Abstract
The commitment of eukaryotic cells to division normally occurs during the G1 phase of the cell cycle. In mammals D-type cyclins regulate the progression of cells through G1 and therefore are important for both proliferative and developmental controls. Plant CycDs (D-type cyclin homologs) have been identified, but their precise function during the plant cell cycle is unknown. We have isolated three tobacco (Nicotiana tabacum) CycD cyclin cDNAs: two belong to the CycD3 class (Nicta;CycD3;1 and Nicta;CycD3;2) and the third to the CycD2 class (Nicta;CycD2;1). To uncouple their cell-cycle regulation from developmental control, we have used the highly synchronizable tobacco cultivar Bright Yellow-2 in a cell-suspension culture to characterize changes in CycD transcript levels during the cell cycle. In cells re-entering the cell cycle from stationary phase, CycD3;2 was induced in G1 but subsequently remained at a constant level in synchronous cells. This expression pattern is consistent with a role for CycD3;2, similar to mammalian D-type cyclins. In contrast, CycD2;1 and CycD3;1 transcripts accumulated during mitosis in synchronous cells, a pattern of expression not normally associated with D-type cyclins. This could suggest a novel role for plant D-type cyclins during mitosis.
Progression through the eukaryotic cell cycle is largely regulated at two principal control points, during the late G1 phase and at the G2/M boundary. Transit through these control points requires activated kinase complexes consisting of a cyclin-dependent Ser/Thr kinase bound to a regulatory cyclin (for review, see Pines, 1995). cdk activity is dependent on cyclin binding, which also determines the substrate specificity and subcellular localization of the cdk complex. Cell-cycle progression depends on the sequential activation and deactivation of distinct cdk-cyclin complexes. This is controlled in part by the stage-specific synthesis and destruction of the components, although their activity is also regulated by activating or inhibitory phosphorylations and the binding of inhibitory proteins (for review, see Pines, 1995).
Animal cyclins are classified as mitotic (A and B types) or G1 (D and E types) cyclins, based on sequence characteristics and analysis of their role and time of action in the cell cycle (Pines, 1995). A- and B-type cyclins accumulate periodically during the S, G2, and early M phases (A types) or G2 and early M phases (B types) before being destroyed later in mitosis (Pines and Hunter, 1989, 1990). The G1 cyclins, D and E, have distinct roles in controlling progression through the G1 into the S phase. The late G1 restriction (R) point is of particular significance, since cells must interpret extracellular signals and either commit to a further round of division or adopt alternative differentiation pathways (Pardee, 1989). This process is mediated by D-type cyclins, unstable proteins whose transcription is absolutely dependent on the presence of serum growth factors (Matsushime et al., 1991; Ajchenbaum et al., 1993; Ando et al., 1993; for review, see Sherr, 1993, 1994). The essential action of cyclin D-dependent kinases is to direct phosphorylation of the Rb protein in the mid-to-late G1 phase, thereby driving cells through the R point. Cyclin E then accumulates transiently in late G1, forming kinase complexes that accelerate the phosphorylation of Rb, irreversibly driving cells across the G1/S boundary (for review, see Sherr, 1996).
Multiple cdk and cyclin homologs have also been identified in plants (Francis and Halford, 1995; Jacobs, 1995). However, little is known about their function during the cell cycle. Most plant cyclins isolated to date are related to animal A and B types and have been named CycA and CycB cyclins, respectively (Renaudin et al., 1996). CycA cyclins are expressed in the S, G2, and M phases and CycB is expressed in the late G2 and M phases, as are their animal equivalents (Hirt et al., 1992; Setiardy et al., 1995; Reichheld et al., 1996; Ito et al., 1997; for review, see Jacobs, 1995; Renaudin et al., 1996). A small number of cyclins with sequence homologies to animal D-type cyclins have been cloned and analyzed in Arabidopsis and alfalfa (Dahl et al., 1995; Soni et al., 1995; Fuerst et al., 1996; for review, see Murray et al., 1998). These CycD cyclins form three distinct groups (CycD1, CycD2, and CycD3) based on sequence criteria (Renaudin et al., 1996; Murray et al., 1998). The limited analysis to date suggests that they show expression patterns reminiscent of animal D-type cyclins, including rapid transcript accumulation upon stimulation of quiescent suspension-cultured cells with growth-promoting substances (Dahl et al., 1995; Soni et al., 1995; Fuerst et al., 1996; Murray et al., 1998). Plant CycD cyclins have been shown to interact with maize Rb proteins (Ach et al., 1997; Huntley et al., 1998), leading to the suggestion that plant CycD cyclins may, like animal D-type cyclins, regulate G1 controls through interactions with plant Rb proteins. Since the G1-control point in plants appears to be central to decisions relating to the commitment to further proliferation or differentiation (Murray, 1994; Hirt, 1996), understanding its regulation is a prerequisite to establishing the relationship between cell division and plant development. To this end, our goal was to clarify the role of CycD cyclins in the cell cycle.
A detailed understanding of how cell-cycle-control proteins regulate the mammalian cell cycle has come mainly from studies with well-characterized cell culture systems. One of the most widely used and best-characterized plant cell culture systems is the tobacco (Nicotiana tabacum) Bright Yellow-2 (BY-2) cell line (Nagata et al., 1992). This line is particularly suitable for cell-cycle studies, since it can be highly synchronized. Here we report the isolation of a tobacco CycD2 and two CycD3 cyclin cDNAs and describe their expression in tobacco BY-2 cells. We show that CycD3;2 is induced in G1 as stationary cells re-enter the cell cycle, a pattern of expression that is consistent with this cyclin having a G1 role. CycD3;2 RNA then remains at a constant level throughout the cell cycle in synchronous cells. In contrast, transcripts from the other two genes, CycD2;1 and CycD3;1, show greatest abundance in mitotic cells, a pattern of expression not previously reported, to our knowledge, for any plant CycD cyclin.
