, as the relative fitnesses of genotypes will vary depending on the pathogen community faced by the host [30]. For Atlantic salmon, evidence for the occurrence of temporal variation in pathogen-driven selection was previously suggested in Dionne et al. [48], where striking seasonal differences in microbial infections were reported in juvenile fish. Our examination of MHC population genetic differentiation further supports a role for temporally variable natural selection pressures across years. We found that genetic differentiation at the MHC locus was significantly higher than at the microsatellite loci, particularly between years 1+ and 2+ and 0+ and 2+, indicating that the MHC genetic variation across years is probably being driven by diverging selection processes. A recent study of guppies (Poecilia reticulata) also showed evidence of temporally variable selection operating on the MHC class II [32]. Interestingly, Fraser et al. [32] found that the magnitude of genetic differentiation observed at the MHC was related to the difference in the proportion of individuals infected by Gyrodactylus in the populations, suggesting that selection pressures imposed by the parasites could be driving MHC evolution. Similarly, a 13-year study of Soay sheep (Ovis aries) and a 9-year study of Great reed warblers (Acrocephalus arundinaceus) indicated strong patterns of temporally variable selection on the MHC class II [31], and class I [33], respectively.
Our results examining variation in allele frequencies across years also suggest a likely role for directional selection on MHC class II evolution. We found that alleles 5 and 7 decreased and increased in frequency, respectively, among years. For allele 7, a previous study conducted on Atlantic salmon revealed a similar pattern of seasonally increasing frequency, and suggested that this allele may confer resistance against myxozoa infections, a potentially emerging parasitic infection in this region [48]. Thus, in addition to providing some of the first evidence that the long-term survival of Atlantic salmon offspring depends on MHC genetic variation, our results implicate both temporally variable and directional selection pressures in MHC evolution. Further studies are needed to detail how these within-population fluctuations in the parasite community could be driving the evolution of the MHC.
This study is among the first to examine the interplay between natural selection pressures and the evolution of MHC-dependent mating preferences. While evidence of mate choice for MHC genetic diversity is mounting, we have been largely unable to interpret the fitness consequences of these preferences because data on offspring survival in the wild are difficult to obtain. We suggest that mate choice for MHC diversity in the Atlantic salmon is adaptive because it represents a type of bet-hedging strategy. Indeed, our examination of the relationships between reproductive success and MHC genetic variation indicates that this type of mate choice does not necessarily maximize fitness [22], but may nevertheless be adaptive when offspring are faced with temporally variable natural selection pressures. Long-term studies are needed to concurrently examine variation in mating preferences and natural selection pressures (pathogen communities) in the wild as our results point to a critical role for both processes in understanding adaptive evolution in wild populations.
Acknowledgements
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant and Canada Research Chair Grant to L. Bernatchez, an NSERC CGS D award to M. Dionne, and an NSERC PDF award to M. Evans. We thank M. Valentine from the Ministère des Ressources Naturelles et de la Faune and D. Garant from Université de Sherbrooke for their support and for providing access to the samples, and T. Ming and K. Giguère for their assistance in the laboratory. Thanks to Bryan Neff for suggestions on the use of the unbiased reproductive success measure. Gary Carvalho, Jacek Radwan and two anonymous reviewers provided valuable comments on this manuscript.
