Abstract
Plant studies that have investigated the fitness consequences of growing with siblings have found conflicting evidence that can support different theoretical frameworks. Depending on whether siblings or strangers have higher fitness in competition, kin selection, niche partitioning and competitive ability have been invoked. Here, we bring together these processes in a conceptual synthesis and argue that they can be co-occurring. We propose that these processes can be reconciled and argue for a trait-based approach of measuring natural selection instead of the fitness-based approach to the study of sibling competition. This review will improve the understanding of how plants interact socially under competitive situations, and provide a framework for future studies.
Keywords: plant, fitness, kin selection, niche partitioning, sibling competition
1. Introduction
Most plants are closely surrounded by other plants [1]. Neighbouring plants interact intensely with each other by altering the local microclimate [2], providing cues and signals that elicit phenotypic responses [3], exuding chemicals into the soil [4] and reducing the availability of resources, particularly light, water and mineral nutrients [5]. Negative interactions between plants resulting from the depletion of resources constitute exploitation competition (hereafter, competition) [1]. In plants, the scale of competition is local, with the nearest neighbours dominating interactions [6]. Depending on dispersal mechanisms, germination timing, mechanisms of vegetative reproduction and chance, the nearest neighbours may be another species, strangers of the same species, relatives or clones [7]. Relatedness among neighbours of the same species is thought to affect competition by at least two processes: (i) kin selection theory predicts that relatives may cooperate with each other [8] and (ii) the elbow room hypothesis, or the niche partitioning hypothesis predicts that relatives overlap more in their niche use and compete more with each other compared with unrelated conspecifics [9]. Strikingly, these processes predict opposing outcomes for sibling competition. Kin cooperation predicts that groups of competing siblings will outperform groups of competing strangers, while niche partitioning predicts the opposite (figure 1).
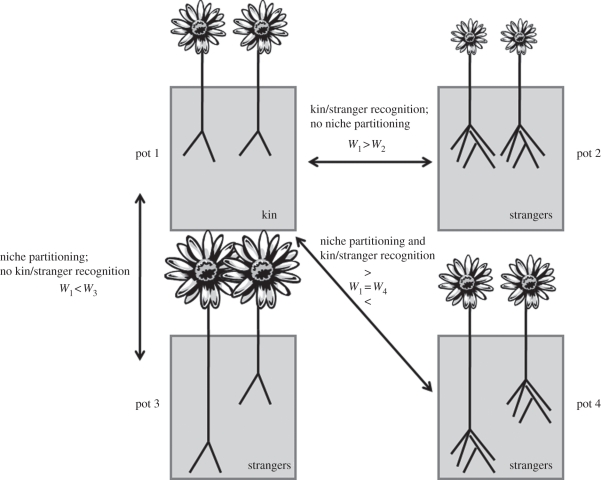
Figure 1.
In this cartoon, two siblings or two strangers compete in a pot. These plants vary in two traits—root proliferation, a competitive behaviour, and rooting depth, a niche-use trait. Fitness is indicated by flower size. Pot 1 is a sibling treatment with two plants of genotype A, while pots 2–4 are stranger treatments with both genotypes A and B represented. Pots 2–4 illustrate the different hypothesized scenarios involving strangers. The comparison between pots 1 and 2 reflects the kin selection and kin recognition hypothesis: strangers but not siblings increase root proliferation in response to competitors, and this costly competitive response reduces their fitness. The comparison between pots 1 and 3 reflects the niche partitioning hypothesis: strangers differ more in rooting depth and therefore compete less, performing better than siblings. The difference between pots 4 and 1 indicates the hypothesis of both processes occurring independently: strangers increase root proliferation but also differ in rooting depth, so the fitness outcome of competition depends on the relative impact of the two processes.
Several researchers have tested these predictions in plants, measuring traits associated with performance and fitness consequences. These empirical studies of fitness in plant sibling competition (hereafter called group studies) have yielded variable results (see electronic supplementary material, table S1): in nine group studies, siblings outperformed strangers [10–16], in 11 group studies strangers outperformed siblings [14,17–25] and in 21 group studies there were no differences [10,17,20,22,26–37]. Moreover, variation in the consequences of sibling competition was found among traits [11], between environments [14] and among families [12,23,24]. Individually, the findings of group studies have been interpreted as an indirect demonstration of kin selection or niche partitioning in plants depending on the direction of the results. But as a whole, the results are equivocal.
A recent approach to plant kin cooperation measures plant responses to their relatives [38–41]. Phenotypic plasticity in morphology and allocation traits to the neighbour identity [42] indicate that plants actively participate in social interactions by recognizing their neighbours. Responses to presence/absence, self/non-self, species and kin/stranger have been found. In these studies, only the identity of the neighbour or neighbouring root differs between treatments. Phenotypic plasticity to the identity of neighbours is then measured. Most identity studies have demonstrated root responses to other roots (reviewed in [41]), although one study has demonstrated responses from an aboveground signal [13]. Aboveground [39] and reproductive traits [43] have also been shown to respond to the identity of neighbouring roots.
Kin recognition studies are extensions of the phenotypic plasticity approach. When plants are grown in high density with the same average soil volume per plant, competitive plasticity of root allocation is conditional on whether those neighbours are siblings or strangers of the same species [38–40]. Plant exudates cue these responses [41,44]. The response to kin versus stranger varies in ways that match the ecology of the species. For example, Cakile edentula grows on the dunes of sandy beaches where water and soil nutrients are limiting and light is abundant [45]. Thus, it is not surprising to find responses to relatedness of neighbours in belowground, competitive traits. When C. edentula is grown with strangers, plants increase fine root allocation compared to when they are grown with siblings [38], probably in order to acquire the limiting resources of the environment, nutrients and water. In contrast, Impatiens pallida grows in the forest understory where light is limited. When grown with strangers, they decrease root allocation and increase allocation to leaves, allowing them to increase their light reception at the cost of belowground resource acquisition [39].
The finding of plant kin recognition has renewed interest in the fitness consequences of sibling competition [24,30,37,46], including three new group studies [24,30,37], discussion of whether group studies should accompany kin recognition studies [24,46] and a critique of the group study methodology [30]. But rather than carrying out more group studies, we advocate that researchers must measure how the traits of individuals and their neighbours affect fitness.
Here, we review aspects of kin selection and niche partitioning theory relevant to plants of the same species growing together, with a narrow focus on why fitness could differ between kin and strangers. We discuss the disadvantages of the group study approach, including how other processes create fitness differences between kin and strangers. With kin selection and niche partitioning in the same theoretical framework, that of phenotypic selection analysis [47], we can then compare and contrast how traits of individuals and their neighbours affect inclusive fitness [48] for these processes. We bring together the empirical evidence from phenotypic selection analysis for kin selection and niche partitioning. From this starting point, we discuss how phenotypic selection analysis could be implemented.
2. Kin selection and plants
Social interactions among members of the same species can result in kin selection. Hamilton's rule provides a summation of kin selection that highlights its most important result, an explanation of altruism towards relatives. Because genes are shared between close relatives, a trait or behaviour that does not directly increase the fitness of an individual may still spread if it sufficiently increases the fitness of a close relative [8]. Kin selection is most frequently expressed in Hamilton's rule: altruistic traits are favoured when
where C is the cost to the focal individual, B is the benefit to a relative and r is the relatedness between them [8]. Kin selection does not only apply to costly social behaviours that benefit relatives; it is evolution through natural selection for any trait of an individual that affects the fitness of that individual's relatives [49].