MATERIALS AND METHODS
Isolation of Tobacco Cyclins
Randomly primed α-32P-labeled cDNA probes were used to screen 7.5 × 105 clones from a cDNA library constructed in a Lambda Zap Express vector (Stratagene) with poly(A+) RNA isolated from exponentially growing tobacco (Nicotiana tabacum cv BY-2; Nagata et al., 1992) cells. For isolation of CycD3 cDNAs, a mixed probe consisting of the Arabidopsis Arath;CycD3;1 cDNA (Soni et al., 1995) and the snapdragon (Antirrhinum majus) Antma;CycD3;1 and Antma;CycD3;2 cDNAs (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data) was used. Hybridizations were carried out at 55°C and the membranes were washed at room temperature for 10 min three times, twice in 2× SSC and 0.1% SDS, and once in 0.1× SSC and 0.1% SDS. For the isolation of the tobacco CycD2 cDNA the Arabidopsis Arath;CycD2;1 (Soni et al., 1995) cDNA was used as a probe. Hybridizations were at 48°C with three 10-min washes at room temperature in 2× SSC and 0.1% SDS. Selected positive clones were purified, and the cDNA inserts were excised in vivo according to the manufacturer's instructions and analyzed by restriction mapping and DNA sequencing. Other positive clones were analyzed without further rounds of purification by PCR using primers specific for the tobacco CycD cDNAs already identified. Thermocycle conditions consisted of 25 cycles of 95°C (1 min), 55°C (1 min), and 72°C (1 min). Sequence analysis was carried out using the Genetics Computer Group package (Madison, WI) and the Sequencher 3.0 software program (Gene Codes Corp., Ann Arbor, MI). Potential PEST sequences were identified using the PESTFIND program (Rogers et al., 1986) available on the European Molecular Biology Network (EMBnet) Austria server (http:// www.at.embnet.org/embnet/tools/bio/PESTfind/). This program uses a corrected hydrophobicity value for Tyr of 58, not 32 as in the original program (Stellwagen correction). The program was run using the default setting of five for the minimum number of amino acids between positive flanks.
Culture of Tobacco BY-2 Cells and Experimental Treatments
Tobacco BY-2 cells were maintained as previously described (Nagata et al., 1992). For the growth-cycle experiments, 2 mL of stationary-phase culture (7 d after subculture) was incubated in 95 mL of fresh medium for 7 d. Cell density was determined after maceration (Brown and Rickless, 1949) using a modified Fuchs-Rosenthal counting chamber. Cells were suspended in a 15% (w/v) CrO3 solution and dispersed by repeated passage through a 1-mL disposable pipette tip. Cell number measurements were performed in triplicate for each of three samples removed each day. Cells were synchronized using 3 to 6 μg/mL aphidicolin (Sigma) essentially as described by Nagata et al. (1992). Measurements of DNA synthesis and mitotic index were according to the method of Reichheld et al. (1995). For the inhibitor treatments, 10 mL of stationary-phase culture was incubated in 90 mL of fresh medium containing 15 μg/mL aphidicolin or 15 μm oryzalin (Riedel-de Haën) for 24 h. For the propyzamide (Riedel-de Haën) treatment, cells were treated as previously described (Nagata et al., 1992). For the induction experiment, 10-mL aliquots of stationary-phase culture were incubated in 90 mL of fresh medium for 10 h.
RNA Extraction and RNA Gel-Blot Analysis
Total RNAs (30–40 μg) were extracted as described by Verwoerd et al. (1989) or Goodall et al. (1990), separated on formaldehyde-agarose gels, and blotted onto nylon membranes. In some experiments equal loading was confirmed prior to hybridization by staining the membranes in 0.02% (w/v) methylene blue/0.5 m sodium acetate (pH 5.2) for 5 to 10 min and then destaining in 1× SSPE (150 mm NaCl, 10 mm NaH2PO4, and 1 mm EDTA, pH 7.4) for 10 to 15 min. The complete tobacco CycD cDNAs were used as probes; other probes used were as described by Reichheld et al. (1996). α-32P-labeled probes were generated by single- or double-nucleotide random-prime labeling and hybridized in a buffer containing 50% (v/v) formamide at 42°C.
RESULTS
Isolation of Tobacco CycD Cyclins
A cDNA library made from exponentially dividing tobacco BY-2 cells was screened with Arabidopsis (Soni et al., 1995) and snapdragon (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data) CycD cDNA probes. When screened with a mixed probe consisting of the Arabidopsis CycD3;1 cDNA and the snapdragon CycD3;1 and CycD3;2 cDNAs, 110 positive clones were identified. An analysis of 30 of these clones by sequencing, restriction digestion, or PCR revealed two families of cDNAs. One cDNA from each family was completely sequenced. The cDNA (designated CycD3;1) from the first family was 1679 bp in length with an ORF encoding a polypeptide 373 amino acids in length. The cDNA (designated CycD3;2) from the second family was 1431 bp in length and contained an ORF encoding a 367-amino-acid-long polypeptide. A screen with the Arabidopsis CycD2;1 cDNA yielded two cDNA clones belonging to the same family. One of these cDNAs (designated CycD2;1) was completely sequenced and found to be 1284 bp in length, with an ORF encoding a polypeptide 354 amino acids long. The complete tobacco CycD2;1, CycD3;1, and CycD3;2 cDNAs gave discrete patterns of hybridization when used to probe BY-2 genomic DNA gel blots, suggesting that they do not cross-hybridize under standard conditions (data not shown).
Sequence Relationships
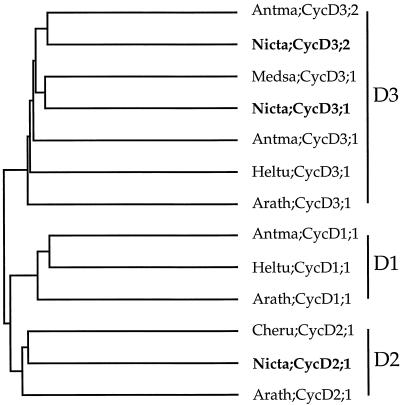
Comparison of the tobacco cyclins with sequence databases using the software program BLAST (Altschul et al., 1990) showed that they have the closest similarity with plant and mammalian D-type cyclins (data not shown). A detailed comparison of the tobacco cyclins with other plant CycD cyclins, over the full length of their predicted polypeptides, using the pairwise comparison program PileUp, is shown graphically in Figure 1. This analysis supported previous observations that CycD cyclins fall into three distinct groups, CycD1, CycD2, and CycD3, originally defined in Arabidopsis (Soni et al., 1995; Renaudin et al., 1996; Murray et al., 1998). Tobacco CycD3;1 and CycD3;2 fell into the CycD3 group, showing highest similarity to the alfalfa CycD3;1 (CycMs4; 66% identity) and the snapdragon CycD3;2 (61% identity), respectively. CycD2;1 fell into the CycD2 class, having the highest similarity with the C. rubrum CycD2 (55% identity). The tobacco CycD cyclins showed much less similarity to tobacco mitotic cyclins (25%–31% identity).