References
- 1.Fisher R. A. 1930. The genetical theory of natural selection. London, UK: Oxford University Press [Google Scholar]
- 2.Candolin U., Heuschele J. 2008. Is sexual selection beneficial during adaptation to environmental change? Trends Ecol. Evol. 23, 446–452 10.1016/j.tree.2008.04.008 (doi:10.1016/j.tree.2008.04.008) [DOI] [PubMed] [Google Scholar]
- 3.Neff B. D., Pitcher T. E. 2005. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol. Ecol. 14, 19–38 10.1111/j.1365-294X.2004.02395.x (doi:10.1111/j.1365-294X.2004.02395.x) [DOI] [PubMed] [Google Scholar]
- 4.Penn D., Potts W. K. 1998. Chemical signals and parasite-mediated sexual selection. Trends Ecol. Evol. 13, 391–396 10.1016/S0169-5347(98)01473-6 (doi:10.1016/S0169-5347(98)01473-6) [DOI] [PubMed] [Google Scholar]
- 5.Milinski M. 2006. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 37, 159–186 10.1146/annurev.ecolsys.37.091305.110242 (doi:10.1146/annurev.ecolsys.37.091305.110242) [DOI] [Google Scholar]
- 6.Penn D., Potts W. K. 1999. The evolution of mating preferences and major histocompatibility genes. Am. Nat. 153, 145–164 10.1086/303166 (doi:10.1086/303166) [DOI] [PubMed] [Google Scholar]
- 7.Janeway C. A., Travers P., Walport M., Shlomchik M. J. 2001. Immunobiology: the immune system in health and disease, 5th edn New York, NY: Garland Science [Google Scholar]
- 8.Bernatchez L., Landry C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 10.1046/j.1420-9101.2003.00531.x (doi:10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- 9.Hedrick P. W. 1999. Balancing selection and MHC. Genetica 104, 207–214 10.1023/A:1026494212540 (doi:10.1023/A:1026494212540) [DOI] [PubMed] [Google Scholar]
- 10.Piertney S. B., Oliver M. K. 2006. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 [DOI] [PubMed] [Google Scholar]
- 11.Freeman-Gallant C. R., Meguerdichian M., Wheelwright N. T., Sollecito S. V. 2003. Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Mol. Ecol. 12, 3077–3083 10.1046/j.1365-294X.2003.01968.x (doi:10.1046/j.1365-294X.2003.01968.x) [DOI] [PubMed] [Google Scholar]
- 12.Neff B. D., Garner S. R., Heath J. W., Heath D. D. 2008. The MHC and non-random mating in a captive population of Chinook salmon. Heredity 101, 175–185 10.1038/hdy.2008.43 (doi:10.1038/hdy.2008.43) [DOI] [PubMed] [Google Scholar]
- 13.Olsson M., Madsen T., Nordby J., Wapstra E., Ujvari B., Wittsell H. 2003. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. Lond. B 270, S254–S256 10.1098/rsbl.2003.0079 (doi:10.1098/rsbl.2003.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans M. L., Neff B. D. 2009. MHC heterozygote advantage and widespread bacterial infections in populations of Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. 18, 4716–4729 10.1111/j.1365-294X.2009.04374.x (doi:10.1111/j.1365-294X.2009.04374.x) [DOI] [PubMed] [Google Scholar]
- 15.Penn D. J., Damjanovich K., Potts W. K. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11 260–11 264 10.1073/pnas.162006499 (doi:10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eizaguirre C., Yeates S. E., Lenz T. L., Kalbe M., Milinski M. 2009. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329 10.1111/j.1365-294X.2009.04243.x (doi:10.1111/j.1365-294X.2009.04243.x) [DOI] [PubMed] [Google Scholar]
- 17.Kalbe M., Eizaguirre C., Dankert I., Reusch T. B. H., Sommerfeld R. D., Wegner M., Milinski M. 2009. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc. R. Soc. B 276, 925–934 10.1098/rspb.2008.1466 (doi:10.1098/rspb.2008.1466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milinski M., Griffiths S., Wegner K. M., Reusch T. B. H., Haas-Assenbaum A., Boehm T. 2005. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA 102, 4414–4418 10.1073/pnas.0408264102 (doi:10.1073/pnas.0408264102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reusch T. B. H., Haberli M. A., Aeschlimann P. B., Milinski M. 2001. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature 414, 300–302 10.1038/35104547 (doi:10.1038/35104547) [DOI] [PubMed] [Google Scholar]
