Abstract
Motivated by the need to solve ecological problems (climate change, habitat fragmentation and biological invasions), there has been increasing interest in species distribution models (SDMs). Predictions from these models inform conservation policy, invasive species management and disease-control measures. However, predictions are subject to uncertainty, the degree and source of which is often unrecognized. Here, we review the SDM literature in the context of uncertainty, focusing on three main classes of SDM: niche-based models, demographic models and process-based models. We identify sources of uncertainty for each class and discuss how uncertainty can be minimized or included in the modelling process to give realistic measures of confidence around predictions. Because this has typically not been performed, we conclude that uncertainty in SDMs has often been underestimated and a false precision assigned to predictions of geographical distribution. We identify areas where development of new statistical tools will improve predictions from distribution models, notably the development of hierarchical models that link different types of distribution model and their attendant uncertainties across spatial scales. Finally, we discuss the need to develop more defensible methods for assessing predictive performance, quantifying model goodness-of-fit and for assessing the significance of model covariates.
Keywords: distribution model, bioclimate envelope, spatial analysis, niche model, process-based model, climate impacts
1. Introduction
The spatial (and temporal) distribution of a species is one of the most fundamental pieces of ecological information. Knowing what drives or constrains geographical distribution is pivotal for many purposes: for example, for many macro-ecological questions searching for pattern and process in aggregate distribution statistics [1]; knowledge of the distribution of disease vectors for targeting of health programmes [2]; predicting spread of invasive species [3] and development of successful conservation plans [4,5]. From origins in conceptual models based on expert opinion and reasoned extrapolation, the development of formal species distribution modelling (SDM) using a variety of statistical and machine-learning techniques has formalized the process. Distribution modelling is now a major field of ecological research: between 2005 and 2010, there were over 850 publications in the Thomson Reuters web of knowledge database (found by a search for ‘species distribution mode*’ or ‘niche mode*’) compared with only 79 between 1999 and 2004.
Many recent uses of SDMs are to make two types of predictions of practical use to conservation: predictions of where species may be present but unrecorded (or indeed, where they might be found if human activities had not eliminated them [6]), and predictions of where species may be in the future as environmental change alters distributions [7]. The first predictions are essentially spatial interpolation and extrapolation, whereas the second uses a space for time substitution to make extrapolations into the future, based on projections of climate (and/or land-use) change. Both predictions have important uses, particularly for conservation where predictions have been used to identify spatial priorities for conservation [8,9] and in attempts to assess risks from climate change to particular species [10,11]. Given the interest in predicting future distributions, it is unsurprising that many methods have been developed. As with all ecological models, different methods have advantages and disadvantages, and are appropriate for different questions. All methods, however, are subject to uncertainty; an issue which, while not overlooked [12–15], is often only partially considered.
Identifying uncertainty is important for two main reasons. Firstly, without acknowledging sources of uncertainty, it is hard to see where future improvements can be made. For example, if there is uncertainty in a particular parameter estimate, or if one covariate is poorly measured, then future work on this will improve prediction. Equally important is the confidence or scepticism of the general public: many activities that rely on SDMs are of high profile [16] and if scientists are perceived as overstating problems or downplaying uncertainty, then the public is likely to further lose confidence in scientists [17]. Indeed, although minimizing uncertainty (increasing precision) is worthwhile, a whole field of theory has been developed to help make decisions in the face of uncertainty, but this is possible only when uncertainties are acknowledged and quantified [18]. Instead of trying to eliminate uncertainty completely, we suggest a pragmatic approach that embraces uncertainty and seeks to make accurate predictions with correctly identified precision.
Here, we review methods used to predict species distributions and focus particularly on their associated uncertainty, identifying where methodological development can reduce prediction uncertainty, and other areas where uncertainty is inherent.