Figure 1.
The relationship between tobacco CycD cyclins (bold letters) and other plant CycD cyclins was determined using the pairwise comparison program PileUp in the Genetics Computer Group package. Cyclin nomenclature is according to Renaudin et al. (1996). Antma, A. majus (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data); Arath, Arabidopsis (Soni et al., 1995); Cheru, Chenopodium rubrum (Renz et al., 1997) Heltu, Helianthus tuberosus (D. Freeman and J.A.H. Murray, unpublished data); Medsa, Medicago sativa (Dahl et al., 1995); Nicta, N. tabacum. The three distinct groups of CycD cyclins, CycD1, CycD2, and CycD3, are indicated on the right.
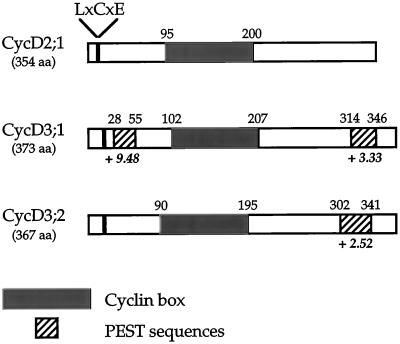
All three tobacco CycDs contain the conserved cyclin box region that was approximately 100 amino acids long, which is involved in cdk binding (Fig. 2; Jeffrey et al., 1995) and is present in all cyclins. In addition, they contain sequence motifs characteristic of animal and plant D-type cyclins (Fig. 2). They lack the N-terminal ”destruction box,” a motif (RxxL[x]2–4xxN) in mitotic cyclins required for their ubiquitin-mediated degradation during mitosis (Glotzer et al., 1991). Instead, animal and plant D-cyclins contain PEST sequences, regions rich in these four residues, which are characteristic of many rapidly turned over proteins (Rogers et al., 1986; Rechsteiner and Rogers, 1996). Using the software program PESTFIND (Rogers et al., 1986; Materials and Methods) we identified potential PEST sequences in both CycD3 cyclins (Fig. 2). In contrast, no potential PEST sequences were found in CycD2;1. A distinguishing feature of mammalian D-type cyclins and plant CycD cyclins is the presence of an LxCxE (x is any amino acid) Rb protein-binding motif near their N terminus. This motif is present in the N terminus of all three tobacco CycD cyclins (Fig. 2). In common with animal D-type cyclins, all CycD cyclins isolated to date have at least one acidic residue (D or E) at positions -1 or -2 relative to the LxCxE motif (Renaudin et al., 1996). Both tobacco CycD3s have a D residue at the -2 position, but, in contrast, CycD2 lacks a preceding acidic residue (data not shown).
Figure 2.
Tobacco CycD cyclin structural features. Schematic representation of the putative tobacco CycD cyclin proteins showing the relative positions of the LxCxE motif, cyclin box, and PEST sequences. The software program PESTFIND (Rogers et al., 1986; Methods) was used to identify potential PEST sequences (defined as giving a positive score, which is shown in boldface italics). The total lengths of the putative proteins are given. aa, Amino acids.
Tobacco CycD Cyclins Are Expressed in a Growth-Phase-Dependent Manner
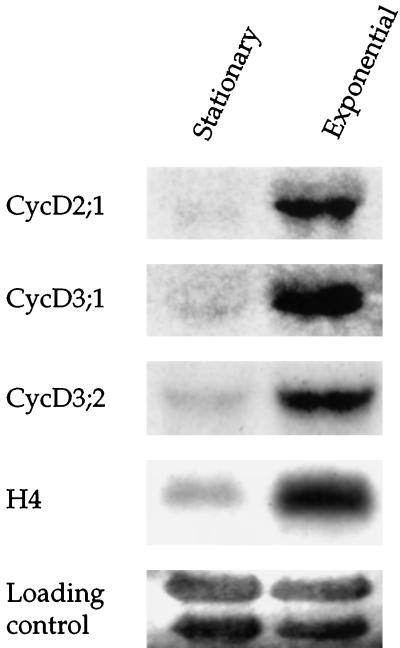
We compared the expression of the three CycD cyclins in stationary tobacco BY-2 cells (7 d after previous subculture) with exponentially dividing cells (3 d after subculture) by RNA gel-blot analysis. CycD2;1 and CycD3;2 were represented by single transcripts of about 1.9 kb, and CycD3;1 was represented by a single transcript of approximately 1.75 kb. All three cyclins showed the highest level of expression in exponentially growing cells (Fig. 3). This pattern of expression was mirrored by histone H4, whose expression has previously been shown to be strongly associated with the exponential phase of growth in Arabidopsis cells (Fig. 3; Chaubet et al., 1996). CycD3;2 transcripts were the most abundant, with the other two cyclins being approximately 10- to 20-fold less abundant.
Figure 3.
Growth-phase-dependent expression of tobacco CycD cyclins. Total RNA isolated from stationary-phase BY-2 cells (7 d after previous subculture) and exponentially growing cells (3 d after subculture) was blotted and hybridized to the indicated cDNA probes. The membrane was stained in methylene blue prior to hybridization to visualize the principal rRNA bands to control for loading (Methods). Note that CycD3;2 is approximately 10- to 20-fold more abundant than the other two cyclins.
