Abstract
The S-like ribonucleases (RNases) RNS1 and RNS2 of Arabidopsis are members of the widespread T2 ribonuclease family, whose members also include the S-RNases, involved in gametophytic self-incompatibility in plants. Both RNS1 and RNS2 mRNAs have been shown previously to be induced by inorganic phosphate (Pi) starvation. In our study we examined this regulation at the protein level and determined the effects of diminishing RNS1 and RNS2 expression using antisense techniques. The Pi-starvation control of RNS1 and RNS2 was confirmed using antibodies specific for each protein. These specific antibodies also demonstrated that RNS1 is secreted, whereas RNS2 is intracellular. By introducing antisense constructs, mRNA accumulation was inhibited by up to 90% for RNS1 and up to 65% for RNS2. These plants contained abnormally high levels of anthocyanins, the production of which is often associated with several forms of stress, including Pi starvation. This effect demonstrates that diminishing the amounts of either RNS1 or RNS2 leads to effects that cannot be compensated for by the actions of other RNases, even though Arabidopsis contains a large number of different RNase activities. These results, together with the differential localization of the proteins, imply that RNS1 and RNS2 have distinct functions in the plant.
Plants contain a large number of different RNase activities (for review, see Bariola and Green, 1997; Parry et al., 1997). By far the best-characterized group of plant RNases is that of the T2 family, designated as such because the fungal RNase T2 is the prototype enzyme of the class. First isolated from fungi, proteins in this family have subsequently been identified in a wide variety of organisms ranging from viruses and bacteria to mammals, making it the most broadly distributed family of RNA-degrading enzymes known (for review, see Irie, 1997; Trubia et al., 1997). In particular, this family has a much broader distribution than the extensively described RNase A superfamily, the occurrence of which is limited to vertebrates. The only enzymes in the T2 family for which the in vivo role is known are the S-RNases, involved in gametophytic self-incompatibility in plants. It has been shown that S-RNase expression is sufficient to function as the stylar component of self-incompatibility in some Solanaceous plants (Lee et al., 1994; Murfett et al., 1994), and that the RNase activity of the proteins is required in this process (Huang et al., 1994).
Much less is known about the roles of the other major group of plant RNases in the T2 family, the S-like RNases. Although they are close molecular relatives to the S-RNases, the S-like RNases have important differences in structure, expression, and function (for review, see Bariola and Green, 1997). Most notably, they do not participate in the control of self-incompatibility and their genes can be induced in response to specific stimuli. To our knowledge, S-like RNase genes have been found in all plants that have been examined for their presence, indicating that they constitute a major family of RNA-degrading enzymes in plants. In contrast to the S-RNase genes, whose expression is generally restricted to the style, S-like RNase genes are often expressed in other organs under certain environmental conditions. For example, these genes are induced by senescence in Arabidopsis (Taylor et al., 1993; Bariola et al., 1994) and tomato (Lers et al., 1998), and by Pi starvation in several different plant species (Taylor et al., 1993; Bariola et al., 1994; Köck et al., 1995; Dodds et al., 1996). This suggests that S-like RNases may participate in the related processes of nutrient recycling during senescence and scavenging Pi sequestered in RNA, in combination with the actions of phosphatases during starvation for Pi. S-like RNase genes are induced in zinnia by tracheal-element differentiation and wounding (Ye and Droste, 1996), processes that may also have nutrient-recycling aspects.
The plant species that have been investigated thus far all contain multiple, highly regulated S-like RNase genes. Gene-specific probes have made it possible to study the expression of individual S-like RNase genes at the RNA level. However, the lack of specific antibodies has limited the analysis of individual RNases at the protein level for studies on regulation and localization. Another limitation has been the lack of mutant plants altered specifically in the expression of individual S-like RNase genes, plants that could provide useful functional insights.
In Arabidopsis, the system in which S-like RNase genes have been most extensively characterized, the S-like RNase family consists of three genes, RNS1, RNS2, and RNS3, each with a unique pattern of expression (Taylor et al., 1993; Bariola et al., 1994). All three genes are induced by senescence, although to varying extents. Among the three, RNS1 and RNS2 are induced by Pi starvation, whereas RNS3 is not. RNS2 in general has a higher level of basal expression than the other two genes, and its transcript is more widely distributed. These studies have been carried out almost exclusively at the RNA level. The RNS proteins were postulated to have different subcellular locations: RNS1 and RNS3 were predicted to be extracellular proteins, whereas RNS2 may be vacuolar due to the presence of a C-terminal extension not present in RNS1 and RNS3 (Taylor et al., 1993; Bariola et al., 1994).
We describe the analyses of RNS1 and RNS2 using specific antibodies. We demonstrate that production of both proteins is induced during Pi starvation, but that RNS1 is secreted, whereas RNS2 remains intracellular. We also demonstrate that decreasing the expression of these genes individually in transgenic antisense plants is sufficient to induce anthocyanin overproduction, indicating that decreases in the levels of the corresponding proteins lead to physiological effects that may not be compensated for by the actions of other RNases.
MATERIALS AND METHODS
Antibody Preparation
RNS1 protein for use as an antigen was heterologously produced in yeast as described previously (Bariola et al., 1994). Liquid, minimal-dextrose, low-Pi medium (250 mL; Thill et al., 1983) was inoculated with yeast (Saccharomyces cerevisiae strain BJ2168) containing an RNS1 expression construction (Bariola et al., 1994) and grown until saturation (about 48 h). Cells were removed by centrifugation, the supernatant was concentrated to 0.5% of its original volume in Centriprep-10 units (Amicon, Beverly, MA), and the buffer was replaced with 20 mm Mes, pH 6.0. The supernatant was loaded onto a Mono-Q HR 5/5 column (Pharmacia) to which RNS1 adhered at pH 6.0. Bound proteins were eluted with a gradient of 0 to 0.25 m NaCl, and eluted fractions were analyzed by SDS-PAGE and silver staining.
RNS1 eluted between 0.15 and 0.18 m NaCl. Pooling of the RNS1-containing fractions resulted in a preparation >95% pure in RNS1, as monitored by SDS-PAGE and silver staining (data not shown). The absence of major contaminating proteins was also confirmed by IEF and silver staining (data not shown). A synthetic peptide, PG1, was prepared as an antigen to produce specific anti-RNS2 antibodies with the sequence CYRSDFKEKE. Peptide PG1 was prepared at the Michigan State University Department of Biochemistry Macromolecular Structure Facility. The peptide was coupled to maleimide-activated KLH (keyhole limpet hemocyanin) carrier protein (Pierce).
Samples of RNS1 protein or peptide PG1-KLH complex were emulsified with TiterMax adjuvant (CytRx, Norcross, GA) and injected subcutaneously into two female New Zealand White rabbits following collection of preimmune serum. Initial injections of RNS1 contained 80 μg of protein, followed approximately 3 weeks later by 300-μg boosts. Peptide PG1 coupled to KLH was administered at 270 μg in the first injection and 400 μg in the boosts. Blood was collected 10 d after the boosts and at 2-week intervals thereafter. Sera were screened for anti-RNS1- or anti-RNS2-binding ability by testing various dilutions on immunoblots containing yeast-produced RNS1 or RNS2 as appropriate (described below) or, in the case of anti-RNS2 sera, on dot blots containing spotted peptides.