For relatives to affect each other's performance, individuals need to be close enough to interact. For plants, the proximity of relatives will depend on mechanisms of seed and pollen dispersal and vegetative reproduction [50], the community of other species [51] and chance. Dispersal traits affect whether seedlings are likely to emerge near to relatives [50]. But because seed dispersal is stochastic, and conspecifics and interspecific competitors may be present, proximity to kin is not certain. Even for species that frequently grow in structured monospecific stands [52] with a high likelihood of kin interactions, some seeds will disperse into competition with strangers and other species.
For both plants and animals, proximity of relatives provides the opportunity for relatives to cooperate with each other, but also creates conditions where relatives compete for scarce resources. This kin competition is often argued to constrain the benefits of cooperation from kin selection [53]. If circumstances that increase association with kin, i.e. population viscosity, increase the overall level of competition that individuals experience, then kin competition is a constraint for the evolution of kin cooperation [54]. However, for plants, the absence of kin does not mean the absence of competition, because plants tend to grow closely associated with other plants. Consequently, for most plants, the population viscosity can increase the opportunities for competition with kin without changing the overall degree of competition that individuals experience [54].
The ability to recognize relatives is not required for kin selection to occur. But the evolution of indiscriminate altruism is only likely when individuals predictably interact with relatives [8]. Since seed dispersal is stochastic, kin recognition is probably crucial for plants to evolve altruism towards relatives. In an environment where the relatedness of neighbours varies, kin recognition reduces the net cost of altruism, because altruistic behaviours can be preferentially directed at relatives [55]. While in the short term, population viscosity can create an environment where plants interact primarily with relatives, plant communities vary in composition and numbers over time. A criterion for stable coexistence within a community is the ability to increase in numbers from low density [56], and an indiscriminately altruistic genotype is unlikely to be able to do so.
Animals often learn who their kin are from context, e.g. individuals in the same nest are relatives [57]. But reliable context is lacking for plants. Moreover, plants recognize the identity of neighbours and neighbouring roots in experiments where researchers eliminated context [38–41]. The only plant-identity recognition system whose mechanism has been established are the self-incompatibility systems. These act by preventing mating with pollen from the same plant, and may function to reduce mating with relatives.
Hamilton's rule is usually invoked to explain why one individual might sacrifice its interests to benefit a relative, such as why ground squirrels produce warning calls to alert others about predators [57]. However, kin selection can also explain the avoidance of aggressive behaviours with relatives [58]. For plants, the most likely traits in which to see kin recognition, and consequently kin selection, are competitive responses to other plants. Competitive phenotypic plasticity allows plants to pre-empt resources aboveground and belowground [5] through changes in morphology and allocation to increase resource uptake or reduce their neighbour's ability to access resources [3]. If relatives do not display competitive behaviour towards each other, then they benefit by gaining indirect fitness, and not by investing in costly competitive behaviour. A game theory approach helps in understanding plant competition in a kin selection context (see electronic supplementary material; Does the neighbour's phenotype matter?, and electronic supplementary material, figure S1).
3. Niche partitioning and plants
A common definition of niche partitioning is differential resource use by individuals in close proximity. Individuals with slightly different niches may access limiting resources in a different manner, thus reducing or avoiding competition [59]. The link between the individual's genotype and its niche is the trait that allows the individual to specialize in acquiring a certain portion of a limiting resource. For Galapagos finches, beak size is a well-known trait that allows an organism to specialize on a particular size range of seeds [60]. For plants, a candidate trait is rooting depth to acquire belowground resources that vary with soil depth [61] (see electronic supplementary material, figure S2 for an example). A payoff matrix represents how the fitness of individuals in a competing population is affected by trait differences rather than trait values themselves (see electronic supplementary material; Does the neighbour's phenotype matter?, and electronic supplementary material, figure S3).
Niche partitioning is a simple concept with broad implications. Niche partitioning occurs among or within species and evolves during speciation, i.e. character displacement. Among-species niche partitioning underlies community assembly rules theory, which suggests that communities are structured to increase functional diversity [62], affecting ecosystem function [63]. The Tangled Bank theory indicates that niche partitioning provides a possible explanation for why organisms have sex [64]. Compared with asexual offspring, sexually produced offspring are more genetically diverse, likely have less niche overlap and thus, compete less for limiting resources [19,65]. Character displacement resulting from niche partitioning explains patterns of speciation in fish [66] and oaks [67,68].
For all its broad implications, niche partitioning is difficult to study. Niche partitioning is a non-additive mechanism because the fitness of individuals in competition with phenotypically and genetically diverse neighbours cannot be predicted by their performance in a genetically homogeneous environment [69]. Niche partitioning can be directly demonstrated when the niche trait is known, e.g. if fitness is higher when individuals in a group differ [70]. When the niche trait is unknown, niche partitioning is inferred from performance measures [59]. In plants, niche partitioning is primarily inferred from segregation among species along niche axes, trade-offs among potential niche traits, niche shifts in the presence of competitors and from the outcome of intraspecific versus interspecific competition [59]. There is, as yet, no evidence for niche shifts in response to same species competitors.
4. More reasons to move beyond the fitness-based approach
Conflicting fitness outcomes for sibling competition are predicted by at least two major processes: kin cooperation and niche partitioning. However, fitness consequences in group studies may result from competition differences among genotypes rather than niche partitioning or kin cooperation processes. The usual practice of creating stranger groups comprising several families or genotypes creates potential for misleading results [30]. If a few families are more strongly competitive than other families, then these competitive plants could suppress the fitness of other plants in the stranger groups. Consequently, the fitness of the other sibling groups will be relatively high when compared with the stranger groups without any cooperation among kin [30]. Consequently, a pairwise design with stranger groups of two families provides more valid results because competitive ability can be measured and the effects of rare, competitive families are not weighted disproportionately [30].
Another reason that fitness consequences may not indicate niche partitioning or kin cooperation processes is that frequency-dependent selection can affect the fitness differences between sibling and stranger groups. Positive frequency-dependent natural selection will favour phenotypic similarity to neighbours. For example, predator swamping can favour synchrony in events such as emergence, fruiting and hatching [71]. Because siblings should be more similar than strangers, selection for synchrony will increase the fitness of sibling groups. Negative frequency-dependent selection could give better performance in stranger groups. Common phenotypes can be targeted by predators [72] or be more susceptible to disease [73], so that a more diverse group of strangers will outperform a group of similar siblings. Thus, the fitness outcome of frequency-dependent selection is indistinguishable from niche partitioning during competition for scarce resources or altruism towards relatives.
A growing body of literature shows that the microbes in the soil interact with plants and affect plant community structure. Positive feedbacks, where the microbial population becomes increasingly favourable to a species over time, can favour monospecific stands, while negative feedbacks, where the microbial populations become less favourable over time, increase biodiversity (see reviews [74,75]). Though much of the plant–microbe literature focuses on how feedback affects interspecific plant dynamics, there is evidence for positive and negative microbial feedback for genotypes within species [76,77]. It is very possible that microbes are drivers of intraspecific processes favouring sibling or stranger groups, but more studies are needed.