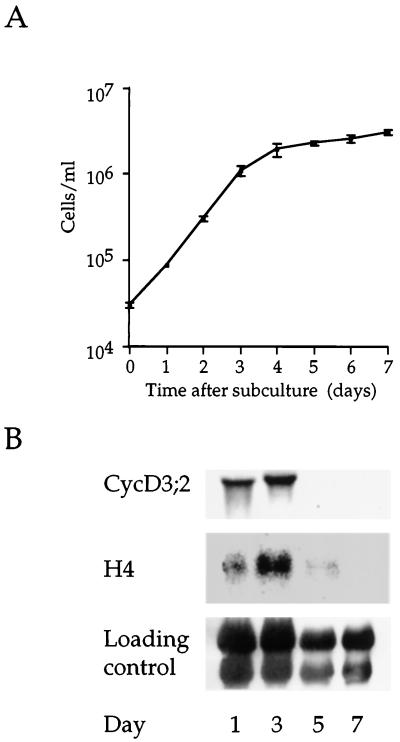
To confirm the relationship between CycD3;2 expression and growth phase, we examined transcript abundance during the complete tobacco BY-2 cell-growth cycle. Stationary-phase cells (7 d after subculture) were inoculated into fresh medium and samples were obtained during the following 7 d. Changes in cell number and in CycD3;2 and histone H4 transcript levels were monitored (Fig. 4). CycD3;2 was highly expressed at 1 and 3 d, which fall within the exponential phase of growth (d 1–4), the highest expression being observed at d 3. By d 5, transcript levels had declined rapidly as cells exited the cell cycle and entered the stationary phase. This pattern of expression was mirrored by histone H4.
Figure 4.
Growth-phase-dependent expression of CycD3;2. A, Growth curve of tobacco BY-2 cells in batch-suspension culture. Stationary-phase cells (7 d after subculture) were subcultured into fresh medium and incubated for 7 d. The mean cell number was determined daily; the error bars represent the se determined from three samples. B, Total RNA isolated from samples taken at the indicated times was blotted and hybridized to the indicated cDNA probes. The loading control was by methylene blue staining of the membrane.
These results suggest that tobacco CycD cyclins are expressed in a growth-phase-dependent manner in BY-2 cells.
CycD3;2 Is Induced in the Late G1 Phase as Stationary BY-2 Cells Re-Enter the Cell Cycle
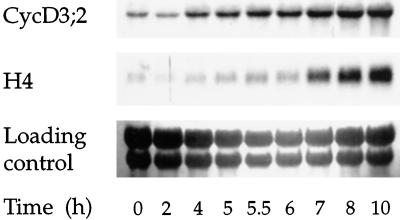
We then investigated the timing of CycD3;2 accumulation when cells exit from the stationary phase when diluted into fresh medium. Stationary BY-2 cells have a G1 nuclear DNA content (Planchais et al., 1997), indicating that they exited the cell cycle in G1 phase. By subculturing stationary cells into fresh medium, we could examine the induction of CycD3;2 expression during G1, as cells re-entered the cell cycle from a state of quiescence. Changes in the expression of CycD3;2 and the S-phase marker gene histone H4 (Reichheld et al., 1995) during a 10-h period are shown in Figure 5. Histone H4 transcripts started accumulating rapidly between 6 and 7 h, with the highest levels reached at 10 h. This indicated that the majority of cells started to enter the S phase in a fairly synchronous manner at approximately 7 h. By 4 h CycD3;2 transcript levels had started to accumulate, increasing steadily up to 10 h. The level of CycD3;2 transcripts at 10 h was similar to that seen in cells 24 h after subculture (data not shown). This indicates that CycD3;2 was induced at least 3 h before histone H4, between 2 and 4 h after stimulation, in mid-G1 phase.
Figure 5.
Expression of CycD3;2 in stationary BY-2 cells re-entering the cell cycle. Stationary BY-2 cells were incubated in fresh medium for 10 h. Total RNA from samples taken at the indicated times was blotted and hybridized with the indicated probes. Entry into the S phase was monitored by following changes in histone H4 expression. To control for loading the membrane was stained with methylene blue.
CycD Cyclin Expression Is Differentially Regulated in Synchronous BY-2 Cells
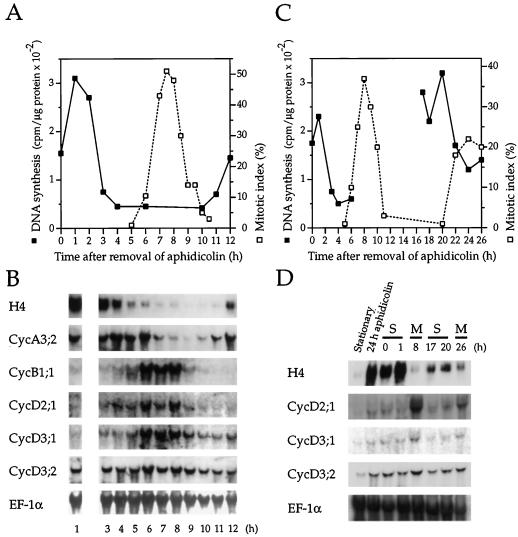
The G1 induction of CycD3;2 could be characteristic only of quiescent cells re-entering the cell cycle or could also occur in every cell cycle in rapidly dividing cells. We therefore investigated the expression of the CycDs during the cell cycle in synchronously cycling BY-2 cells. Cells were synchronized (see Methods) with the drug aphidicolin, an inhibitor of DNA polymerase activity that blocks BY-2 cells in the early S phase (Nagata et al., 1992). After the synchronous cohort of cells was released from the block, the progress was followed by monitoring changes in DNA synthesis, mitotic index, and the expression of the S-phase marker gene histone H4. Changes in steady-state mRNA levels were compared over one complete cell cycle for CycD cyclins and the mitotic cyclins CycA3;2 and CycB1;1 (Fig. 6, A and B). As previously described, CycA3;2 had high transcript levels during the S and G2 phases, and CycB1;1 cyclin transcripts were most abundant in late G2 and M phases (Reichheld et al., 1996). In contrast to expectations, both CycD2;1 and CycD3;1 transcript levels appeared to peak during mitosis, like CycB1;1. However, unlike CycB1;1 transcripts, which increased dramatically at the G2/M boundary, CycD2;1 and CycD3;1 transcripts showed a more gradual accumulation during the S and G2 phases. As cells exited mitosis, CycD2;1 transcripts declined rapidly to the levels found during the S phase. In contrast, CycD3;1 levels declined more gradually, stabilizing at a level higher than observed in the first S phase. CycD3;2 transcripts, which are induced in G1 phase in stimulated cells, were present at a constant level throughout the cell cycle. The constitutively expressed translation elongation factor mRNA transcript was used to demonstrate equal loading between lanes.