Plant Material and Protein Sample Preparation
All Arabidopsis tissues described in this report were of the Columbia ecotype. Roots, stems, leaves, flowers, and total aboveground tissues from soil-grown plants were grown and harvested as described previously (Bariola et al., 1994). Seedlings grown on Pi-rich and -deficient media were grown and harvested as described previously (Bariola et al., 1994), except that harvesting took place 2 d following transfer to the media instead of 3 d. Green siliques 5 to 15 mm in length were collected from 4-week-old plants. Seeds used for protein extraction were viable, dry seeds that had been stored at room temperature for approximately 1 year. Proteins were extracted from harvested tissues as described previously (Bariola et al., 1994).
The Arabidopsis liquid cell culture line T87-C33 of ecotype Columbia (Axelos et al., 1992) was grown either as described previously (Bar-Peled and Raikhel, 1997) or dark grown at 26°C with 150 rpm shaking. After 5 d of growth following subculturing, cells and culture medium were separated by centrifugation at 200g for 5 min. Cells were frozen at −80°C and were later ground to a fine powder under liquid nitrogen and mixed with an approximately equal volume of extraction buffer that has been described previously (Bariola et al., 1994). The supernatant resulting from centrifugation of this slurry at 15,000g for 10 min at 4°C was concentrated in a Centricon 10 unit (Amicon) to one-half of the original volume. The culture medium from which the cells were harvested was dialyzed against a buffer containing 25 mm Hepes, pH 7.5, 40 mm KCl, and 0.1 mm EDTA, using Spectra-Por tubing (Spectrum Medical Industries, Los Angeles, CA) with a Mr cutoff of 3,500. Subsequently, the sample was lyophilized to dryness, redissolved in a small volume of buffer, redialyzed against the same buffer, and finally concentrated in a Centricon-10 unit to 0.4% of the original volume. Protoplasts were prepared from cell cultures essentially as described previously (Bar-Peled and Raikhel, 1997). Protoplast extracts were obtained by adding the previously described extraction buffer (Bariola et al., 1994) to 10% (v/v) and lysing the protoplasts by passing them several times through a pipette tip, followed by centrifugation to remove cell debris.
RNS1, RNS2, and RNS3 proteins were produced heterologously in yeast as described previously (Taylor et al., 1993; Bariola et al., 1994), and concentrated in Centricon-10 units to approximately 0.5% of the original volumes. Glycerol was added to 10% (v/v) in all protein extracts, and samples were stored in small aliquots at −80°C.
Protein Gels and Immunoblot Analysis
After quantitation using the Bradford assay (Bio-Rad), protein extracts were mixed with sample buffer and boiled for 5 min before being separated on 11% SDS-PAGE gels (Laemmli, 1970). Proteins were then either visualized using Coomassie-blue staining, or blotted onto PVDF membranes (Immobilon-P, Millipore) using semidry electrophoretic transfer, as described previously (Harlow and Lane, 1988). After transfer but before drying, membranes destined for incubation with anti-RNS2 antibodies were autoclaved in transfer buffer for 20 min at 120°C, as described previously (Swerdlow et al., 1986), because autoclaving increases the signal strength of RNS2 detection by the anti-RNS2 antibodies described above (data not shown).
Blots were processed for RNS2 signal detection using one of two protocols: one with TBS-Tween buffer (Birkett et al., 1985) and another with Blotto/Tween buffer (Harlow and Lane, 1988) as the blocking agent. For signal detection anti-RNS1 serum was diluted 1:1000 and anti-PG1 (anti-RNS2) serum was diluted 1:2000. Goat-anti-rabbit IgG:alkaline phosphatase conjugate (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was used as the secondary antibody in both cases. Signals were developed by using nitroblue tetrazolium/5-chloro-4-bromo-3-indolyl phosphate as the substrates (Harlow and Lane, 1988) and incubating in development buffer for 10 min. Dot blots with peptides were prepared by spotting 1 μL of 5 mg mL−1 solutions of peptide onto strips of nitrocellulose membrane. For signal detection the dot blots were incubated with various dilutions of sera and processed as described above.
Plasmid Constructions
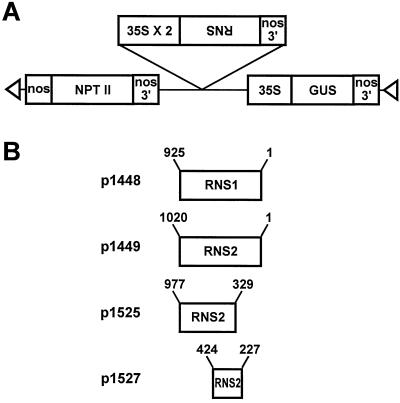
To construct the RNS1 and RNS2 antisense plant-transformation plasmids, RNS1 or RNS2 cDNA fragments were first fused in antisense orientation between a doubly-enhanced cauliflower mosaic virus 35S promoter and a nos 3′ end in plasmid p1079 (Diehn et al., 1998). The cauliflower mosaic virus 35S-antisense RNS-nos cassette was then excised and inserted into the HindIII site of pBI121 (Jefferson, 1987) so that the cassette was oriented in the same direction relative to the GUS cassette of pBI121. This resulted in the construction of four antisense plasmids that were used for plant transformation: p1448, a BamHI-SalI fragment encoding the entire RNS1 cDNA (Taylor and Green, 1991); p1449, a BamHI-KpnI fragment encoding the entire RNS2 cDNA (Taylor and Green, 1991); p1525, a 648-bp ScaI-EcoRV fragment (nucleotides 329–977; Taylor et al., 1993) of the RNS2 cDNA; and p1527, a 208-bp EcoRI-XhoI fragment (PCR product corresponding to nucleotides 227–424; Taylor and Green, 1991; Taylor et al., 1993) of the RNS2 cDNA. Additional details of plasmid construction are available upon request.
Generation of Transgenic Plants
Plasmids p1448, p1449, p1525, and p1527 were transformed into Agrobacterium tumefaciens strain GV3101 C58C1 Rifr (pMP90) (Koncz and Schell, 1986) via electroporation using a Gene-Pulser apparatus (Bio-Rad) according to the manufacturer's recommendations. The T-DNA regions of the plasmids were inserted into Arabidopsis with the vacuum-infiltration method of A. tumefaciens-mediated transformation. Previous protocols for this method (Bechtold et al., 1993; Bent et al., 1994; T. Araki, personal communication) were modified or combined. Rosettes of 4-week-old plants with bolts of 5 to 15 cm were submerged in a solution of A. tumefaciens containing the plasmid of interest and subjected to a vacuum of 400 mm Hg for 5 min. The vacuum was quickly broken and the plants were allowed to recover and set seed under normal growth conditions. Details of this protocol can be viewed on the Web at http://www.bch.msu.edu/pamgreen/green.htm#prot. Seeds from these plants were plated on solid Arabidopsis growth medium (Taylor et al., 1993) containing 50 μg mL−1 kanamycin. One antibiotic-resistant seedling from each plant that had originally been infiltrated was transferred to soil to be certain that all plants analyzed were the result of unique integration events. To ensure unbiased selection of transformants to be transferred to soil, the antibiotic-resistant plant closest to the edge of each plate was selected.
Analyses of Transgenic Plants
Antisense RNS1 Plants
One-hundred-twenty independent kanamycin-resistant transformants of p1448 were analyzed for reduced RNS1 activity. Original transformants (T1 generation) and wild-type plants were grown in soil under conditions described previously (Bariola et al., 1994). Flowers were collected from 4- to 5-week-old plants on 1 d only per plant; all flowers on the plant that were in the range of development from buds with petals showing to fully open flowers that did not yet have developing siliques protruding were collected. Flowers were frozen on dry ice and stored at −80°C. Flower proteins were extracted as described for other tissues (Bariola et al., 1994).