Moreover, even when kin selection is present, it will not always result in higher fitness in sibling groups. Kin selection may favour indiscriminate altruism in highly viscous populations [8]. That is, if dispersal is local, neighbours are likely to be relatives, and therefore indiscriminate cooperation can evolve. However, even if kin selection favoured indiscriminate altruism or less competition, no difference in fitness between siblings and strangers would be visible in group studies. It is specifically altruism directed at kin only that would generate increased fitness with siblings.
The focus on kin selection versus niche partitioning has resulted in little consideration of alternative processes. The usual discussion concerns whether the fitness outcome indicates kin selection or niche partitioning, or a lack of evidence for any process. But the either/or nature of the interpretation seems to have resulted in the implicit or explicit assumption that only one process can occur. At the extreme, Milla et al. [24] explicitly state their expectation that all plants will demonstrate the same process [24]. They conclude that the body of evidence from group studies provides more support for niche partitioning than kin selection [24] as more studies have results consistent with niche partitioning than kin selection (electronic supplementary material, table S1).
Another assumption, implicit in most group studies, is that only one process is occurring in a given species or study. However, it is possible that both processes can happen simultaneously. Fitness measures are confounded if multiple processes of niche partitioning, kin selection and competitive ability [30] can co-occur. Credible evidence for their co-occurrence has been found in morning glory [37]. In this study, three inbred lines were grown in sibling and stranger groups, and measures of morphology, allocation and fitness were made at harvest. Though sibling and stranger groups did not differ in fitness, the three lines differed in how fitness responded to neighbour relatedness, with one showing better performance with siblings, one better with strangers and one not differing. Biernaskie [37] found greater root allocation in stranger groups than in sibling groups, a result consistent with kin recognition. Surprisingly, the greatest fitness was found in the most unequal groups for height, consistent with niche partitioning [37]. These results further motivate examining the processes involved in sibling versus stranger competition.
5. Moving forward
The conclusions from the group studies are not as clear-cut as usually assumed, and they do not advance the pursuit of the mechanism. However, the question they raise remains unanswered: does the neighbours' phenotype matter in same-species competition, and if so, why? Answering this question requires actually examining processes. The commonality between kin selection and niche partitioning is that fitness depends on the phenotype of the focal individual and the phenotypes of its neighbours. The difference between these processes lies in the mechanisms by which the individual trait and the neighbour trait affect fitness. The payoff matrices provide a clear demonstration of this difference (see electronic supplementary material; Does the neighbour's phenotype matter?).
We advocate measuring natural selection on candidate traits [47] to determine how traits of focal plants and neighbours affect fitness, and whether the effects correspond to the predictions for kin selection or niche partitioning. The measurement of natural selection on continuous, correlated traits [47] is a powerful tool in understanding microevolution. Here, we bring together the extension of selection theory to evolution in variable environments [78], multi-level selection [48] and frequency-dependent disruptive selection [79] that are needed to ask whether and how the phenotype of neighbours affects performance.
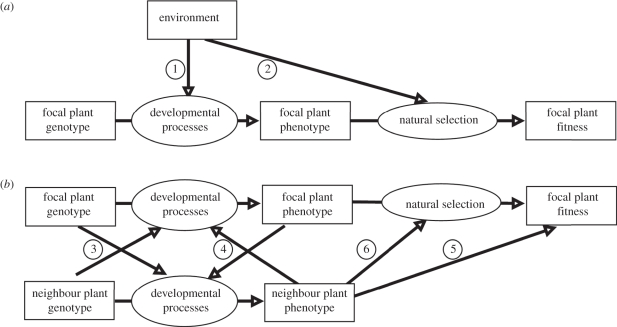
A fundamental evolutionary concept is that the genotype affects the phenotype and the phenotype affects fitness. Phenotypic plasticity theory [80] recognizes that environment can affect both the phenotype (figure 2a, arrow 1) and the natural selection on that phenotype (figure 2a, arrow 2). Here, we define the environment as aspects of the plant neighbours, including their density, phenotype and identity (figure 2b). Kin recognition is first incorporated as phenotypic plasticity to the relatedness of neighbours (figure 2b, arrow 3). Plasticity to neighbours may be incorporated as a direct response to their phenotype (figure 2b, arrow 4). Whether kin recognition is in a competitive or cooperative trait then needs to be determined. While Hamilton's rule [8] has proved useful in developing theory, the crucial fitness parameters, the cost and benefits of an altruistic act, are hard to measure in natural populations [81]. Another definition of kin selection, evolution through natural selection for any trait of an individual that affects the fitness of that individual's relatives [49], provides a practical approach for empirical work. The effect of neighbour traits on the target individual's fitness is equivalent to selection at the level of the group, which can be estimated through multi-level selection analysis [52,82,83]. In this conceptual model, the fitness of the target individual is determined by natural selection on its own traits, i.e. individual selection (figure 2b: natural selection), and natural selection on its neighbour traits, i.e. group selection, (figure 2b, arrow 5). If the trait under selection is altruistic, then a negative individual selection gradient will measure the cost of cooperation, while a positive group selection gradient measures the benefit to the individual of cooperation by its neighbours. If a trait is selfish or competitive, the individual selection gradient is positive, while the group selection coefficient is negative [83]. Whether or not the plant is growing with kin, the fitness of an individual is increased by having a more competitive value for a trait, and decreased by having a less competitive trait value, i.e. natural selection at the individual level favours competitiveness and selects against cooperation [52].
Figure 2.
(a) Path model of the fitness consequences of phenotypic plasticity to some aspect of the environment (adapted from Scheiner [80]). (b) Potential mechanisms by which neighbour phenotype and identity could affect an individual's fitness. Circled numbers on the figure indicate the following processes: (1) phenotypic plasticity to the environment; (2) natural selection on a trait can differ between environments; (3) kin recognition, as phenotypic plasticity to relatedness; (4) phenotypic plasticity to the presence and phenotype of neighbours; (5) effect of neighbour traits on focal plant fitness (selection on group-level traits); and (6) effects of neighbour traits on natural selection for the focal plant trait (frequency-dependent disruptive selection induced by competition for a limiting resource).
In the conceptual model, the trait of the neighbour affects natural selection on the individual's trait (figure 2b, arrow 6), indicating niche partitioning. The appropriate theory for natural selection on continuous niche-partitioning traits is usually identified by other names [84–86]. In theoretical models, the phenotype of the individual is linked to its niche use. For any two individuals, greater phenotypic similarity results in more competition for the same resources. Since these resources are limited and depleted by use, competition is then linked with fitness through a Lotka–Volterra model [79,84]. When the models are applied to one species, the most common result is frequency-dependent, disruptive selection [87] on the phenotype related to resource use. Both the frequency dependence and disruptive selection result from intraspecific competition. Consider our example (electronic supplementary material, figure S2) of rooting depth (phenotype) to acquire belowground resources that vary with soil depth (niche parameter). In the absence of competition, natural selection favours a character value that allows an individual to access the resource where it is most abundant. The presence of competitors reduces the availability of resources used by the population, e.g. resources become depleted at the 8 cm depth when competitors are present. Consequently, the population experiences frequency-dependent, disruptive selection on the character. Selection is disruptive because the pool of resources corresponding to the phenotype at the population mean is reduced while resources corresponding to more extreme phenotypes remain abundant. Selection is frequency-dependent because resources used by the more abundant character class are reduced [87]. In sum, the selective regime on the extant population is created by resource availability, and availability of resources depends on how the resources are supplied and how the extant population uses them (see electronic supplementary material, figure S2). Thus, the presence and the phenotype of neighbours affect the direction and strength of natural selection on the target plant's phenotype. The simple prediction is that plants with phenotypes that differ from those of the group will have higher fitness.