Figure 6.
Expression of CycD cyclins during the cell cycle in synchronous BY-2 cells. Cells were synchronized in the early S phase with aphidicolin (Methods) in two independent experiments. Progress of cells through the cell cycle was monitored by following changes in DNA synthesis and mitotic index. A, In the first experiment, progress of cells was followed for one complete cell cycle. B, Cells were harvested at the indicated times for RNA analysis and blots were hybridized with the probes indicated. Note that no sample was taken at 2 h. EF-1α, Elongation factor 1α. C, In a second synchrony experiment, progress through two cell cycles was followed. Changes in DNA synthesis were monitored between 0 to 6 and 17 to 26 h, the times when the two S phases were predicted to occur. D, Cells in the S phase (S) or mitosis (M) were collected at the indicated times for RNA analysis. Samples from stationary cells and cells treated with aphidicolin for 24 h (before washing and release into synchrony) are included for comparison. RNA gel blots were hybridized to the probes indicated.
To confirm these observations, we compared CycD transcript levels during the S phase and mitosis on RNA gel blots originating from a separate synchrony experiment using transgenic BY-2 cells containing an Arabidopsis H4 gene promoter fused to the GUS reporter gene. We believe it unlikely that this construct perturbs expression of the CycD cyclins. Cell samples were taken at various times during two complete cell cycles for RNA gel-blot analysis (Fig. 6, C and D). The level of DNA synthesis and H4 expression in samples taken at 0, 1, 17, and 20 h indicated that these cells were engaged in S phase. The mitotic index at 8 and 26 h showed that cells sampled at these times were engaged in mitosis; some H4 transcript was apparent at 26 h, reflecting the decay in synchrony associated with the second cycle. CycD2;1 and CycD3;1 transcript levels were more abundant in mitosis compared with S phase. CycD3;2 transcript levels were the same in both S and M phases. These data confirm the timing of CycD cyclin expression observed during the cell cycle in the first synchrony experiment.
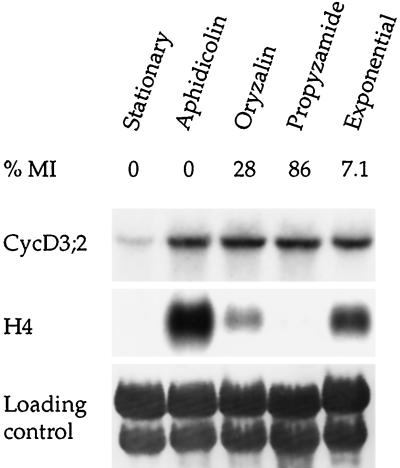
We demonstrated further that CycD3;2 transcript levels remain unchanged during the cell cycle by examining transcript abundance in cells blocked in S phase with aphidicolin or in mitosis using the antitubulin drugs oryzalin or propyzamide. Stationary cells were diluted in fresh medium containing aphidicolin or oryzalin and cultured for 24 h (see Methods). For the propyzamide treatment, G2/M cells obtained 4 to 5 h after release from an aphidicolin block were treated with the drug for 5 h (Nagata et al., 1992). Figure 7 shows the transcript abundance in blocked, stationary, and exponentially growing (3 d after subculture) cells. The mitotic index and histone H4 transcript levels confirmed that aphidicolin blocked cells in S phase, and oryzalin or propyzamide blocked cells in mitosis. The oryzalin mitotic block was less efficient than the propyzamide block, as indicated by the 3-fold lower mitotic index and higher histone H4 transcript abundance. However, the 28% mitotic index in these oryzalin-treated cultures is consistent with that in other reports (Shaul et al., 1996) and is indicative of a substantial fraction of cells being in mitosis compared with stationary, exponentially growing or S-phase-blocked cells. CycD3;2 transcripts were detected in the same abundance in S- and M-phase-blocked cultures and in exponentially growing cells. In addition, CycD3;2 transcripts remained at a constant level in highly synchronous cells during progression through the G1 and into the S phase after release from a sequential aphidicolin-propyzamide treatment (data not shown). These data are consistent with the conclusion that CycD3;2 transcripts are present at constant levels during the cell cycle.
Figure 7.
Expression of CycD3;2 in cells treated with cell-cycle inhibitors. Cells were arrested in the S phase with aphidicolin or in mitosis with oryzalin or propyzamide (Methods) and harvested for RNA analysis. Cell-cycle arrest was confirmed by determining the mitotic index (MI) and the level of histone H4 transcripts. Samples from stationary and exponentially dividing (3 d after subculture) cells are included for a comparison. The loading control was by methylene blue staining of the membrane.
We conclude that in cycling cells CycD3;2 transcripts are present at a constant level throughout the cell cycle, whereas CycD2;1 and CycD3;1 levels fluctuate, with a peak during mitosis.
DISCUSSION
All tobacco cyclins isolated to date fall into the CycA and CycB classes of plant mitotic cyclins (Qin et al., 1995; Setiady et al., 1995; Reichheld et al., 1996; Renaudin et al., 1996). To our knowledge we describe the first isolation of tobacco CycD cyclin genes. Based on a comparison with other CycD cyclins, two of these tobacco cyclins belong to the CycD3 group of plant cyclins, and the third belongs to the CycD2 group. In accordance with revised plant gene nomenclature (Renaudin et al., 1996), we propose to formally designate the two CycD3 cyclins as Nicta;CycD3;1 and Nicta;CycD3;2, and the CycD2 cyclin as Nicta;CycD2;1.