RNS1 activity was analyzed by electrophoresing 20 μg of flower proteins from each transformed plant on RNase activity gels (Yen and Green, 1991). The intensity of the RNase activity band corresponding to RNS1 was compared in transformed lines and in the wild type. For lines with decreased flower RNS1 activity, seeds were collected, one randomly selected kanamycin-resistant progeny plant (T2 generation) was grown in soil, and flowers from each plant were analyzed for RNS1 activity as for the T1 generation. Seeds from lines that still appeared to have lowered flower RNS1 activity were once again selected for kanamycin resistance, and several resistant plants (the T3 generation) were moved to soil and flowers were screened for lowered RNS1 activity. In some cases the T4 generation was screened in the same manner.
Seeds of T3 or T4 lines were plated on mesh circles on Arabidopsis growth medium containing kanamycin, moved after 2 d to kanamycin-containing Pi-rich or -deficient media, and harvested 7 d later, as previously described for wild-type seeds (Bariola et al., 1994). Control lines included in these experiments were the wild type (grown on medium without kanamycin) and transgenic lines containing the pBI121 vector. Any kanamycin-sensitive seedlings were removed before harvesting. Harvested seedlings were frozen in liquid nitrogen for RNA extraction and on dry ice for protein extraction. Total RNA was isolated as described previously (Newman et al., 1993). RNA gels were prepared and blotted to nylon membrane.
The RNA gel blots were probed first with a 32P-labeled eIF4A probe as described previously (Taylor et al., 1993). The blots were then stripped as described previously (Bariola et al., 1994) and reprobed with a 32P-labeled RNS1 antisense RNA probe such that only sense RNA strands would be detected. The antisense RNA probe was made using a kit (Riboprobe, Promega) and corresponded to the entire RNS1 cDNA. For this probe, a different hybridization buffer was used, increasing the formaldehyde concentration to 50% and decreasing the SSC concentration to 1×, and hybridization was overnight at 65°C. Quantitation of signal in RNS1 and eIF4A bands was achieved using a phosphor imager (Molecular Dynamics, Sunnyvale, CA). Proteins were extracted from seedlings and electrophoresed on RNase activity gels as described previously (Bariola et al., 1994), and electrophoresed and blotted to a PVDF membrane for immunoblots as described above.
Antisense RNS2 Plants
For the first strategy, 119 independent kanamycin-resistant transformants of p1449 were analyzed for decreased amounts of RNS2 in leaves using immunoblots, as described above. In addition, 74 independent kanamycin-resistant transformants of p1525 and 63 of p1527 were screened for decreased RNS2 mRNA levels by RNA gel-blot analysis. As described above, the original transformants (T1 generation) and wild-type plants were grown in soil. Several healthy, nonsenescing leaves were collected from 4- to 5-week-old plants on 1 d only per plant, frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted from leaves, and RNA gel blots were prepared and probed as described above, except that the antisense RNS1 probe was replaced by an antisense RNS2 probe corresponding to the entire RNS2 cDNA. Levels of RNS2 and eIF4A mRNA were measured with a phosphor imager, the RNS2 to eIF4A ratio was calculated for each line, and these results were divided by the RNS2 to eIF4A ratio of the wild-type sample on the same blot to calculate the relative level of RNS2 mRNA in putative antisense RNS2 lines. Lines in which the level of RNS2 mRNA was less than or equal to 70% of that in the wild type were selected for rescreening. Several kanamycin-resistant progeny (T2 generation) were grown in soil, the leaves were harvested, RNA gel-blot analysis was performed, and relative RNS2 mRNA levels were calculated as they were for the T1 generation.
Anthocyanin Assays
For assay of anthocyanin content in antisense lines, T3 or T4 seedlings (for antisense RNS1 lines) or T3 seedlings (for antisense RNS2 lines) were grown on Pi-rich or -deficient media and harvested 7 d after transfer, as described above. Fresh weight was recorded for each sample, and ranged from 0.08 to 0.3 g per sample. Seedlings were frozen in liquid nitrogen, lyophilized in 13-mL plastic test tubes (Sarstedt, Nümbrecht Germany), and pulverized with 3-mm-diameter glass beads (Fisher Scientific). Anthocyanin content from each line was measured using a procedure based on the methods of Rabino and Mancinelli (1986), Feinbaum and Ausubel (1988), and Kubasek et al. (1992). Ground tissue was gently shaken in 2.5 mL of 1% HCl/methanol for 2 h at room temperature, 2 mL of chloroform was added, the mixture was vortexed, 5 mL of water was added, and the vortex step was repeated.
After separating the phases by centrifugation, 1 mL of the aqueous/methanol phase was assayed. A530 minus A657 was used as a measure of anthocyanin content; values were normalized to the fresh weight of each sample. For antisense RNS1 plants, two separate trials were performed, each including four plates for each line (two on Pi-rich and two on Pi-deficient media) so four readings were incorporated for each data point. For antisense RNS2 plants, three to four trials were performed, incorporating six to eight readings for each data point. Within each trial results were normalized such that one plate of wild-type seedlings grown on Pi-rich medium was assigned a value of 1, and the other data were adjusted proportionately. Data from the different experiments were averaged to arrive at the final values.
RESULTS
Production of Anti-RNS1 and Anti-RNS2 Antibodies
Although the expression of the RNS1 and RNS2 genes has been analyzed extensively at the RNA level (Taylor et al., 1993; Bariola et al., 1994), generating antibodies specific for RNS1 and RNS2 became necessary to obtain further information about the regulation of the proteins. The heterologous yeast expression system previously used to produce the RNS proteins (Bariola et al., 1994) was used to obtain RNS1 for use as an antigen. In this system amounts of RNS1 up to 9 mg L−1 are secreted into the medium, from which the protein can be easily purified. RNS1 produced in this manner was prepared in highly purified form and injected into rabbits.
The resulting antiserum contained a high titer of antibodies that recognize yeast-produced RNS1 and a protein of about 25 kD, which is close to the predicted size of 23 kD for RNS1 in extracts of Pi-starved Arabidopsis seedlings (Fig. 1A). These antibodies are not entirely specific for RNS1; RNS3 protein produced in yeast is also detected at an approximately 10-fold lower efficiency (data not shown); the RNS3 band is faintly visible in Figure 1A. However, RNS3 has a slightly greater electrophoretic mobility than RNS1, so these antibodies are adequate for the specific detection of RNS1 in immunoblots of Arabidopsis tissues. In addition, efforts to detect RNS3 in plant extracts using these antibodies have been unsuccessful. Preimmune serum reacts very little with proteins in Arabidopsis extracts (data not shown).
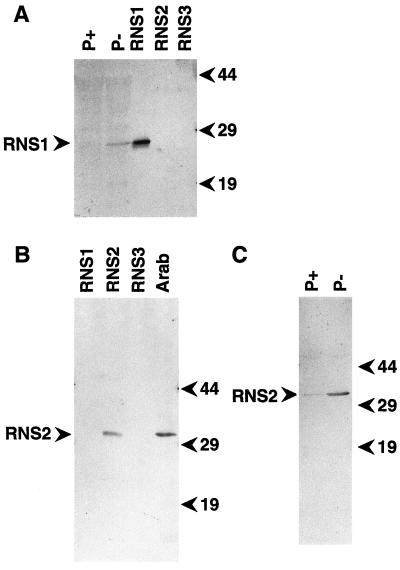
Figure 1.