Multi-level selection (also called group selection or contextual selection) has been measured several times in plants and related to kin selection. The traits measured tended to be size and morphology traits that are easily measured. All of the studies found selection at the group level, though many traits showed only individual selection or group selection. The most common result was that a measure of size appears as a competitive/selfish trait, favoured in individual selection but selected against in group selection [45,88–91]. This is the expected pattern for a kin-selected trait, with the direction of selection reversed between individual and group selection, for size or mass. Elongation [90], height [12,45], production of cleistogamous buds [89], lifespan [90] and herbivore resistance [88] were also found to show contrasting group and individual selection gradients typical of kin selection. Group selection was in the same direction as individual selection for height in Silene [91], and for mass in Cakile sibling groups [12]. In Cakile, sibling groups had significant selection on group traits, while stranger groups did not [12].
Manipulative multi-level studies more directly demonstrate a causal response to group selection and the potential for kin selection in plants. Kelly [52] manipulated phenotype in natural population by removing the apical meristem. This manipulation changed a suite of traits he termed ‘bushy’; more bushy plants were taller, with more leaves held lower on the plant. Bushiness responses to this manipulation varied across years and treatments, indicating the complexity of plant phenotypic plasticity. Consistently, though, plants performed better if their neighbours were more bushy, demonstrating that the bushiness phenotype met the requirements for kin selection.
Goodnight [92] did a selection experiment over nine generations. Selection lines were subject to both individual selection for leaf area (higher, lower and control) and group selection for leaf area (higher, lower and control). The lines responded rapidly to group selection, with the expected responses. The responses to individual selection were slower, and not as expected: those selected for both higher and lower leaf area showed reduced leaf area when compared with the control until the final generation. Thus, natural selection on plant size causes indirect selection on the social interactions: individual selection for increased size favours a more competitive phenotype, but group selection for increased size favours a more cooperative phenotype.
When phenotypic selection analysis is used to study niche partitioning, multiple lines of evidence are needed to establish niche partitioning by demonstrating that a trait is under disruptive selection that is negatively frequency-dependent, with competition for scarce resources as the agent of selection [87]. Several animal studies have directly demonstrated the impact of traits on fitness resulting from competition for limiting resources [85,93]. These demonstrations include showing that selection on a trait depends on the nature of a competing taxa, that fitness of pairs of competitors are highest when their resource-use traits are most dissimilar, that disruptive selection is present with high density but absent with low density, or that negative frequency-dependence of polymorphic traits is present. In these systems, there is often a continuous trait that ranges from suitable to specialization on resource A to generalizing on both A and B to specialization on resource B. Researchers typically use multiple lines of evidence to demonstrate that natural selection resulting from niche partitioning is acting.
In plants, a considerable challenge in finding niche differentiation is that putative traits and resource axes are difficult to identify. All plants need the same resources of water, mineral nutrients and light, limiting in the degree to which they specialize and making it difficult to determine which traits to measure a priori [59]. Traits or axes that have been shown to be important for interspecific niche partitioning include drought and flood resistance, height (reviewed by Silvertown [59]), root depth [94], nitrogen source [95] and timing of growth [61]. Cavender-Bares [67] found that soil moisture, nutrient availability and fire regime gradients were important resource axes for congeneric oak species that would normally competitively exclude one another. In one of the few studies that has empirical evidence for within-species variation, Michaels & Bazzaz [96] measured the degree of individual specialization on proposed plant niche axes of light and nutrients. Competition versus colonization is recognized as a niche axis for long-term coexistence [59], but not immediate competition.
6. Research programme
By measuring how neighbours affect natural selection on a trait and fitness, measurement of natural selection in a subdivided population will determine if kin selection or niche partitioning is occurring. The measurement of selection is done independently of the source of phenotypic variation, so that measuring kin selection does not require that kin recognition be present or that groups of siblings be measured [48]. Rather, the important requirement is the construction of many groups that vary in group means and in the relative frequency of different phenotypes [48]. Constructing groups with extreme phenotypes, use of genetic mutants or phenotypic manipulation of cues can be used to extend the phenotypic variation and get a better measure of natural selection [97].
Using multi-level selection analysis to estimate the effect of individual and group level traits on individual fitness, and apply the results to kin selection, is an established procedure [48,83]. The classic expectations for competitive and cooperative traits are that individual and group selection components will be of opposing signs. An altruistic trait will be under negative individual selection, but positive group selection. A competitive trait will be under positive individual selection, but negative group selection. Any significant selection on group means, however, will indicate that kin selection is acting. For kin selection, the important indicator is significant selection on group means with consistent selection gradients for individual selection across differing subpopulations.
Using multi-level selection analysis to assess the niche partitioning process is a more subtle problem. If the trait is part of a niche partitioning process, then natural selection on that trait is predicted to be affected by the population composition in the smaller subpopulations, with rarer trait values favoured in each population [87]. This effect will not be measured by nonlinear selection on individual and group mean traits. Instead, nonlinear selection on the deviation from the group mean will need to be included, to measure whether there is frequency-dependent disruptive selection.
Carrying out studies with plant competition as the environment offers unique challenges. The presence of other plants presents a complex environment both aboveground and belowground. The relative importance of aboveground versus belowground competition varies with species and environment [98,99], while competitive responses to neighbours show tradeoffs and opportunity constraints between aboveground and belowground traits [100,101]. Because plants show phenotypic plasticity to the presence of neighbours [3], traits expressed in the presence of neighbours will differ from traits expressed in isolated plants. A particular problem in carrying out a selection study is that the root traits implicated in niche partitioning and kin selection are usually measured destructively and ideally at an early age, making it difficult to assess lifetime fitness. Though some studies measured root traits and fitness at harvest [37,102], the cause and effect relationship is not established, as performance can affect the trait rather than the usual assumption of natural selection that the trait affects fitness.
In this approach, multiple, correlated traits can be measured, so it can be determined if kin recognition and niche partitioning are mutually exclusive or can occur concurrently (figure 1). If different traits are subject to different processes, then the question of whether these processes show trade-offs and constraints can be addressed. To assess the fitness consequences for a given trait, population parameters and plasticity to neighbours need to be measured. To assess the fitness consequences of niche partitioning, the genotypic and phenotypic variances are needed. Natural selection can be estimated and integrated with the genetic variance and covariances, and plasticity to competitors and relatedness, using models of natural selection. The challenge for modelling long-term evolution is that spatial structure among interacting individuals needs to be directly accounted for [103,104].