- 20.Woelfing B., Traulsen A., Milinski M., Boehm T. 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Phil. Trans. R. Soc. B 364, 117–128 10.1098/rstb.2008.0174 (doi:10.1098/rstb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsberg L. A., Dannewitz J., Petersson E., Grahn M. 2007. Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout females fishing for optimal MHC dissimilarity. J. Evol. Biol. 20, 1859–1869 10.1111/j.1420-9101.2007.01380.x (doi:10.1111/j.1420-9101.2007.01380.x) [DOI] [PubMed] [Google Scholar]
- 22.Waser N. M. 1993. Population structure, optimal outbreeding, and assortative mating in angiosperms. In The natural history of inbreeding and outbreeding (ed. Thornhill N. W.), pp. 173–199 Chicago, IL: University of Chicago Press [Google Scholar]
- 23.Ekblom R., Sæther A., Grahn M., Fiske P., Kålås A., Höglund J. 2004. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media). Mol. Ecol. 13, 3821–3828 10.1111/j.1365-294X.2004.02361.x (doi:10.1111/j.1365-294X.2004.02361.x) [DOI] [PubMed] [Google Scholar]
- 24.Croisetière S., Tarte P. D., Bernatchez L., Belhumeur P. 2008. Identification of MHC Class IIB resistance/susceptibility alleles to Aeromonas salmonicida in brook charr (Salvelinus fontinalis). Mol. Immun. 45, 3107–3116 10.1016/j.molimm.2008.03.007 (doi:10.1016/j.molimm.2008.03.007) [DOI] [PubMed] [Google Scholar]
- 25.Harf R., Sommer S. 2005. Association between major histocompatibility complex class II DRB alleles and parasite load in the hairy-footed gerbil, Gerbillurus paeba, in the southern Kalahari. Mol. Ecol. 14, 85–91 10.1111/j.1365-294X.2004.02402.x (doi:10.1111/j.1365-294X.2004.02402.x) [DOI] [PubMed] [Google Scholar]
- 26.Hill A. V. S., et al. 1991. Common west African HLA antigens are associated with protection from severe malaria. Nature 352, 595–600 10.1038/352595a0 (doi:10.1038/352595a0) [DOI] [PubMed] [Google Scholar]
- 27.Langefors A., Lohm J., Grahn M., Andersen O., von Schantz T. 2001. Association between major histocompatibility complex class IIB alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc. R. Soc. Lond. B 268, 479–485 10.1098/rspb.2000.1378 (doi:10.1098/rspb.2000.1378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen T., Ujvari B. 2006. MHC class I variation associates with parasite resistance and longevity in tropical pythons. J. Evol. Biol. 19, 1973–1978 10.1111/j.1420-9101.2006.01158.x (doi:10.1111/j.1420-9101.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 29.Eizaguirre C., Lenz T. L. 2010. Major histocompatibility complex polymorphism: dynamics and consequences of parasite-mediated local adaptation in fishes. J. Fish Biol. 77, 2023–2047 10.1111/j.1095-8649.2010.02819.x (doi:10.1111/j.1095-8649.2010.02819.x) [DOI] [PubMed] [Google Scholar]
- 30.Ingleby F. C., Hunt J., Hosken D. J. 2010. The role of genotype-by-environment interactions in sexual selection. J. Evol. Biol. 23, 2031–2045 10.1111/j.1420-9101.2010.02080.x (doi:10.1111/j.1420-9101.2010.02080.x) [DOI] [PubMed] [Google Scholar]
- 31.Charbonnel N., Pemberton J. 2005. A long-term genetic survey of an ungulate population reveals balancing selection acting on MHC through spatial and temporal fluctuations in selection. Heredity 95, 377–388 10.1038/sj.hdy.6800735 (doi:10.1038/sj.hdy.6800735) [DOI] [PubMed] [Google Scholar]
- 32.Fraser B. A., Ramnarine I. W., Neff B. D. 2010. Temporal variation at the MHC class IIB in wild populations of the guppy (Poecilia reticulata). Evolution 64, 2086–2096 10.1111/j.1558-5646.2010.00965.x (doi:10.1111/j.1558-5646.2010.00965.x) [DOI] [PubMed] [Google Scholar]
- 33.Westerdahl H., Hansson B., Bensch S., Hasselquist D. 2004. Between-year variation of MHC allele frequencies in great reed warblers: selection or drift? J. Evol. Biol. 17, 485–492 10.1111/j.1420-9101.2004.00711.x (doi:10.1111/j.1420-9101.2004.00711.x) [DOI] [PubMed] [Google Scholar]