Like other CycDs isolated, the tobacco CycDs contain an N-terminal LxCxE motif (Renaudin et al., 1996; Murray et al., 1998). This motif enables animal D-type cyclin-associated kinases to bind and subsequently phosphorylate Rb proteins (Dowdy et al., 1993; Kato et al., 1993). The presence of this motif in plant CycDs, the recent isolation of plant Rb homologs from maize (Xie et al., 1996; Grafi et al., 1996; Ach et al., 1997), and the in vitro binding of CycDs to maize Rb proteins (Ach et al., 1997; Huntley et al., 1998) suggest that this interaction is also important for the regulation of the plant cell cycle.
Mammalian D-type cyclins and other animal and yeast G1 cyclins are rapidly degraded because of the presence of PEST sequences that target the protein for ubiquitin-mediated degradation (Rechsteiner and Rogers, 1996). All CycD cyclins isolated to date contain PEST regions, suggesting that they are labile proteins degraded by a similar pathway to animal D-type cyclins (Murray et al., 1998). The predicted amino acid sequences for the tobacco CycD3;1 and CycD3;2 cyclins contain PEST sequences. However, no prominent PEST sequences could be detected in the CycD2;1 sequence. We do not believe that this was due to the putative ORF being incomplete, because an in-frame stop codon is present upstream of the predicted translation start. Moreover, based on homology, the regions where PEST sequences are located in the Arabidopsis and C. rubrum CycD2 sequences are present in the tobacco CycD2 sequence. Therefore, to our knowledge, this is the first D-type cyclin in any organism reported not to contain a prominent PEST sequence. We cannot rule out the presence of weak PEST sequences that were undetected by the PESTFIND software program. Like other plant and animal D-type cyclins, CycD2;1 lacks the ”destruction box” motif characteristic of mitotic cyclins (Glotzer et al., 1991). Therefore, it is possible that the degradation of CycD2;1 may be regulated in a way different from other cyclins, although this feature appears so far limited to tobacco CycD2;1 and is not characteristic of the CycD2 group in general. We also note that tobacco CycD2;1 lacks an acidic residue preceding the L of the LxCxE motif, a characteristic of all other animal and plant cyclins (Renaudin et al., 1996). The isolation of further CycDs and biochemical studies will reveal whether these features have any functional significance.
Individual plant species often have more than one cyclin within a given CycA or CycB group (Renaudin et al., 1996, 1998). For example, the CycA3 group of plant cyclins contains three tobacco cyclins (Reichheld et al., 1996). The isolation of two distinct CycD3 cyclin cDNAs from tobacco and snapdragon (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data) suggests that the CycD class will be of a similar complexity in terms of the number of cyclins and relationships between cyclins, as is the case for the CycA and CycB classes. Moreover, the differential expression of the two tobacco CycD3 cyclins in synchronized cells and the two snapdragon CycD3s in meristems (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data) suggests that these cyclins may have distinct functions. This suggestion is supported by sequence comparisons showing that both tobacco CycD3 cyclins have higher sequence similarity with CycD3s from other species than they do to each other. We note that tobacco CycD3;1 is most similar to alfalfa CycD3;1 (CycMs4; Dahl et al., 1995), whereas tobacco CycD3;2 is most similar to snapdragon CycD3;2 (V. Gaudin, P. Forbert, T. Lunness, C. Riou-Khamlichi, J.A.H. Murray, E. Coen, and J.H. Doonan, unpublished data). This suggests the probable existence of at least three subgroups within the CycD3 group, the first typified by alfalfa CycD3;1 and tobacco CycD3;1, the second by tobacco CycD3;2 and snapdragon CycD3;2, and a third, for which snapdragon CycD3;1 is to date the only member. It remains unclear whether the Arabidopsis CycD3;1 and Jerusalem artichoke CycD3;1 belong to an additional group(s). The isolation and functional characterization of more CycDs in the future will allow the complexity of the CycD class to be fully assessed.
Mammalian D-type cyclins are predominantly regulated by transcription, with changes in protein levels generally mirroring changes in transcript levels (Matsushime et al., 1991; Ajchenbaum et al., 1993; Ando et al., 1993). In the absence of serum growth factors, many mammalian cell types exit the cell cycle during G1, entering a quiescent (G0) state. Upon readdition of serum growth factors, D-type cyclins are rapidly synthesized, forming active complexes with their cognate cdk partners, which phosphorylate the Rb protein during the G1 phase (for review, see Sherr, 1994, 1996).
We investigated the function of tobacco CycD cyclins by examining transcript levels in the tobacco BY-2 cell line, a model system for studying the plant cell cycle (Nagata et al., 1992). Stationary BY-2 cells (7 d after subculture) have a G1 nuclear content, indicating that they exited the cell cycle from G1 (Planchais et al., 1997). Tobacco CycD cyclin transcript levels were found to be low in stationary cells, which is analogous to the absence or low levels of D-type cyclin transcripts in growth-factor-deprived, quiescent mammalian cells. We examined CycD3;2 transcript levels in stationary BY-2 cells stimulated to re-enter the cell cycle by the addition of fresh medium. This is a situation that parallels the re-entry of quiescent mammalian cells into the cell cycle upon serum stimulation. We found that CycD3;2 was induced in G1 at least 3 h prior to the onset of the S phase, which is consistent with this cyclin having a role in G1 similar to the role of mammalian D types. The Arabidopsis CycD2;1 and CycD3;1, and the alfalfa CycD3;1 are also induced in G1 in cells re-entering the cycle (Dahl et al., 1995; Soni et al., 1995; Fuerst et al., 1996; C. Riou-Khamlichi and J.A.H. Murray, unpublished data).
The consensus view, based on studies with a variety of mammalian cell lines, is that cyclin D transcripts show minimal oscillations during the cell cycle, in contrast to the periodicity of cyclin A, B, and E transcript levels (Matsushime et al., 1991; Ajchenbaum et al., 1993; Sewing et al., 1993; Sherr, 1994). We studied the behavior of the tobacco CycD cyclins during the cell cycle by examining transcript levels in synchronized BY-2 cells and cells arrested at various stages of the cell cycle by treatment with chemical inhibitors. We found that CycD3;2 transcripts remained at a remarkably constant level throughout the cell cycle. A similar pattern of expression has been suggested for the Arabidopsis CycD2;1 and CycD3;1 cyclins (Fuerst et al., 1996), although the synchrony in the system used was significantly less than that achieved with BY-2 cells, which could possibly obscure cell-cycle variations. The expression of CycD3;2 is also consistent with the behavior of mammalian D-type cyclins. We also note that alfalfa CycD3;1 showed some phase-dependent expression in alfalfa cells, and their transcript levels peaked at the G1/S transition, which is consistent with a role in late G1 (Dahl et al., 1995).