7. Conclusion and future directions
At one time, fitness consequences studies were the only approach to kin selection and niche partitioning in plants. While this approach has been valuable in providing evidence that both niche partitioning and kin selection may occur in plants, it is fundamentally limited. The theoretical synthesis and the empirical data support both processes. Now, researchers need to follow putative niche partitioning and competition traits through plasticity and contextual selection to their fitness implications.
Acknowledgements
The authors thank S. Balshine for helpful discussions on cooperative behaviour, J. Bever for introducing us to literature on microbial feedback at the genotype level, H. Maherali for guidance on the direction of the paper, L. Birch, J. Dushoff, A. Khandelwal, M. Bhatt, A. May, A. Khan and several anonymous reviewers for their helpful comments on previous versions of this manuscript. The research was supported by an NSERC discovery grant to S.A.D.
References
- 1.Casper B. B., Jackson R. B. 1997. Plant competition underground. Ann. Rev. Ecol. Syst. 28, 545–570 10.1146/annurev.ecolsys.28.1.545 (doi:10.1146/annurev.ecolsys.28.1.545) [DOI] [Google Scholar]
- 2.Jones H. G. 1992. Plant and microclimate: a quantitative approach to environmental plant physiology. Cambridge, UK: Cambridge University Press [Google Scholar]
- 3.Novoplansky A. 2009. Picking battles wisely: plant behaviour under competition. Plant Cell Environ. 32, 726–741 10.1111/j.1365-3040.2009.01979.x (doi:10.1111/j.1365-3040.2009.01979.x) [DOI] [PubMed] [Google Scholar]
- 4.Bais H. P., Weir T. L., Perry L. G., Gilroy S., Vivanco J. M. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266 10.1146/annurev.arplant.57.032905.105159 (doi:10.1146/annurev.arplant.57.032905.105159) [DOI] [PubMed] [Google Scholar]
- 5.Grace J. B. 1990. On the relationship between plant traits and competitive ability. In Perspectives on plant competition (eds Grace J. B., Tilman D.), pp. 51–65 NY, USA: Academic Press [Google Scholar]
- 6.Milbau A., Reheul D., De Cauwer B., Nijs I. 2007. Factors determining plant–neighbour interactions on different spatial scales in young species-rich grassland communities. Ecol. Res. 22, 242–247 10.1007/s11284-006-0018-8 (doi:10.1007/s11284-006-0018-8) [DOI] [Google Scholar]
- 7.Wade M. J. 1980. An experimental study of kin selection. Evolution 34, 844–855 10.2307/2407991 (doi:10.2307/2407991) [DOI] [PubMed] [Google Scholar]
- 8.Hamilton W. D. 1964. The genetical evolution of social behavior, I & II. J. Theor. Biol. 7, 1–52 10.1016/0022-5193(64)90038-4 (doi:10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 9.Young J. P. W. 1981. Sib competition can favour sex in two ways. J. Theor. Biol. 88, 755–756 10.1016/0022-5193(81)90249-6 (doi:10.1016/0022-5193(81)90249-6) [DOI] [PubMed] [Google Scholar]
- 10.Willson M. F., Hoppes W. G., Goldman D. A., Thomas P. A., Katusic-Malmborg P. L., Bothwell J. L. 1987. Sibling competition in plants: an experimental study. Am. Nat. 129, 304–311 10.1086/284636 (doi:10.1086/284636) [DOI] [Google Scholar]
- 11.Tonsor S. J. 1989. Relatedness and intraspecific competition in Plantago lanceolata. Am. Nat. 134, 897–906 10.1086/285020 (doi:10.1086/285020) [DOI] [Google Scholar]
- 12.Donohue K. 2003. The influence of neighbor relatedness on multilevel selection in the Great Lakes sea rocket. Am. Nat. 162, 77–92 10.1086/375299 (doi:10.1086/375299) [DOI] [PubMed] [Google Scholar]
- 13.Karban R., Shiojiri K. 2009. Self-recognition affects plant communication and defense. Ecol. Lett. 12, 502–506 10.1111/j.1461-0248.2009.01313.x (doi:10.1111/j.1461-0248.2009.01313.x) [DOI] [PubMed] [Google Scholar]
- 14.Andalo C., Goldringer I., Godelle B. 2001. Inter- and intragenotypic competition under elevated carbon dioxide in Arabidopsis thaliana. Ecology 82, 157–164 10.1890/0012-9658(2001)082[0157:IAICUE]2.0.CO;2 (doi:10.1890/0012-9658(2001)082[0157:IAICUE]2.0.CO;2) [DOI] [Google Scholar]
- 15.Ronsheim M. L. 1996. Evidence against a frequency-dependent advantage for sexual reproduction in Allium vineale. Am. Nat. 147, 718–734 10.1086/285876 (doi:10.1086/285876) [DOI] [Google Scholar]
- 16.Collins A., Hart E. M., Molofsky J. 2010. Differential response to frequency-dependent interactions: an experimental test using genotypes of an invasive grass. Oecologia 164, 959–969 10.1007/s00442-010-1719-9 (doi:10.1007/s00442-010-1719-9) [DOI] [PubMed] [Google Scholar]
- 17.Boyden S., Binkley D., Stape J. L. 2008. Competition among Eucalyptus trees depends on genetic variation and resource supply. Ecology 89, 2850–2859 10.1890/07-1733.1 (doi:10.1890/07-1733.1) [DOI] [PubMed] [Google Scholar]
- 18.Allard R. W., Adams J. 1969. Population studies in predominantly self-pollinating species. XIII. Intergenotypic competition and population structure in barley and wheat. Am. Nat. 103, 621–645 10.1086/282630 (doi:10.1086/282630) [DOI] [Google Scholar]
- 19.Ellstrand N. C., Antonovics J. 1985. Experimental studies of the evolutionary significance of sexual reproduction. II. A test of the density-dependent selection hypothesis. Evolution 39, 657–666 10.2307/2408660 (doi:10.2307/2408660) [DOI] [PubMed] [Google Scholar]
- 20.Schmitt J., Antonovics J. 1986. Experimental studies of the evolutionary significance of sexual reproduction. IV. Effect of neighbor relatedness and aphid infestation on seedling performance. Evolution 40, 830–836 10.2307/2408467 (doi:10.2307/2408467) [DOI] [PubMed] [Google Scholar]
- 21.Kelley S. E. 1989. Experimental studies of the evolutionary significance of sexual reproduction. V. A field-test of the sib competition lottery hypothesis. Evolution 43, 1054–1065 10.2307/2409585 (doi:10.2307/2409585) [DOI] [PubMed] [Google Scholar]
- 22.Escarre J., Houssard C., Thompson J. D. 1994. An experimental study of the role of seedling density and neighbor relatedness in the persistence of Rumex acetosella in an old-field succession. Can. J. Bot. 72, 1273–1281 10.1139/b94-155 (doi:10.1139/b94-155) [DOI] [Google Scholar]