Immunoblot characterization of anti-RNS1 and anti-RNS2 antibodies and RNS1 and RNS2 regulation in response to Pi starvation. Protein samples were resolved by SDS-PAGE, transferred to a membrane, and immunodetected with the corresponding antiserum. A, Immunoblot with lanes containing approximately 70 ng of RNS1, RNS2, or RNS3 in supernatant from RNase-expressing yeast cells as indicated or 30 μg of total proteins from Arabidopsis seedlings grown on media rich (P+) or deficient (P−) in Pi. The blot was developed using anti-RNS1 antibodies. B, Immunoblot with lanes containing approximately 300 ng of RNS1, RNS2, or RNS3, as indicated, from supernatant from RNase-expressing yeast cells. Arab, Lane containing 50 μg of proteins extracted from aboveground tissues of 5-week-old wild-type Arabidopsis plants. The blot was developed using anti-RNS2 antibodies. C, Increase in RNS2 abundance during Pi starvation. Lanes contain 25 μg of protein extracts from seedlings grown on media rich (P+) or deficient (P−) in Pi. The blot was developed using the same antibody as in B. Positions of molecular mass markers (in kD) are shown to the left of the blots.
A different approach was used for anti-RNS2 antibodies, because in the yeast system RNS2 is not secreted as abundantly as RNS1, making purification from this system impractical. An attempt to use as an antigen RNS2 heterologously expressed and purified from Escherichia coli did not result in sera with a high titer of antibodies (data not shown). Instead, a synthetic peptide encompassing a sequence of the RNS2 protein that differs from the corresponding regions of RNS1 and RNS3 was used (Fig. 2), with the advantage that antibodies recognizing this peptide would likely be specific to RNS2. The sequence of this peptide, PG1, is shown in Figure 2. Injection into rabbits of peptide PG1 coupled to a carrier protein led to the production of sera rich in antibodies that recognize RNS2, both yeast-produced and in Arabidopsis extracts (Fig. 1B). These antibodies do not recognize yeast-produced RNS1 or RNS3 (Fig. 1B), and preimmune serum exhibited very low reactivity with proteins in Arabidopsis extracts (data not shown). The anti-RNS2 antibodies detected a single band of approximately 32 kD in Arabidopsis seedlings (Fig. 1B). Assuming that RNS2 undergoes removal of the putative secretion signal sequence (2 kD) and glycosylation at one or both of the potential N-glycosylation sites (Taylor et al., 1993), this size corresponds well with its predicted molecular mass of 27.2 kD.
Figure 2.
Synthetic peptide used for producing anti-RNS2 antibodies. Deduced amino acid sequences of RNS1, RNS2, and RNS3 are aligned, beginning with residues 7 and 12 of RNS1 and RNS2 proteins, respectively. The region corresponding to peptide PG1 is shaded. Boxes highlight the conserved regions described in Ioerger et al. (1991).
Induction of RNS1 and RNS2 at the Protein Level in Response to Pi Starvation
As shown above, RNS1 protein was detected in immunoblots of extracts of Pi-starved seedlings (Fig. 1A). The protein was not detected in extracts of seedlings grown on a medium rich in Pi (Fig. 1A), nor in extracts of individual organs grown under standard conditions in soil (data not shown). These observations are consistent with the abundance of RNS1 mRNA under the same conditions (Bariola et al., 1994). The abundance of RNS2 protein in seedlings grown on Pi-rich or -deficient media was also investigated, using the anti-RNS2 antibodies described above. As shown in Figure 1C, RNS2 was more abundant in the Pi-deprived seedlings than in seedlings grown on a Pi-rich medium. The increased abundance of RNS2 protein during Pi starvation was similar to what was observed at the mRNA level under the same conditions (Taylor et al., 1993).
RNS2 mRNA is present in all major organs of plants grown in soil under standard long-day conditions (Taylor et al., 1993). To investigate if this distribution extends to the protein level, we carried out immunoblot analysis using anti-RNS2 antibodies. As expected, these studies demonstrated that RNS2 protein was present in roots, stems, leaves, and flowers (Fig. 3). RNS2 was also present in significant amounts in extracts of siliques and seeds.
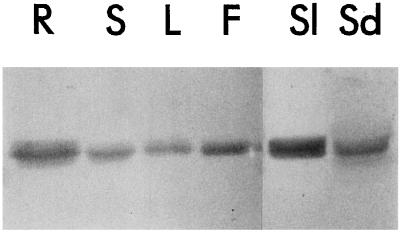
Figure 3.
Distribution of RNS2 among various organs of Arabidopsis. Protein extracts (30 μg per lane) were made from organs of 4- to 5-week-old wild-type plants and resolved by SDS-PAGE. Proteins were then transferred to a membrane and immunodetected with anti-RNS2 serum. R, Roots; S stems; L, leaves; F, flowers; Sl, siliques; Sd, seeds.
Unlike RNS2, RNS1 Is Extracellular
Determining the subcellular location of a protein can give insights into its role. Previously, all three Arabidopsis RNS proteins were predicted to enter the secretory pathway because of the presence of putative N-terminal secretory sequences (Taylor et al., 1993; Bariola et al., 1994). RNS1 and RNS3 were predicted to be extracellular proteins (Bariola et al., 1994); however, the presence of a unique C-terminal extension in the RNS2 sequence suggested that the protein might be targeted to vacuoles (Taylor et al., 1993). An Arabidopsis liquid cell culture (Axelos et al., 1992) was used to prepare samples of extracellular proteins, total cell extracts, and protoplast lysates. Immunoblots containing these samples were subjected to detection with anti-RNS1 and anti-RNS2 antibodies.
As shown in Figure 4, RNS1 was present exclusively in the extracellular fraction, confirming the prediction that RNS1 is an extracellular protein. The RNS1 band detected in the extracellular protein sample is somewhat broader than that seen in extracts of Pi-starved seedlings in Figure 1A, possibly the result of two closely spaced proteins. As mentioned above, the anti-RNS1 antibodies are slightly cross-reactive for RNS3, and RNS3 has a slightly greater electrophoretic mobility than RNS1. It is possible that the anti-RNS1 antibodies detect RNS3 in the extracellular fraction as well as RNS1, or the minor band may correspond to a modified form of RNS1.
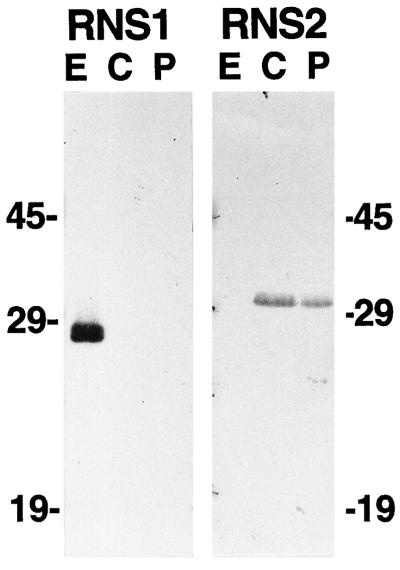
Figure 4.
Extra- and intracellular localization of RNS1 and RNS2. Protein samples were prepared from Arabidopsis cell cultures as described in Methods. Seventy-five micrograms of protein per lane was separated on SDS-PAGE gels, transferred to a membrane, and immunodetected with anti-RNS1 or anti-RNS2 serum. E, Extracellular fraction; C, whole-cell lysate; P, protoplast lysate.
In contrast to RNS1, RNS2 is undetectable in the same extracellular sample, but is strongly detected in cell extracts. RNS2 is also abundant in protoplast lysates, eliminating the possibility that the enzyme is associated with the cell wall. Results were identical whether cells were grown at 26°C in darkness or at 22°C in continuous light. It is evident that RNS2 has an intracellular location, most likely in an organelle targeted by the secretory system (such as the vacuole or the ER). The divergent in vivo locations of RNS1 and RNS2 imply that these related proteins have different functions in the plant.