In contrast, tobacco CycD2;1 and CycD3;1 transcripts accumulated in mitosis in BY-2 cells, a pattern of expression not normally associated with D-type cyclins. One interpretation for this is that these cyclins may be required for entry into or progression through mitosis. In proliferating mammalian cells, Rb proteins are further phosphorylated in G2/M, prior to being dephosphorylated in the latter stages of mitosis (DeCaprio et al., 1992; Ludlow et al., 1993; Taya, 1997). This, together with functional studies showing that the overexpression of Rb after the G1/S boundary causes mammalian cells to arrest in G2, has led to the suggestion that Rb proteins may have secondary functions during later stages of the cell cycle in addition to their G1 function, although the exact identity of the kinases that phosphorylate Rb in the mammalian G2/M is far from clear (Karantza et al., 1993; Sterner et al., 1995; Taya, 1997). Therefore, CycD2;1 and CycD3;2 may play a role in phosphorylating or maintaining the phosphorylation of plant Rb-like proteins during entry into or progression through the M phase as a normal part of the plant cell cycle. It remains to be seen whether this is a tobacco-specific phenomenon or if it is more widespread in the CycD genes of other species. It is possible that this pattern of expression is exhibited by some of the CycDs already analyzed, but, because of the absence of cell systems as highly synchronized as the BY-2 cells, it has gone undetected.
An alternative interpretation of this mitotic accumulation is that it is a BY-2 cell-specific phenomenon and not a normal feature of plant cell-cycle progression. Human cyclin D1 transcripts have been reported to accumulate during the G2/M phase in the long-established HeLa tumor cell line (Motokura et al., 1991, 1992). This is a pattern of expression not observed in other cell lines or in primary cultures (Motokura et al., 1992; Sewing et al., 1993), which has led to the suggestion that selection may have occurred for deregulated or altered expression of cyclin D1 in HeLa cells as a consequence of extended proliferation in culture (Sewing et al., 1993). The ability of some plant cell cultures to proliferate indefinitely is not well understood, and it is unclear whether this ability arises through a process analogous to the transformation of mammalian cells. Tobacco BY-2 cells have been grown in vitro for more than 30 years and exhibit an exceptional ability for rapid cell proliferation (Nagata et al., 1992). It is therefore possible that the G2/M accumulation of CycD2;1 and CycD3;1 transcripts represents a deregulation of CycD expression, which perhaps assists in the rapid transit of the ensuing G1 phase, resulting either from the acquisition of indefinite proliferation potential or as consequence of long-term culture. Further studies are required to compare the expression of these cyclins in BY-2 cells with their expression in alternative cell systems and in planta.
The accession numbers for the sequences reported in this article are AJ011892 for Nicta;CycD2;1, AJ011893 for Nicta;CycD3;1, and AJ011894 for Nicta;CycD3;2.
ACKNOWLEDGMENTS
We thank our colleagues for helpful comments concerning the manuscript, Dr. Donna Freeman for advice regarding library screening, Wen Hui Shen for providing the cDNA library, and Alison Inskip and Martine Flénet for technical assistance.
Abbreviations:
- cdk
cyclin-dependent kinase
- ORF
open reading frame
- Rb
retinoblastoma
Footnotes
This work was supported in part by a Biotechnology and Biological Sciences Research Council (BBSRC) grant (no. P01552) to J.A.H.M. D.A.S. was the recipient of a special studentship from the BBSRC.
LITERATURE CITED
- Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W. RRB1 and RRB2encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajchenbaum F, Ando K, DeCaprio JA, Griffin JD. Independent regulation of human D-type cyclin gene expression during G1 phase in primary human T-lymphocytes. J Biol Chem. 1993;268:4113–4119. [PubMed] [Google Scholar]
- Ando K, Ajchenbaum-Cymbalista F, Griffin JD. Regulation of G1/S transition by cyclins D2 and D3 in hematopoietic cells. Proc Natl Acad Sci USA. 1993;90:9571–9575. doi: 10.1073/pnas.90.20.9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brown R, Rickless P. A new method for the study of cell division and cell extension with some preliminary observations on the effect of temperature and of nutrients. Proc R Soc Lond Ser B Biol Sci. 1949;136:110–125. doi: 10.1098/rspb.1949.0008. [DOI] [PubMed] [Google Scholar]
- Chaubet N, Flénet M, Clément B, Brignon P, Gigot C. Identification of cis-elements regulating the expression of an Arabidopsishistone H4 gene. Plant J. 1996;10:425–435. doi: 10.1046/j.1365-313x.1996.10030425.x. [DOI] [PubMed] [Google Scholar]
- Dahl M, Meskiene I, Bögre L, Ha DTC, Swoboda I, Hubmann R, Hirt H, Heberle-Bors E. The D-type alfalfa cyclin gene cycMs4complements G1 cyclin-deficient yeast and is induced in the G1 phase of the cell cycle. Plant Cell. 1995;7:1847–1857. doi: 10.1105/tpc.7.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Furukawa Y, Ajchenbaum F, Griffin JD, Livingstone DM. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc Natl Acad Sci USA. 1992;89:1795–1798. doi: 10.1073/pnas.89.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- Francis D, Halford NG. The plant cell cycle. Physiol Plant. 1995;93:365–374. [Google Scholar]
- Fuerst RAUA, Soni R, Murray JAH, Lindsey K. Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 1996;112:1023–1033. doi: 10.1104/pp.112.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goodall G, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG. A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA. 1996;93:8962–8967. doi: 10.1073/pnas.93.17.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt H. In and out of the plant cell cycle. Plant Mol Biol. 1996;31:459–464. doi: 10.1007/BF00042220. [DOI] [PubMed] [Google Scholar]