- 23.Cheplick G. P., Kane K. H. 2004. Genetic relatedness and competition in Triplasis purpurea (Poaceae): resource partitioning or kin selection? Int. J. Plant Sci. 165, 623–630 10.1086/386556 (doi:10.1086/386556) [DOI] [Google Scholar]
- 24.Milla R., Forero D. M., Escudero A., Iriondo J. M. 2009. Growing with siblings: a common ground for cooperation or for fiercer competition among plants? Proc. R. Soc. B 276, 2531–2540 10.1098/rspb.2009.0369 (doi:10.1098/rspb.2009.0369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delesalle V. R. A., Mazer S. J. 2002. The neighborhood matters: effects of neighbor number and sibling (or kin) competition on floral traits in Spergularia marina (Caryophyllaceae). Evolution 56, 2406–2413 10.1111/j.0014-3820.2002.tb00166.x (doi:10.1111/j.0014-3820.2002.tb00166.x) [DOI] [PubMed] [Google Scholar]
- 26.Argyres A. Z., Schmitt J. 1992. Neighborhood relatedness and competitive performance in Impatiens capensis (Balsaminaceae): a test of the resource partitioning hypothesis. Am. J. Bot. 79, 181–185 10.2307/2445106 (doi:10.2307/2445106) [DOI] [Google Scholar]
- 27.Burt A., Bell G. 1992. Tests of sib diversification theories of outcrossing in Impatiens capensis: effects of inbreeding and neighbor relatedness on production and infestation. J. Evol. Biol. 5, 575–588 10.1046/j.1420-9101.1992.5040575.x (doi:10.1046/j.1420-9101.1992.5040575.x) [DOI] [Google Scholar]
- 28.Cheplick G. P., Salvadori G. M. 1991. Intraclonal and interclonal competition in the cleistogamous grass Amphibromus scabrivalvis. Am. J. Bot. 78, 1494–1502 10.2307/2444974 (doi:10.2307/2444974) [DOI] [Google Scholar]
- 29.Karron J. D., Marshall D. L. 1993. Effects of environmental variation on fitness of singly and multiply sired progenies of Raphanus sativus (Brassicaceae). Am. J. Bot. 80, 1407–1412 10.2307/2445669 (doi:10.2307/2445669) [DOI] [Google Scholar]
- 30.Masclaux F., Hammond R. L., Meunier J., Gouhier-Darimont C., Keller L., Reymond P. 2010. Competitive ability not kinship affects growth of Arabidopsis thaliana accessions. New Phytol. 185, 322–331 10.1111/j.1469-8137.2009.03057.x (doi:10.1111/j.1469-8137.2009.03057.x) [DOI] [PubMed] [Google Scholar]
- 31.McCall C., Mitchell-Olds T., Waller D. M. 1989. Fitness consequences of outcrossing in Impatiens capensis—tests of the frequency-dependent and sib-competition models. Evolution 43, 1075–1084 10.2307/2409587 (doi:10.2307/2409587) [DOI] [PubMed] [Google Scholar]
- 32.Monzeglio U., Stoll P. 2007. Effects of spatial pattern and relatedness in an experimental plant community. Evol. Ecol. 22, 723–741 10.1007/s10682-007-9197-1 (doi:10.1007/s10682-007-9197-1) [DOI] [Google Scholar]
- 33.Puustinen S., Koskela T., Mutikainen P. 2004. Relatedness affects competitive performance of a parasitic plant (Cuscuta europaea) in multiple infections. J. Evol. Biol. 17, 897–903 10.1111/j.1420-9101.2004.00728.x (doi:10.1111/j.1420-9101.2004.00728.x) [DOI] [PubMed] [Google Scholar]
- 34.Rautiainen P., Koivula K., Hyvarinen M. 2004. The effect of within-genet and between-genet competition on sexual reproduction and vegetative spread in Potentilla anserina ssp egedii. J. Ecol. 92, 505–511 10.1111/j.0022-0477.2004.00878.x (doi:10.1111/j.0022-0477.2004.00878.x) [DOI] [Google Scholar]
- 35.Schmitt J., Ehrhardt D. W. 1987. A test of the sib-competition hypothesis for outcrossing advantage in Impatiens capensis. Evolution 41, 579–590 10.2307/2409259 (doi:10.2307/2409259) [DOI] [PubMed] [Google Scholar]
- 36.Willis C. G., Brock M. T., Weinig C. 2010. Genetic variation in tolerance of competition and neighbour suppression in Arabidopsis thaliana. J. Evol. Biol. 23, 1412–1424 10.1111/j.1420-9101.2010.02003.x (doi:10.1111/j.1420-9101.2010.02003.x) [DOI] [PubMed] [Google Scholar]
- 37.Biernaskie M. 2011. Evidence for competition and cooperation among climbing plants. Proc. R. Soc. B 278, 1989–1996 10.1098/rspb.2010.1771 (doi:10.1098/rspb.2010.1771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudley S. A., File A. L. 2007. Kin recognition in an annual plant. Biol. Lett. 3, 435–438 10.1098/rsbl.2007.0232 (doi:10.1098/rsbl.2007.0232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy G. P., Dudley S. A. 2009. Kin recognition: competition and cooperation in Impatiens (Balsaminaceae). Am. J. Bot. 96, 1990–1996 10.3732/ajb.0900006 (doi:10.3732/ajb.0900006) [DOI] [PubMed] [Google Scholar]
- 40.Bhatt M. V., Khandelwal A., Dudley S. A. 2010. Kin recognition, not competitive interactions, predicts root allocation in young Cakile edentula seedling pairs. New Phytol. 189, 1135–1142 10.1111/j.1469-8137.2010.03458.x (doi:10.1111/j.1469-8137.2010.03458.x) [DOI] [PubMed] [Google Scholar]
- 41.Biedrzycki M., Jilany T., Dudley S., Bais H. 2010. Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 3, 1–8 10.4161/cib.3.1.9694 (doi:10.4161/cib.3.1.9694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Kroon H. 2007. Ecology: how do roots interact? Science 318, 1562–1563 10.1126/science.1150726 (doi:10.1126/science.1150726) [DOI] [PubMed] [Google Scholar]
- 43.Lankinen A. 2008. Root competition influences pollen competitive ability in Viola tricolor: effects of presence of a competitor beyond resource availability? J. Ecol. 96, 756–765 10.1111/j.1365-2745.2008.01387.x (doi:10.1111/j.1365-2745.2008.01387.x) [DOI] [Google Scholar]
- 44.Biedrzycki M. L., Venkatachalam L., Bais H. P. 2011. The role of ABC transporters in kin recognition in Arabidopsis thaliana. Plant Signall. Behav. 6, 1154–1161 10.4161/psb.6.8.15907 (doi:10.4161/psb.6.8.15907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donohue K. 2004. Density-dependent multilevel selection in the Great Lakes sea rocket. Ecology 85, 180–191 10.1890/02-0767 (doi:10.1890/02-0767) [DOI] [Google Scholar]
- 46.Callaway R. M., Mahall B. E. 2007. Plant ecology: family roots. Nature 448, 145–147 10.1038/448145a (doi:10.1038/448145a) [DOI] [PubMed] [Google Scholar]
- 47.Lande R., Arnold S. J. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226 10.2307/2408842 (doi:10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 48.Bijma P., Wade M. J. 2008. The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. J. Evol. Biol. 21, 1175–1188 10.1111/j.1420-9101.2008.01550.x (doi:10.1111/j.1420-9101.2008.01550.x) [DOI] [PubMed] [Google Scholar]