Antisense Inhibition of RNS1 Expression Elevates Anthocyanin Levels
To obtain antisense RNS1 transgenic plants, Arabidopsis was transformed with p1448 (Fig. 5), which contains the entire RNS1 cDNA fused between the cauliflower mosaic virus 35S promoter and the nos 3′ end. Transgenic control lines were made by transforming plants with empty pBI121 vector. On RNase activity gels the band corresponding to RNS1 activity has been clearly identified (Bariola et al., 1994) and has the advantage that it is separated from other Arabidopsis RNase activity bands. Although this assay measures RNase activity only semiquantitatively, relative levels of activity can be easily compared for individual RNase activities, so this method was chosen to screen individual transformants for decreased RNS1 activity. Under normal growth conditions, the flower is the only major organ of the plant in which RNS1 mRNA is abundant (Bariola et al., 1994). For this reason, flowers were selected as the organ used for assay of RNS1 activity.
Figure 5.
Structure of RNS1 and RNS2 antisense constructs p1448, p1449, p1525, and p1527. A, General structure of the T-DNA region of the antisense RNS transformation vectors. B, RNS cDNA fragments fused in the antisense orientation between the doubly enhanced cauliflower mosaic virus 35S promoter and the nos 3′ end. Sequences flanking the 35S-antisense RNS-nos cassette are derived from the plasmid pBI121, as described previously (Jefferson, 1987). Numbers refer to nucleotide positions in the cDNA sequences. Triangles indicate the right and left borders of the T-DNA.
The initial generation of putative antisense RNS1 transgenic plants was in soil, and proteins extracted from flowers gathered from independently transformed plants were resolved on RNase activity gels. Of the 120 individual transformants screened in this way, 13 plants appeared to have diminished RNS1 activity compared with control plants. Of these lines, three continued to display decreased RNS1 activity in the T2 and T3 generations, but one was discarded due to kanamycin sensitivity. To monitor the effects of Pi starvation in these lines, T3 and T4 seedlings were grown on media rich or deficient in Pi. Both lines segregated as heterozygotes.
The RNase activity profiles of the antisense RNS1 lines are shown in Figure 6A. The antisense effect in these lines is more apparent in extracts of seedlings grown under Pi-deficient conditions, because RNS1 activity is low in both antisense and control seedlings grown on media rich in Pi. As shown in Figure 6A, lines 8d.5.2 and 8d.5.3 (two progeny lines from T2 line 8d.5) and line 23g.4 all have significantly lower RNS1 activity than the wild type and the vector (34d.3) controls when grown on Pi-deficient medium. As expected for sibling lines, 8d.5.2 and 8d.5.3 displayed an antisense effect of similar magnitude. Line 23g.4 exhibited even lower RNS1 activity under Pi-deficient conditions and also appeared to have less activity in high-Pi conditions than all of the other lines shown in Figure 6A. Line 36h.3.2, which carries the antisense RNS1 construct but exhibits only a weak antisense effect, was included for comparison.
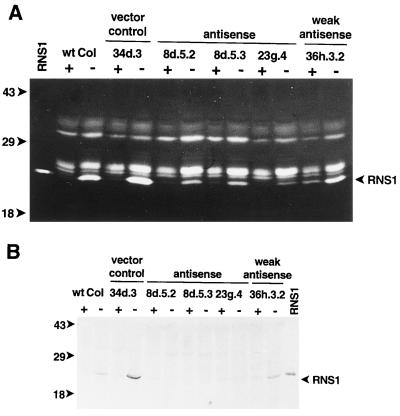
Figure 6.
Decreased RNS1 activity and protein in RNS1 antisense lines. T3 or T4 seedlings of antisense RNS1 lines were germinated on AGM medium, transferred 2 d after germination to media rich (+) or deficient (−) in Pi, and grown for an additional 7 d. Protein extracts were prepared from all kanamycin-resistant seedlings and each sample was resolved on RNase activity gels (A, 50 μg per lane) or SDS-PAGE gels for immunoblots (B, 100 μg per lane). wt Col, Columbia wild type; vector control, transgenic line containing pBI121 vector; weak antisense, transgenic line containing construction p1448 but with a near-normal amount of RNS1 activity; RNS1, RNS1 protein produced in yeast. The bands of RNS1 activity are indicated. Positions of molecular mass standards (in kD) are shown to the left of the gels.
To determine whether the amounts of RNS1 protein in the lines described above would reflect the amount of RNS1 activity seen on activity gels, we used immunoblots to investigate levels of the protein. Using the anti-RNS1 antiserum described above, the reduction in RNS1 protein levels appeared as expected in the antisense RNS1 lines. RNS1 protein was clearly detected in Pi-starved extracts of the wild type as well as in the 34d.3 (vector control) and the 36h.3.2 (weak antisense) lines (Fig. 6B), all of which have more RNS1 activity (Fig. 6A) and RNS1 mRNA (see below and Fig. 7) than the antisense lines. However, little if any RNS1 appeared in extracts of the three antisense lines (Fig. 6B), in contrast to the reduced but visible levels of RNS1 activity observed in Figure 6A, which was probably due to the differences in sensitivity between RNase activity gels and immunoblots. In any case, it is clear that the antisense lines had markedly reduced amounts of RNS1 protein.
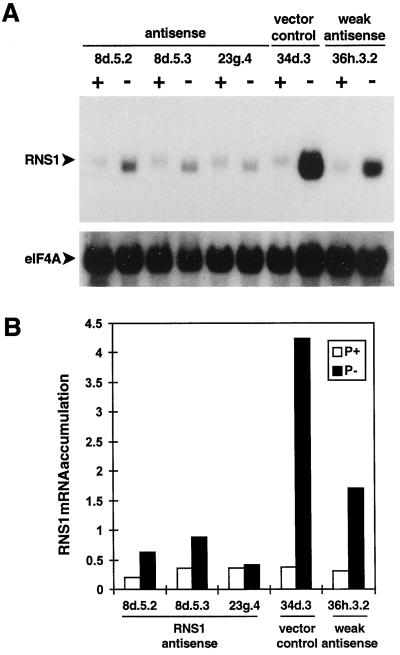
Figure 7.
Decreased RNS1 mRNA levels in RNS1 antisense lines. Seedlings of antisense RNS1 lines were grown as described in Figure 6, and total RNA was isolated from all kanamycin-resistant seedlings. A, RNA gel blots containing 10 μg of sample per lane were hybridized to an eIF4A probe and subsequently to the antisense RNS1 probe. Labels are the same as in Figure 6. B, RNS1 and eIF4A counts were quantitated with a phosphor imager, RNS1 counts were divided by eIF4A counts for each lane, and the results were plotted to represent RNS1 mRNA accumulation in each line.
RNS1 gene expression in the above lines was examined by hybridizing total RNA isolated from the same batches of tissue harvested for Figure 6 with an RNS1 RNA probe that hybridizes only with the sense RNS1 mRNA (Fig. 7A). Again, the antisense effect was most visible in Pi-deprived seedlings. As RNS1 mRNA levels were similar under high-Pi conditions in all of the lines shown, we quantitated antisense suppression of RNS1 by examining RNS1 transcript levels during Pi starvation following normalization with eIF4A (Taylor et al., 1993) (Fig. 7B). The 34d.3 vector control line was induced 11.3-fold by this stimulus, whereas the 8d.5.2, 8d.5.3, and 23g.4 lines were induced only 3.1-, 2.4-, and 1.1-fold, respectively. Although RNS1 expression was not completely abolished in line 23g.4, its RNS1 mRNA level during Pi starvation was only 10% of that of the 34d.3 control. The levels of RNS2 and RNS3 mRNA and the induction of RNS2 by Pi starvation were normal in the antisense RNS1 plants (data not shown), indicating that the antisense effect is specific to RNS1.