2. Distribution modelling methods
There are many distribution models in current use [19–22]. Here, we seek not to review individual methods, but to identify classes of distribution models, discuss the assumptions they make and explore predictions that are possible with each class. In classifying distribution models, we recognize an axis from the purely statistical that seek through correlation to identify process (ecological niche) from pattern (geographical distribution), to methods that try to determine directly the processes and ecological mechanisms that underlie pattern. Although individual models fall anywhere on this continuum, for the purposes of this review we split models into three classes: statistical (and machine-learning) methods, called niche models; process-based models at the opposite end of the continuum, and an intermediate class such as metapopulation models that typically combine statistical associations between climate and habitat variables with demographic components to identify distribution and range dynamics. Along this axis, there is a rapid increase in the need for data and a concomitant decrease in the practicality of application across many species.
Uncertainty in predictions originates from many different sources, and we follow others in identifying three main areas where uncertainty occurs: data, model and prediction uncertainty (table 1). Although uncertainty from each area occurs in all models, not all sources are relevant to all model types. In the sections that follow, we start with a brief description of the model class, describe relevant sources of uncertainty and conclude with our view of where future development in each model class should focus to adequately model uncertainty.
Table 1.
Sources of uncertainty relevant to classes of species distribution model.
| uncertainty class | uncertainty source | niche-based models | demographic models | process-based models |
|---|---|---|---|---|
| data | observed distribution data (including recorder effort) | relevant both to model building and model fit assessment | relevant to model fit assessment | relevant to model fit assessment |
| current covariate (habitat and/or climate) data | relevant to model building and model fit assessment | may be relevant to model building (if spatial information on demography is required), always to model fit assessment | relevant to model fit assessment | |
| other data sources (e.g. noise in demographic data) | not relevant | relevant to demographic data for model building | relevant to model building, e.g. through observations on phenology | |
| model | model fitting methods (generalized-linear model, generalized-additive model, etc.) | highly relevant for niche identification | relevant for, e.g. identification of demographic links to weather | not (or minimally) relevant |
| structural model misspecification | highly relevant | highly relevant | highly relevant | |
| parameter estimation | relevant, but often fewer in number than more complex models | highly relevant | highly relevant and often involving large numbers of parameters | |
| uncertain model fit to true niche/distribution | highly relevant | highly relevant | highly relevant | |
| prediction | uncertainties in covariate data | highly relevant | highly relevant | highly relevant |
| no-analogue conditions | highly relevant | highly relevant | not important |
3. Niche-based distribution models
The majority of papers describing predicted species distributions fall into this class. A huge range of methods have been developed to fit these models from statistical regression methods, such as generalized-linear models and generalized-additive models to machine-learning approaches, including artificial neural networks and implementations of genetic or other learning algorithms [20]. Reviews of these methods that describe their strengths and weakness are available and we do not seek to repeat this information here. Instead, we focus on the common properties of this class of method and describe the uncertainties involved in predictions from their application.
In general, niche-based models are straightforward and efficient to fit. They rely on simple (primarily presence/absence records) and relatively little data (as few as five observed data points have been suggested as a minimum for some methods [23,24]); some methods address cases where no information is available on species absence. With such little information required to generate output and the widespread availability of software to fit even sophisticated models, it is unsurprising that many studies use these methods, including numerous high-profile publications [8,16]. However, the same reasons should provide warning that there is considerable uncertainty in predictions from these methods: all models are only as good as the data upon which they are built.
As the name suggests, niche-based distribution models implicitly focus on estimation of a species' niche from the geographical distribution of species [22,25]. Once the niche is estimated, then predictions (in space and time) can be made by re-projection from niche- to geographical space, provided data are available on the important niche dimension for the locations or time for which predictions are required.
Data quality is an obvious source of uncertainty and much can be undertaken to improve matters. Unfortunately, knowledge of species distributions is incomplete: many of the most biodiverse regions remain poorly known, with discoveries of new species regularly reported [26,27]. Species misidentifications can also be common and individual observers vary in their error rates (C. Beale 2011, personal observation). Even when species distributions are perfectly observed, the raw data may still be a source of uncertainty, e.g. some presence records refer to vagrants that have dispersed away from their usual range, adding to the error in modelling. Similarly, presence records may refer to sink populations [28,29]: while including such presence records is inevitable, it raises the unwelcome possibility that the populations predicted to be present at some time in the future may consist only of potential sink populations that would not be viable on their own.