- 49.West Eberhard M. 1975. The evolution of social behavior by kin selection. Q. Rev. Biol. 50, 1–33 10.1111/j.1469-185X.1975.tb00988.x (doi:10.1111/j.1469-185X.1975.tb00988.x) [DOI] [Google Scholar]
- 50.Cheplick G. P. 1992. Sibling competition in plants. J. Ecol. 80, 567–575 10.2307/2260699 (doi:10.2307/2260699) [DOI] [Google Scholar]
- 51.Thorpe A. S., Aschehoug E. T., Atwater D. Z., Callaway R. M. 2011. Interactions among plants and evolution. J. Ecol. 99, 729–740 10.1111/j.1365-2745.2011.01802.x (doi:10.1111/j.1365-2745.2011.01802.x) [DOI] [Google Scholar]
- 52.Kelly J. K. 1996. Kin selection in the annual plant Impatiens capensis. Am. Nat. 147, 899–918 10.1086/285885 (doi:10.1086/285885) [DOI] [Google Scholar]
- 53.Platt T. G., Bever J. D. 2009. Kin competition and the evolution of cooperation. Trends Ecol. Evol. 24, 370–377 10.1016/j.tree.2009.02.009 (doi:10.1016/j.tree.2009.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Dyken J. D. 2010. The components of kin competition. Evolution 64, 2840–2854 10.1111/j.1558-5646.2010.01033.x (doi:10.1111/j.1558-5646.2010.01033.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gardner A., West S. A. 2010. Greenbeards. Evolution 64, 25–38 10.1111/j.1558-5646.2009.00842.x (doi:10.1111/j.1558-5646.2009.00842.x) [DOI] [PubMed] [Google Scholar]
- 56.Chesson P. L., Ellner S. 1989. Invasibility and stochastic boundedness in monotonic competition models. J. Math. Biol. 27, 117–138 10.1007/BF00276099 (doi:10.1007/BF00276099) [DOI] [PubMed] [Google Scholar]
- 57.Waldman B. 1988. The ecology of kin recognition. Annu. Rev. Ecol. Syst. 19, 543–571 10.1146/annurev.es.19.110188.002551 (doi:10.1146/annurev.es.19.110188.002551) [DOI] [Google Scholar]
- 58.Pfennig D. W., Reeve H. K., Sherman P. W. 1993. Kin recognition and cannibalism in Spadefoot Toad tadpoles. Anim. Behav. 46, 87–94 10.1006/anbe.1993.1164 (doi:10.1006/anbe.1993.1164) [DOI] [Google Scholar]
- 59.Silvertown J. 2004. Plant coexistence and the niche. Trends Ecol. Evol. 19, 605–611 10.1016/j.tree.2004.09.003 (doi:10.1016/j.tree.2004.09.003) [DOI] [Google Scholar]
- 60.Price T. 1987. Diet variation in a population of Darwin's finches. Ecology 68, 1015–1028 10.2307/1938373 (doi:10.2307/1938373) [DOI] [Google Scholar]
- 61.Parrish J. A. D., Bazzaz F. A. 1976. Underground niche separation in successional plants. Ecology 57, 1281–1288 10.2307/1935052 (doi:10.2307/1935052) [DOI] [Google Scholar]
- 62.Stegen J. C., Swenson N. G. 2009. Functional trait assembly through ecological and evolutionary time. Theoret. Ecol. 2, 239–250 10.1007/s12080-009-0047-3 (doi:10.1007/s12080-009-0047-3) [DOI] [Google Scholar]
- 63.Diaz S., Cabido M. 1997. Plant functional types and ecosystem function in relation to global change. J. Veg. Sci. 8, 463–474 [Google Scholar]
- 64.Song Y., Drossel B., Scheu S. 2011. Tangled Bank dismissed too early. Oikos 120, 1601–1607 10.1111/j.1600-0706.2011.19698.x (doi:10.1111/j.1600-0706.2011.19698.x) [DOI] [Google Scholar]
- 65.Antonovics J., Ellstrand N. C. 1984. Experimental studies of the evolutionary significance of sexual reproduction. I. A test of the frequency-dependent selection hypothesis. Evolution 38, 103–115 10.2307/2408550 (doi:10.2307/2408550) [DOI] [PubMed] [Google Scholar]
- 66.Svanbäck R., Bolnick D. I. 2007. Intraspecific competition drives increased resource use diversity within a natural population. Proc. R. Soc. B 274, 839–844 10.1098/rspb.2006.0198 (doi:10.1098/rspb.2006.0198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cavender-Bares J., Kitajima K., Bazzaz F. A. 2004. Multiple trait associations in relation to habitat differentiation among 17 Floridian oak species. Ecol. Monogr. 74, 635–662 10.1890/03-4007 (doi:10.1890/03-4007) [DOI] [Google Scholar]
- 68.Cavender-Bares J., Ackerly D. D., Baum D. A., Bazzaz F. A. 2004. Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843 10.1086/386375 (doi:10.1086/386375) [DOI] [PubMed] [Google Scholar]
- 69.Hughes A. R., Inouye B. D., Johnson M. T. J., Underwood N., Vellend M. 2008. Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623 10.1111/j.1461-0248.2008.01179.x (doi:10.1111/j.1461-0248.2008.01179.x) [DOI] [PubMed] [Google Scholar]
- 70.Pfennig D. W. 2009. Disruptive selection in natural populations: the roles of ecological specialization and resource competition. Am. Nat. 174, 268–281 10.1086/600090 (doi:10.1086/600090) [DOI] [PubMed] [Google Scholar]
- 71.Kerkhoff A. J. 2004. Expectation, explanation and masting. Evol. Ecol. Res. 6, 1003–1020 [Google Scholar]
- 72.Xiao Z., Gao X., Steele M. A., Zhang Z. 2009. Frequency-dependent selection by tree squirrels: adaptive escape of non-dormant white oaks. Behav. Ecol. 21, 169–175 10.1093/beheco/arp169 (doi:10.1093/beheco/arp169) [DOI] [Google Scholar]
- 73.Chaboudez P. B. 1995. Frequency-dependent selection in a wild plant-pathogen system. Oecologia 102, 490–493 10.1007/BF00341361 (doi:10.1007/BF00341361) [DOI] [PubMed] [Google Scholar]
- 74.Bever J. D., et al. 2010. Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 25, 468–478 10.1016/j.tree.2010.05.004 (doi:10.1016/j.tree.2010.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reynolds H. L., Packer A., Bever J. D., Clay K. 2003. Grassroots ecology: plant–microbe–soil interactions as drivers of plant community structure and dynamics. Ecology 84, 2281–2291 10.1890/02-0298 (doi:10.1890/02-0298) [DOI] [Google Scholar]
- 76.Ronsheim M. L., Anderson S. E. 2001. Population-level specificity in the plant-mycorrhizae association alters intraspecific interactions among neighboring plants. Oecologia 128, 77–84 10.1007/s004420000625 (doi:10.1007/s004420000625) [DOI] [PubMed] [Google Scholar]
- 77.Bever J. D., Westover K. M., Antonovics J. 1997. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85, 561–573 10.2307/2960528 (doi:10.2307/2960528) [DOI] [Google Scholar]