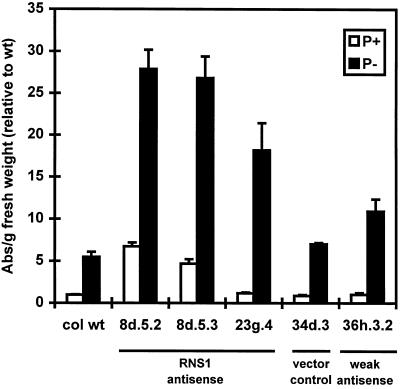
A striking phenotype displayed by the three antisense RNS1 lines was the increased accumulation of anthocyanins. This phenomenon was most evident when the plants were deprived of Pi. When grown for 7 d on selective medium deficient in Pi, seedlings of the antisense plants contained 2.6 to 4.0 times the amount of anthocyanins present in the wild-type and vector control (34d.3) plants (Fig. 8). In addition, lines 8d.5.2 and 8d.5.3 grown on Pi-rich medium contained amounts of anthocyanins comparable to those seen in Pi-deprived control plants. Although there was not a precise quantitative correlation between inhibition of RNS1 transcript levels and anthocyanin content, seedlings of line 36h.3.2 (weak antisense line) starved for Pi had RNS1 mRNA levels between those of the antisense lines and the control lines (Fig. 7), and had anthocyanin levels between those of the two groups.
Figure 8.
Quantitation of anthocyanin levels in seedlings of RNS1 antisense lines. Seedlings of antisense RNS1 lines were grown as described in Figure 6, and anthocyanins were extracted as described in Methods. Each bar represents four independent plates. A530 minus A657 was taken as a measure of anthocyanin content, and for each sample the absorbance (Abs) reading was divided by the fresh weight of the sample in grams. The results were normalized using an arbitrary value of 1 for the wild-type line (col wt) grown on Pi-rich medium (1 = 0.14 absorbance units per gram fresh weight). Labels are the same as in Figure 6. Error bars correspond to ±se.
Line 23g.4 displayed an additional unique phenotype. In the original T1 plant, as well as in some of the descendants of this plant, the following traits were observed in comparison with wild type when the plants were grown in soil: smaller leaves, shorter and thinner stems, reduced seed set, and decreased apical dominance. This phenotype had incomplete penetrance in the 23g descendancy, being evident in only one-half to one-third of the progeny of each generation. This phenotype is similar to that of the pho1 mutant of Arabidopsis, which is impaired in uptake of Pi (Poirier et al., 1991), although the symptoms in line 23g.4 and its progeny are less severe than in pho1.
Anthocyanin Content Is Also Elevated by Antisense Inhibition of RNS2
The same approach used to produce antisense RNS1 plants was used in the initial attempt to obtain antisense RNS2 plants. As shown in Figure 5, the entire RNS2 cDNA was fused in reverse orientation into a T-DNA construction to form p1449, a plasmid analogous to the antisense RNS1 construction. By assaying leaf-protein extracts of soil-grown plants, 119 independent transformants of this plasmid were screened for decreased RNS2 levels using immunoblots probed with the anti-RNS2 antibodies described earlier. This method was used because the high basal level of RNS2 protein in plants allowed this manner of screening to be carried out easily; in addition, RNS2 has not yet been identified in the RNase activity profile of Arabidopsis, so screening using the same method as for the antisense RNS1 plants was not possible. In this population of transgenic plants, RNS2 levels varied to a small extent, but no plants with dramatically decreased amounts of RNS2 were observed.
There is no consensus in the literature as to which portions of plant genes give the best results with antisense techniques (Bourque, 1995); the size and portion of a given cDNA that results in the greatest antisense effect appears to be gene dependent. For this reason, another attempt was made to obtain antisense RNS2 plants by introducing smaller portions of the RNS2 cDNA into transformation vectors and generating putative antisense plants for screening. Two more transformation vectors were constructed. The first construction, p1525 (Fig. 5), included the final two-thirds of the RNS2 cDNA, a fragment of about 650 bp. The second vector, p1527 (Fig. 5), incorporated a fragment of approximately 200 bp between and including the conserved active site regions (for review, see Irie, 1997). Transformants were screened by analyzing RNA from leaves of soil-grown plants for decreased amounts of RNS2 mRNA on gel blots relative to the internal standard eIF4A. We used this method instead of screening RNase activities on immunoblots, as we had done for the first set of transformants, because it offers a greater ease of quantitation and normalization of the RNS2 signal.
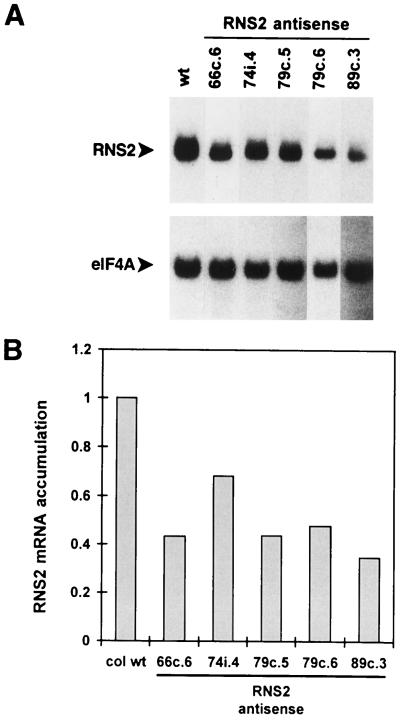
Using this approach, 74 independent transformants of p1525 and 63 of p1527 were screened. Of the 137 plants screened, only 19 had leaf RNS2 mRNA levels less than or equal to 70% that of wild-type plants, and of the 19, only 4 had less than 35% that of the wild type. We screened several antibiotic-resistant T2 progeny of each of these lines in the same way. Progeny of only 4 of the 19 T1 plants showed diminished RNS2 mRNA levels. RNA gel-blot analysis of several of these plants is shown in Figure 9, including two separate progeny lines of the 79c T1 line. In these plants levels of RNS2 mRNA ranged from 35% to 68% of that in wild-type plants. Three of these four lines (74i.4, the 79c lines, and 89c.3) resulted from transformation with plasmid p1527.
Figure 9.
RNS2 mRNA levels in RNS2 antisense lines. A, Total RNA was extracted from leaves of 4-week-old kanamycin-resistant, soil-grown antisense RNS2 lines. RNA gel blots containing 6 μg of sample per lane were hybridized to an antisense RNS2 probe and subsequently to the eIF4A probe. wt, Wild type. B, RNS2 and eIF4A counts for the bands shown in A were quantitated with a phosphor imager. RNS2 counts were divided by eIF4A counts for each lane, and these results were divided by the RNS2 to eIF4A ratio for the wild-type control lane to show relative differences in RNS2 mRNA levels. col wt, Columbia wild type.