- 34.Oloffson H., Ripa J., Jonzen N. 2009. Bet-hedging as an evolutionary game: the trade-off between egg size and number. Proc. R. Soc. B 276, 2963–2969 10.1098/rspb.2009.0500 (doi:10.1098/rspb.2009.0500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Real L. 1980. Fitness, uncertainty, and the role of diversification in evolution and behavior. Am. Nat. 115, 623–638 10.1086/283588 (doi:10.1086/283588) [DOI] [Google Scholar]
- 36.Beaumont H. J. E., Gallie J., Kost C., Ferguson G. C., Rainey P. R. 2009. Experimental evolution of bet hedging. Nature 462, 90–94 10.1038/nature08504 (doi:10.1038/nature08504) [DOI] [PubMed] [Google Scholar]
- 37.Yasui Y. 2001. Female multiple mating as a genetic bet-hedging strategy when mate choice criteria are unreliable. Ecol. Res. 16, 605–616 10.1046/j.1440-1703.2001.00423.x (doi:10.1046/j.1440-1703.2001.00423.x) [DOI] [Google Scholar]
- 38.Brouwer L., Barr I., van de Pol M., Burke T., Komdeur J., Richardson D. S. 2010. MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Mol. Ecol. 19, 3444–3455 10.1111/j.1365-294X.2010.04750.x (doi:10.1111/j.1365-294X.2010.04750.x) [DOI] [PubMed] [Google Scholar]
- 39.Paterson S., Wilson K., Pemberton J. M. 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a larger unmanaged ungulate population (Ovis aries). Proc. Natl Acad. Sci. USA 95, 3714–3719 10.1073/pnas.95.7.3714 (doi:10.1073/pnas.95.7.3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Eyto E., et al. 2007. Natural selection acts on Atlantic salmon major histocompatibility (MH) variability in the wild. Proc. R. Soc. B 274, 861–869 10.1098/rspb.2006.0053 (doi:10.1098/rspb.2006.0053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans M. L., Neff B. D., Heath D. D. 2010. MHC-mediated local adaption in reciprocally translocated Chinook salmon. Conserv. Genet. 11, 2333–2342 10.1007/s10592-010-0119-3 (doi:10.1007/s10592-010-0119-3) [DOI] [Google Scholar]
- 42.Jacob A., Evanno G., von Siebenthal B., Grossen C., Wedekind C. 2010. Effects of different mating scenarios on embryo viability in brown trout. Mol. Ecol. 19, 5296–5307 10.1111/j.1365-294X.2010.04884.x (doi:10.1111/j.1365-294X.2010.04884.x) [DOI] [PubMed] [Google Scholar]
- 43.Pitcher T. E., Neff B. D. 2006. MHC Class IIB alleles contribute to both additive and nonadditive genetic effects on survival in Chinook salmon. Mol. Ecol. 15, 2357–2365 10.1111/j.1365-294X.2006.02942.x (doi:10.1111/j.1365-294X.2006.02942.x) [DOI] [PubMed] [Google Scholar]
- 44.Wedekind C., Evanno G., Urbach D., Jacob A., Muller R. 2008. ‘Good genes’ and ‘compatible genes’ effects in an Alpine whitefish and the information content of breeding tubercules over the course of the spawning season. Genetica 134, 21–30 10.1007/s10709-008-9251-0 (doi:10.1007/s10709-008-9251-0) [DOI] [PubMed] [Google Scholar]
- 45.Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. 2010. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 19, 3064–3075 10.1111/j.1365-294X.2010.04724.x (doi:10.1111/j.1365-294X.2010.04724.x) [DOI] [PubMed] [Google Scholar]
- 46.Garant D., Dodson J. J., Bernatchez L. 2001. A genetic evaluation of mating system and determinants of individual reproductive success in Atlantic salmon (Salmo salar L.). J. Hered. 92, 137–145 10.1093/jhered/92.2.137 (doi:10.1093/jhered/92.2.137) [DOI] [PubMed] [Google Scholar]
- 47.Landry C., Garant D., Duchesne P., Bernatchez L. 2001. ‘Good genes as heterozygosity’: the major histocompatibility complex and mate choice in Atlantic salmon (Salmo salar). Proc. R. Soc. Lond. B 268, 1279–1285 10.1098/rspb.2001.1659 (doi:10.1098/rspb.2001.1659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dionne M., Miller K. M., Dodson J. J., Bernatchez L. 2009. MHC standing genetic variation and pathogen resistance in wild Atlantic salmon. Phil. Trans. R. Soc. B 364, 1555–1565 10.1098/rstb.2009.0011 (doi:10.1098/rstb.2009.0011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dionne M., Miller K. M., Dodson J. J., Caron F., Bernatchez L. 2007. Clinal variation in MHC diversity with temperature: evidence for the role of host–pathogen interaction on local adaptation in Atlantic salmon. Evolution 61, 2154–2164 10.1111/j.1558-5646.2007.00178.x (doi:10.1111/j.1558-5646.2007.00178.x) [DOI] [PubMed] [Google Scholar]