- 78.Via S., Lande R. 1985. Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522 10.2307/2408649 (doi:10.2307/2408649) [DOI] [PubMed] [Google Scholar]
- 79.Taper M., Case T. 1985. Quantitative genetic models for the coevolution of character displacement. Ecology 66, 355–371 10.2307/1940385 (doi:10.2307/1940385) [DOI] [Google Scholar]
- 80.Scheiner S. M. 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68 10.1146/annurev.es.24.110193.000343 (doi:10.1146/annurev.es.24.110193.000343) [DOI] [Google Scholar]
- 81.Krakauer A. H. 2005. Kin selection and cooperative courtship in wild turkeys. Nature 434, 69–72 10.1038/nature03325 (doi:10.1038/nature03325) [DOI] [PubMed] [Google Scholar]
- 82.Heisler I. L., Damuth J. 1987. A method for analyzing selection in hierarchically structured populations. Am. Nat. 130, 582–602 10.1086/284732 (doi:10.1086/284732) [DOI] [Google Scholar]
- 83.Goodnight C. J. 2005. Multilevel selection: the evolution of cooperation in non-kin groups. Popul. Ecol. 47, 3–12 10.1007/s10144-005-0207-2 (doi:10.1007/s10144-005-0207-2) [DOI] [Google Scholar]
- 84.Abrams P. A., Rueffler C., Kim G. 2008. Determinants of the strength of disruptive and/or divergent selection arising from resource competition. Evolution 62, 1571–1586 10.1111/j.1558-5646.2008.00385.x (doi:10.1111/j.1558-5646.2008.00385.x) [DOI] [PubMed] [Google Scholar]
- 85.Pfennig D. W., Pfennig K. S. 2010. Character displacement and the origins of diversity. Am. Nat. 176, S26–S44 10.1086/657056 (doi:10.1086/657056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ackermann M., Doebeli M. 2004. Evolution of niche width and adaptive diversification. Evolution 58, 2599–2612 10.1554/04-244 (doi:10.1554/04-244) [DOI] [PubMed] [Google Scholar]
- 87.Rueffler C., Van Dooren T., Leimar O., Abrams P. 2006. Disruptive selection and then what? Trends Ecol. Evol. 21, 238–245 10.1016/j.tree.2006.03.003 (doi:10.1016/j.tree.2006.03.003) [DOI] [PubMed] [Google Scholar]
- 88.Boege K. 2010. Induced responses to competition and herbivory: natural selection on multi-trait phenotypic plasticity. Ecology 91, 2628–2637 10.1890/09-0543.1 (doi:10.1890/09-0543.1) [DOI] [PubMed] [Google Scholar]
- 89.Stevens L., Goodnight C. J., Kalisz S. 1995. Multilevel selection in natural populations of Impatiens capensis. Am. Nat. 145, 513–526 10.1086/285753 (doi:10.1086/285753) [DOI] [Google Scholar]
- 90.Weinig C., Johnston J. A., Willis C. G., Maloof J. N. 2007. Antagonistic multilevel selection on size and architecture in variable density settings. Evolution 61, 58–67 10.1111/j.1558-5646.2007.00005.x (doi:10.1111/j.1558-5646.2007.00005.x) [DOI] [PubMed] [Google Scholar]
- 91.Aspi J., Jakalaniemi A., Tuomi J., Siikamaki P. 2003. Multilevel phenotypic selection on morphological characters in a metapopulation of Silene tatarica. Evolution 57, 509–517 10.1554/0014-3820(2003)057[0509:MPSOMC]2.0.CO;2 (doi:10.1554/0014-3820(2003)057[0509:MPSOMC]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 92.Goodnight C. J. 1985. The influence of environmental variation on group and individual selection in a cress. Evolution 39, 545–558 10.2307/2408652 (doi:10.2307/2408652) [DOI] [PubMed] [Google Scholar]
- 93.Dayan T., Simberloff D. 2005. Ecological and community-wide character displacement: the next generation. Ecol. Lett. 8, 875–894 10.1111/j.1461-0248.2005.00791.x (doi:10.1111/j.1461-0248.2005.00791.x) [DOI] [Google Scholar]
- 94.Nobel P. S., Linton M. L. 1997. Frequencies, microclimate and root properties for three codominate perennials in the northwestern Sonoran desert on North- versus South-facing slopes. Ann. Bot. 80, 731–739 10.1006/anbo.1997.0508 (doi:10.1006/anbo.1997.0508) [DOI] [Google Scholar]
- 95.von Felten S., Hector A., Buchmann N., Niklaus P., Schmid B., Scherer-Lorenzen M. 2009. Belowground nitrogen partitioning in experimental grassland communities of varying species richness. Ecology 90, 1389–1399 10.1890/08-0802.1 (doi:10.1890/08-0802.1) [DOI] [PubMed] [Google Scholar]
- 96.Michaels H. J., Bazzaz F. A. 1989. Individual and population responses of sexual and apomictic plants to environmental gradients. Am. Nat. 134, 190–207 10.1086/284975 (doi:10.1086/284975) [DOI] [Google Scholar]
- 97.Schmitt J., Dudley S. A., Pigliucci M. 1999. Manipulative approaches to testing adaptive plasticity: phytochrome-mediated shade-avoidance responses in plants. Am. Nat. 154, S43–S54 10.1086/303282 (doi:10.1086/303282) [DOI] [PubMed] [Google Scholar]
- 98.Cahill J. F. 1999. Fertilization effects on interactions between above-ground and below-ground competition in an old field. Ecology 80, 466–480 10.1890/0012-9658(1999)080[0466:FEOIBA]2.0.CO;2 (doi:10.1890/0012-9658(1999)080[0466:FEOIBA]2.0.CO;2) [DOI] [Google Scholar]
- 99.Wilson S. D., Tilman D. 1995. Competitive responses of eight old-field plant species in four environments. Ecology 76, 1169–1180 10.2307/1940924 (doi:10.2307/1940924) [DOI] [Google Scholar]
- 100.Cipollini D. F., Schultz J. C. 1999. Exploring cost constraints on stem elongation using phenotypic manipulation. Am. Nat. 153, 236–242 10.1086/303164 (doi:10.1086/303164) [DOI] [PubMed] [Google Scholar]
- 101.Maliakal S. K., McDonnell K., Dudley S. A., Schmitt J. 1999. Effects of red to far-red ratio and plant density on biomass allocation and gas exchange in Impatiens capensis. Int. J. Plant Sci. 160, 723–733 10.1086/314157 (doi:10.1086/314157) [DOI] [Google Scholar]
- 102.Gersani M., Brown J. S., O'Brien E. E., Maina G. M., Abramsky Z. 2001. Tragedy of the commons as a result of root competition. J. Ecol. 89, 660–669 10.1046/j.0022-0477.2001.00609.x (doi:10.1046/j.0022-0477.2001.00609.x) [DOI] [Google Scholar]
- 103.Axelrod R., Hammond R. A., Grafen A. 2004. Altruism via kin-selection strategies that rely on arbitrary tags with which they coevolve. Evolution 58, 1833–1838 10.1111/j.0014-3820.2004.tb00465.x (doi:10.1111/j.0014-3820.2004.tb00465.x) [DOI] [PubMed] [Google Scholar]
- 104.Nowak M. A. 2006. Five rules for the evolution of cooperation. Science 314, 1560–1563 10.1126/science.1133755 (doi:10.1126/science.1133755) [DOI] [PMC free article] [PubMed] [Google Scholar]