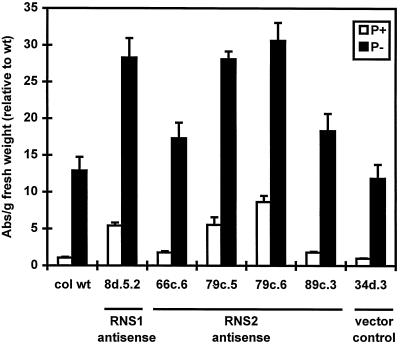
When grown under normal conditions in soil, the RNS2 antisense lines displayed no unusual phenotype, although the leaves of several lines appeared slightly more purple than those of the wild type. For this reason, we tested seedlings of the RNS2 antisense lines for anthocyanin content, as we had for the antisense RNS1 lines. The anthocyanin content of these lines is shown in Figure 10 (except line 74i.4, which could not be measured due to partial kanamycin-sensitivity problems). As with the RNS1 antisense plants, the effect of RNS2 antisense on anthocyanin levels was most evident when the plants were grown on Pi-deficient medium. All of the lines contained elevated amounts of anthocyanins, ranging from 1.5 to 2.6 times that of control plants when grown for 7 d on Pi-deficient medium. Similar to the instance of the RNS1 antisense plants, there was no strict correlation between RNS2 transcript inhibition and anthocyanin levels; the lines with the lowest RNS2 transcript levels did not have the highest anthocyanin content. However, it is clear that reduction of RNS2 transcript levels, even to only 48% (line 79c.6) of that of control plants, was sufficient to elevate anthocyanin levels.
Figure 10.
Seedlings of RNS2 antisense lines grown as described for antisense RNS1 lines in Figure 6. Anthocyanins were extracted as described in Methods. Each bar represents at least six independent plates. Absorbance was measured and results were normalized as in Figure 8 (1 = 0.14 absorbance units per gram fresh weight). An RNS1 antisense line (8d.5.2) was included to facilitate comparison. col wt, Columbia wild type. Error bars correspond to ±se.
DISCUSSION
We examined the regulation and localization of the RNS1 and RNS2 proteins, as well as the effects of diminishing their levels via antisense. First, use of antibodies that detect RNS1 and RNS2 individually showed that RNS1 and RNS2 protein levels increase dramatically during Pi starvation and coordinate with increases previously reported for mRNA accumulation (Taylor et al., 1993; Bariola et al., 1994). These same antibodies demonstrated that RNS1 and RNS2 have different subcellular locations, with RNS1 located extracellularly and RNS2 located intracellularly. Finally, although RNS1 and RNS2 are closely related proteins, diminishing the level of either of these RNases individually leads to elevated levels of anthocyanins in Arabidopsis.
Obtaining antisera that recognize individual RNS proteins is an important first step in dissecting the contributions of individual RNase activities to the physiology of Arabidopsis. Although the deduced amino acid sequences of RNS1 and RNS2 are 37% identical (Bariola et al., 1994), antibodies generated against the RNS1 full-length protein did not cross-react with RNS2. These antibodies cross-reacted only very weakly with RNS3 produced in yeast, despite 61% identity between the RNS1 and RNS3 protein sequences. Immunoblots using these antibodies showed a large increase in abundance of RNS1 protein during Pi starvation in Arabidopsis seedlings, mirroring the increases in RNS1 mRNA abundance and RNS1 activity seen on RNase activity gels in seedlings treated identically.
As expected, the antibodies generated against the PG1 peptide, corresponding to a sequence unique to RNS2, were specific to RNS2. This is particularly important because RNS2 has not yet been identified in the RNase profile of Arabidopsis, as observed on activity gels. Therefore, the RNS2-specific antiserum provides, to our knowledge, the first means for characterizing RNS2 regulation at the protein level. Similar to what was seen with RNS1, the anti-RNS2 antibodies confirmed an increased accumulation of RNS2 during Pi starvation, as expected from the induction of the RNS2 gene during this stimulus (Taylor et al., 1993).
It has been suggested previously that RNS1 is probably an extracellular protein. The RNS1 cDNA encodes an N-terminal signal sequence and has no other features suggesting that the protein would be retained and not secreted from the cell (Bariola et al., 1994). In addition, among the known S-like RNases, RNS1 is most similar to the tomato RNase LE (Bariola and Green, 1997), which has been shown to have an extracellular location (Nürnberger et al., 1990). In this study we used an Arabidopsis liquid cell culture to examine the location of RNS1 in immunoblots with the antibodies described above and RNase activity gels (data not shown). Both techniques showed that RNS1 is exclusively detected in the extracellular fraction.
In contrast, RNS2 was predicted to have an intracellular location. Like RNS1, the deduced RNS2 protein sequence contains an N-terminal signal sequence, and thus presumably enters the secretory pathway of the cell (Taylor et al., 1993). However, an obvious feature of the RNS2 sequence that differs from those of RNS1 and RNS3 is a C-terminal extension of 19 amino acids. This sequence has some features in common with sequences of confirmed C-terminal vacuolar-targeting sequences from plant proteins (Taylor et al., 1993), suggesting that RNS2 may be a vacuolar protein. Evidence for both vacuolar and ER-localized S-like RNases has been obtained in tomato (Löffler et al., 1992, 1993; Köck et al., 1995). The last four residues of the RNS2 protein sequence are REAL, a sequence that bears some resemblance to a motif typical of ER-retention signals known to function in plant and mammalian cells (Gomord and Faye, 1996). The only apparent discrepancy in this sequence, compared with ER-retention signals, is the presence of Ala at the third position, which to our knowledge has not been reported previously in any ER-retention sequence.
Although the experiments presented in this report do not differentiate between an ER and a vacuolar location for RNS2, the intracellular location demonstrated in Figure 4 is consistent with both possibilities. It is also possible that RNS2 is targeted to more than one intracellular compartment, which has been shown to occur in plants with at least one variety of ER-retention signal (Gomord et al., 1997).
To view the effects of diminishing levels of RNS1 and RNS2 individually in Arabidopsis, transgenic plants containing antisense constructions for RNS1 and RNS2 were generated. The low rate of inhibition of RNS1 among transgenic plants carrying the antisense RNS1 construction, of which only 2 out of 120 transgenic plant lines showed an inhibition of more than 80% at the RNA level during starvation for Pi, was not entirely unexpected, because attempts to decrease S3 S-RNase protein levels in Petunia inflata resulted in only 13% of transgenic plants with significantly lowered S3 levels (Lee et al., 1994). RNS2 inhibition was even less efficient: Despite the screening of over 250 transgenic plants containing one of three different antisense RNS2 constructions, the greatest inhibition of RNS2 observed was 35% that of wild type. Why some genes are less sensitive to antisense inhibition than others is unknown at present. It is possible that decreasing RNS2 expression further than 35% of control is lethal to the plant.
It is clear that elevated anthocyanin production is caused by the specific, albeit partial, inhibition of RNS1 and of RNS2. This effect was more pronounced in antisense RNS1 plants, possibly due to the proportionately greater inhibition of RNS1 expression. There are at least two possible explanations for this phenomenon. First, it is known that Pi starvation leads to increased anthocyanin accumulation in Arabidopsis (Trull et al., 1997). It has been proposed that RNS1 and RNS2 function as part of a Pi-starvation rescue system involved in the scavenging of Pi from RNA in the extracellular space or possibly from an intracellular compartment. It is possible that the elevated anthocyanin levels in antisense RNS1 and RNS2 plants are the result of the plants lacking an ability to recycle internal Pi and therefore showing symptoms of Pi starvation.
It is known that anthocyanin production is also triggered by numerous stress conditions such as wounding, low temperature, high light intensity, pathogen attack, and exposure to ozone (for review, see Mol et al., 1996; Trull et al., 1997). It could be that diminishing levels of RNS1 or RNS2 protein subject the plant to stress other than starvation for Pi, thereby increasing anthocyanin accumulation. Examination of the mRNA levels of another Pi-starvation-inducible gene, the PAP1 acid phosphatase gene of Arabidopsis (kindly provided by T. McKnight), in antisense RNS1 and control plants did not reveal increased levels of this transcript (data not shown). However, this observation is insufficient to rule out the first model, because the extent of Pi deprivation required to induce anthocyanin biosynthesis and PAP1 expression may be different.