- 50.Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106 10.1111/j.1365-294X.2007.03089.x (doi:10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 51.Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 52.Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364, 33–39 10.1038/364033a0 (doi:10.1038/364033a0) [DOI] [PubMed] [Google Scholar]
- 53.Manly B. F. J. 1997. Randomization, bootstrapping and Monte Carlo methods in biology. London, UK: Chapman and Hall [Google Scholar]
- 54.Goudet J. 2002. Fstat version 2.9.3.2. Switzerland: UNIL; See http://www2.unil.ch/popgen/softwares/fstat.htm [Google Scholar]
- 55.Hedrick P. W. 2005. A standardized genetic differentiation measure. Evolution 59, 1633–1638 10.1111/j.0014-3820.2005.tb01814.x (doi:10.1111/j.0014-3820.2005.tb01814.x) [DOI] [PubMed] [Google Scholar]
- 56.Garner S. R., Bortoluzzi R., Heath D. D., Neff B. D. 2010. Sexual conflict inhibits female mate choice for MHC dissimilarity in Chinook salmon. Proc. R. Soc. B 277, 885–894 10.1098/rspb.2009.1639 (doi:10.1098/rspb.2009.1639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwensow N., Eberle M., Sommer S. 2008. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proc. R. Soc. B 275, 555–564 10.1098/rspb.2007.1433 (doi:10.1098/rspb.2007.1433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richardson D. S., Komdeur J., Burke T., von Schantz T. 2005. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. B 272, 759–767 10.1098/rspb.2004.3028 (doi:10.1098/rspb.2004.3028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foerster K., Delhey K., Johnsen A., Lifjeld J. T., Kempenaers B. 2003. Females increase offspring heterozygosity and fitness through extra-pair matings. Nature 425, 714–717 10.1038/nature01969 (doi:10.1038/nature01969) [DOI] [PubMed] [Google Scholar]
- 60.Fossoy F., Johnsen A., Lifjeld J. T. 2008. Multiple genetic benefits of female promiscuity in a socially monogamous passerine. Evolution 62, 145–156 10.1111/j.1558-5646.2007.00284.x (doi:10.1111/j.1558-5646.2007.00284.x) [DOI] [PubMed] [Google Scholar]
- 61.Sheldon B. C., Merilä J., Qvarnström A., Gustafsson L., Ellegren H. 1997. Paternal genetic contribution to offspring condition predicted by size of male secondary sexual character. Proc. R. Soc. Lond. B 264, 297–302 10.1098/rspb.1997.0042 (doi:10.1098/rspb.1997.0042) [DOI] [Google Scholar]
- 62.Grimholt U., Larsen S., Nordmo R., Midtlyng P., Kjoeglum S., Storset A., Saebo S., Stet R. J. M. 2003. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics 55, 210–219 10.1007/s00251-003-0567-8 (doi:10.1007/s00251-003-0567-8) [DOI] [PubMed] [Google Scholar]
- 63.Jennions M. D., Petrie M. 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. 75, 21–64 10.1017/S0006323199005423 (doi:10.1017/S0006323199005423) [DOI] [PubMed] [Google Scholar]
- 64.Kokko H., Heubel K. 2008. Condition-dependence, genotype-by-environment interactions and the lek paradox. Genetica 134, 55–62 10.1007/s10709-008-9249-7 (doi:10.1007/s10709-008-9249-7) [DOI] [PubMed] [Google Scholar]
- 65.Bonneaud C., Chastel O., Federici P., Westerdahl H., Sorci G. 2006. Complex MHC-based mate choice in a wild passerine. Proc. R. Soc. B 273, 1111–1116 10.1098/rspb.2005.3325 (doi:10.1098/rspb.2005.3325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richard M., Losdat S., Lecomte J., de Fraipont M., Clobert J. 2009. Optimal level of inbreeding in the common lizard. Proc. R. Soc. B 276, 2779–2786 10.1098/rspb.2009.0319 (doi:10.1098/rspb.2009.0319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neff B. D. 2004. Stabilizing selection on genomic divergence in a wild fish population. Proc. Natl Acad. Sci USA 101, 2381–2385 10.1073/pnas.0307522100 (doi:10.1073/pnas.0307522100) [DOI] [PMC free article] [PubMed] [Google Scholar]