- Hirt H, Mink M, Pfosser M, Bögre L, Györgyey J, Jonak C, Gartner A, Dudits D, Herberle-Bors E. Alfalfa cyclins: differential expression during the cell cycle and in plant organs. Plant Cell. 1992;4:1531–1538. doi: 10.1105/tpc.4.12.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, Lavender P, de Jager S, Greenwood J, Makkerh J, Walker E, Jackman M, Xie Q and others. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Mol Biol. 1998;37:155–169. doi: 10.1023/a:1005902226256. [DOI] [PubMed] [Google Scholar]
- Ito M, Marie-Claire C, Sakabe M, Ohno T, Hata S, Kouchi H, Hashimoto J, Fukuda H, Komamine A, Watanabe A. Cell-cycle-regulated transcription of A- and B-type plant cyclin genes in synchronous cultures. Plant J. 1997;11:983–992. doi: 10.1046/j.1365-313x.1997.11050983.x. [DOI] [PubMed] [Google Scholar]
- Jacobs TW. Cell cycle control. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:317–339. [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP. Mechanism of cdk activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Karantza V, Maroo A, Fay D, Sedivy JM. Overproduction of Rb protein after the G1/S boundary causes G2 arrest. Mol Cell Biol. 1993;13:6640–6652. doi: 10.1128/mcb.13.11.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- Ludlow JW, Glendening CL, Livingston DM, DeCaprio JA. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol Cell Biol. 1993;13:367–372. doi: 10.1128/mcb.13.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime H, Roussel MF, Ashmun RA, Sherr CJ. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- Motokura T, Bloom T, Kim HG, Jüppner H, Ruderman JV, Kronenberg HM, Arnold A. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- Motokura T, Keyomarsi K, Kronenberg HM, Arnold A. Cloning and characterization of human cyclin D3, a cDNA closely related in sequence to the PRAD1/cyclin D1 proto-oncogene. J Biol Chem. 1992;267:20412–20415. [PubMed] [Google Scholar]
- Murray JAH. Plant cell division:the beginning of START. Plant Mol Biol. 1994;26:1–3. doi: 10.1007/BF00039513. [DOI] [PubMed] [Google Scholar]
- Murray JAH, Freeman D, Greenwood J, Huntley R, Makkerh J, Riou-Khamlichi C, Sorrell DA, Cockcroft C, Carmichael JP, Soni R, and others (1998) Plant D cyclins and retinoblastoma protein homologues. In D Francis, D Dudits, D Inzé, eds, Plant Cell Division. Portland Press, London, pp 99–127
- Nagata T, Nemoto Y, Hasezawa S. Tobacco BY-2 cell line as the “HeLa” cell in the cell biology of higher plants. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Pines J. Cyclins and cyclin-dependent kinases:a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- Planchais S, Glab N, Tréhin C, Perennes C, Bureau J-M, Meijer L, Bergounioux C. Roscovitine, a novel cyclin-dependent kinase inhibitor, characterizes restriction point and G2/M transition in tobacco BY-2 cell suspension. Plant J. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- Qin LX, Richard L, Perennes C, Gadal P, Bergounioux C. ) Plant Physiol. 1995;108:425–426. doi: 10.1104/pp.108.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Reichheld J-P, Chaubet N, Shen WH, Renaudin J-P, Gigot C. Multiple A-type cyclins express sequentially during the cell cycle in Nicotiana tabacumBY-2 cells. Proc Natl Acad Sci USA. 1996;93:13819–13824. doi: 10.1073/pnas.93.24.13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J-P, Sonobe S, Clément B, Chaubet N, Gigot C. Cell cycle-regulated histone gene expression in synchronized plant cells. Plant J. 1995;7:245–252. [Google Scholar]
- Renaudin J-P, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, Jacobs T, Kouchi H, Rouzé P, Sauter M and others. Plant cyclins:a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organisation. Plant Mol Biol. 1996;32:1003–1018. doi: 10.1007/BF00041384. [DOI] [PubMed] [Google Scholar]
- Renaudin J-P, Savouré A, Philippe H, Van Montagu M, Inzé D, Rouzé P (1998) Characterization and classification of plant cyclin sequences related to A- and B-type cyclins. In D Francis, D Dudits, D Inzé, eds, Plant Cell Division. Portland Press, London, pp 67–98
- Renz A, Fountain MD, Beck E. Nucleotide sequence of a cDNA encoding a D-type cyclin from a photoautotrophic cell suspension culture of Chenopodium rubrum (L.) (accession no. Y10162) (PGR 97-045) Plant Physiol. 1997;113:1004. [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins:the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Setiady YY, Sekine M, Hariguchi N, Yamamoto T, Kouchi H, Shinmyo A. Tobacco mitotic cyclins: cloning, characterization, gene-expression and functional assay. Plant J. 1995;8:949–957. doi: 10.1046/j.1365-313x.1995.8060949.x. [DOI] [PubMed] [Google Scholar]
- Sewing A, Bürger C, Brüsselbach S, Schalk C, Lucibello FC, Müller R. Human cyclin D1 encodes a labile nuclear protein whose synthesis is directly induced by growth factors and suppressed by cyclic AMP. J Cell Sci. 1993;104:545–555. doi: 10.1242/jcs.104.2.545. [DOI] [PubMed] [Google Scholar]
- Shaul O, Mironov V, Burssens S, Van Montagu M, Inzé D. Two Arabidopsiscyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc Natl Acad Sci USA. 1996;93:4868–4872. doi: 10.1073/pnas.93.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Shah ZH, Murray JAH. A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner JM, Murata Y, Kim HG, Kennett SB, Templeton DJ, Horowitz M. Detection of a novel cell cycle-regulated kinase activity that associates with the amino terminus of the retinoblastoma protein in G2/M phases. J Biol Chem. 1995;270:9281–9288. doi: 10.1074/jbc.270.16.9281. [DOI] [PubMed] [Google Scholar]
- Taya Y. Rb kinases and Rb-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos AP, Hannon GJ, Gutiérrez C. Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]