The phenotypic similarities between the antisense RNS1 line 23g.4 and the pho1 mutant are intriguing. The greater severity of the pho1 phenotype compared with that of 23g.4 could correlate with different extents of Pi starvation: The pho1 mutant is known to have as little as 24% of the Pi content of wild-type plants (Poirier et al., 1991). In any event, our experiments demonstrate that both RNS1 and RNS2 individually provide some physiological function to the plant that cannot be compensated for by the action of other Arabidopsis RNases. This finding is significant because Arabidopsis contains at least 16 individual RNase activities, and probably many more (Yen and Green, 1991), suggesting the existence of a complement of many RNases with specialized functions in plants.
The antibodies described in this paper will be indispensable tools in further characterization of the properties of the RNS proteins in Arabidopsis. For example, it will now be possible to determine in which intracellular compartment(s) RNS2 resides. In addition, the antibodies may be used in screens to isolate mutant plants that lack the enzymes. It would be desirable to obtain plants in which RNS1 and RNS2 expression is completely abolished to advance the studies presented in this report. This is now possible with the development of techniques to isolate plants with T-DNA insertions in the genes of interest (McKinney et al., 1995; Krysan et al., 1996). These developments, together with extensive knowledge of the structures and expression of plant S-like RNases, suggest that plants may be the most fruitful system to use in elucidating the functions of T2 family RNases, a task so far accomplished only for the S-RNases.
ACKNOWLEDGMENTS
We thank Drs. Andrew Bent and David Bouchez for advice on vacuum infiltration of Arabidopsis, Dr. Natasha Raikhel for the gift of the cell-suspension line, Nyerhovwo Tonukari for preparing the protoplasts used in Figure 4, and Don Herrington for expert rabbit care. We also thank Nicole LeBrasseur and Dr. Jay DeRocher for helpful comments on the manuscript.
Footnotes
This research was supported by the National Science Foundation (grant no. IBN-9408052) and the U.S. Department of Energy (grant no. FG0291-ER200210 to P.J.G.).
LITERATURE CITED
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplast isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- Bariola PA, Green PJ (1997) Plant ribonucleases. In G D'Alessio, JF Riordan, eds, Ribonucleases: Structures and Functions. Academic Press, New York, pp 163–190
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSED12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and golgi transport. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Birkett CR, Foster KE, Johnson L, Gull K. Use of monoclonal antibodies to analyze the expression of a multi-tubulin family. FEBS Lett. 1985;187:211–218. doi: 10.1016/0014-5793(85)81244-8. [DOI] [PubMed] [Google Scholar]
- Bourque JE. Antisense strategies for genetic manipulations in plants. Plant Sci. 1995;105:125–149. [Google Scholar]
- Diehn SH, Chiu W-L, De Rocher EJ, Green PJ. Premature polyadenylation at multiple sites within a Bacillus thuringiensis toxin gene-coding region. Plant Physiol. 1998;117:1433–1443. doi: 10.1104/pp.117.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Clarke AE, Newbigin E. Molecular characterization of an S-like RNase of Nicotiana alata that is induced by phosphate starvation. Plant Mol Biol. 1996;31:227–238. doi: 10.1007/BF00021786. [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol. 1988;8:1985–1992. doi: 10.1128/mcb.8.5.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomord V, Denmat L-A, Fitchette-Lainé A-C, Satiat-Jeunemaitre B, Hawes C, Faye L. The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 1997;11:313–325. doi: 10.1046/j.1365-313x.1997.11020313.x. [DOI] [PubMed] [Google Scholar]
- Gomord V, Faye L. Signals and mechanisms involved in intracellular transport of secreted proteins in plants. Plant Physiol Biochem. 1996;34:165–181. [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Huang S, Lee H-S, Karunanandaa B, Kao T-H. Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell. 1994;6:1021–1028. doi: 10.1105/tpc.6.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-H. Primary structural features of the self-incompatible protein in solanaceae. Sex Plant Reprod. 1991;4:81–87. [Google Scholar]
- Irie M (1997) RNase T1/T2 family RNases. In G D'Alessio, JF Riordan, eds, Ribonucleases: Structures and Functions. Academic Press, New York, pp 101–130
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Köck M, Löffler A, Abel S, Glund K. cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol Biol. 1995;27:477–485. doi: 10.1007/BF00019315. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BA, McKillop A, Goodman HM, Briggs W, Ausubel FM. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.1105/tpc.4.10.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Huang S, Kao T-h. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- Lers A, Khalchitski A, Lomaniec E, Burd S, Green PJ. Senescence-induced RNases in tomato. Plant Mol Biol. 1998;36:439–449. doi: 10.1023/a:1005993024161. [DOI] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Beintema JJ, Glund K. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiol. 1992;98:1472–1478. doi: 10.1104/pp.98.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A, Glund K, Irie M. Amino acid sequence of an intracellular, phosphate-starvation-induced ribonuclease from cultured tomato (Lycopersicon esculentum) cells. Eur J Biochem. 1993;214:627–633. doi: 10.1111/j.1432-1033.1993.tb17962.x. [DOI] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2–1 and act4–1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- Mol J, Jenkins G, Schäfer E, Weiss D. Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit Rev Plant Sci. 1996;15:525–557. [Google Scholar]
- Murfett J, Atherton TL, Mou B, Gasser CS, McClure BA. S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature. 1994;367:563–566. doi: 10.1038/367563a0. [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K. Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol. 1990;92:970–976. doi: 10.1104/pp.92.4.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry SK, Liu Y-H, Clarke AE, Newbigin E (1997) S-RNases and other plant extracellular ribonucleases. In G D'Alessio, JF Riordan, eds, Ribonucleases: Structures and Functions. Academic Press, New York, pp 191–211
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabino L, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81:922–924. doi: 10.1104/pp.81.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow PS, Finley D, Varshavsky A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal Biochem. 1986;156:147–153. doi: 10.1016/0003-2697(86)90166-1. [DOI] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, del Cardayré SB, Raines RT, Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Green PJ. Genes with homology to fungal and S-gene RNases are expressed in Arabidopsis thaliana. Plant Physiol. 1991;96:980–984. doi: 10.1104/pp.96.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thill GP, Kramer RA, Turner KJ, Bostian KA. Comparative analysis of the 5′-end regions of two repressible acid phosphatase genes in Saccharomyces cerevisiae. Mol Cell Biol. 1983;3:570–579. doi: 10.1128/mcb.3.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubia M, Sessa L, Taramelli R. Mammalian Rh/T2/S-glycoprotein ribonuclease family genes: cloning of a human member located in a region of chromosome 6 (6q27) frequently deleted in human malignancies. Genomics. 1997;42:342–344. doi: 10.1006/geno.1997.4679. [DOI] [PubMed] [Google Scholar]
- Trull MC, Guiltinan MJ, Lynch JP, Deikman J. The responses of wild-type and ABA mutant Arabidopsis thaliana plants to phosphorus starvation. Plant Cell Environ. 1997;20:85–92. [Google Scholar]
- Ye Z-H, Droste DL. Isolation and characterization of cDNAs encoding xylogenesis-associated and wounding-induced ribonucleases in Zinnia elegans. Plant Mol Biol. 1996;30:697–709. doi: 10.1007/BF00019005. [DOI] [PubMed] [Google Scholar]
- Yen Y, Green PJ. Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol. 1991;97:1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]