Unknown or incompletely known recording effort is a pervasive and an important source of problems for species data, as it is almost always spatially non-random [30–32]. This can have unfortunate consequences, because spatially systematic variation in effort (or indeed habitat-specific differences in detectability) may match some explanatory covariates, resulting in incorrect estimation of niche [31]. Covariate quality is another source of uncertainty. Typically, in large-scale analyses, covariates are themselves predictions from models (e.g. based on reflectance values of satellite-derived data [33]), or interpolation of data measured from fewer sample locations. These covariates are, therefore, uncertain and incorporation of this within the modelling process is desirable—for example, gridded climate data are generated by interpolation from weather stations using modelling methods that give estimates of precision [34]; yet these uncertainty measures are not generally used in SDM.
Because niche-based models focus on identification of that part of a species' ecological niche central to its geographical distribution, modelling this as accurately as possible is important [25]. Before starting to fit a niche model, there is uncertainty over which niche to model: the fundamental niche or the realized niche? With niche-based modelling, data are most directly available on the realized niche (i.e. the environment where the species currently occurs), yet the fundamental niche of a species is often much wider than the realized niche [35]. Using the realized niche to predict distribution is probably appropriate when filling gaps in known distribution owing to lack of observations, but introduces considerable (and unmeasurable) uncertainty when predicting future distributions. Realized niches differ from fundamental niches owing to dispersal limitation and through interactions with other species [25,36,37]. Competitive exclusion of one species by another is common and there are numerous examples of range limits (or holes within distributions) determined by known competitors [38,39]. Competitors, however, may respond to environmental change in different ways; this could result in currently competing species sharing no future locations but instead interacting with other species in different (and currently unobservable) ways [40,41]. When competition (or facilitation) limits distribution, predicting from the realized niche is subject to inestimable errors, making predictions dependent on the unrealistic assumption that newly interacting species will have the same net interactions as current interactions. Furthermore, if dispersal barriers (e.g. sea crossings, mountains) exist and prevent a species from filling its fundamental niche, then future distributions are likely to be equally constrained in geography. Any niche model excluding these constraints fails to identify the limiting process and may incorrectly predict range expansion. Similar arguments apply to problems associated with source–sink dynamics and historical events that have constrained current distributions (e.g. glaciation cycles and volcanism).
These issues notwithstanding, there is information in current distributions that is useful for estimating the fundamental niche—though there is no certainty that sufficient information on niche limits exist in any given dataset. Indeed, it seems unlikely that many species have distributions suitable for full identification of any niche, realized or fundamental: even for taxa with distributions completely within a study region, there is no guarantee that all niche axes have limits within the area. Equally, there is no certainty that species with only partial ranges within a study area do not reach tolerance limits within the area (figure 1, cf. [42,43]): geographical distribution and distribution within niche space are different phenomena. Modelling the fundamental niche from geographical data is harder than modelling the realized niche, but doing so avoids some problems highlighted above. In practice, no matter how sophisticated might the modelling method be, full recovery of the fundamental niche is unlikely. Instead, an approximation somewhere between the realized and fundamental niche is likely, meaning that some issues associated with the realized niche remain in predictions from models of the fundamental niche. For prediction, however, further problems are generated: consider a species where most of its geographical range is limited by competition. For the region of niche space occupied by this competitor, no accurate prediction is possible (though simultaneously modelling both species makes this feasible). Acknowledging this prediction uncertainty is preferable to confidently asserting that areas with such environmental characteristics will always be unsuitable, even if the competitor's distribution changes. Thus, methods that identify the fundamental niche in preference to the realized niche are preferable, despite the greater uncertainty associated with their predictions, because the narrower precision of the realized niche model probably underestimates uncertainty.
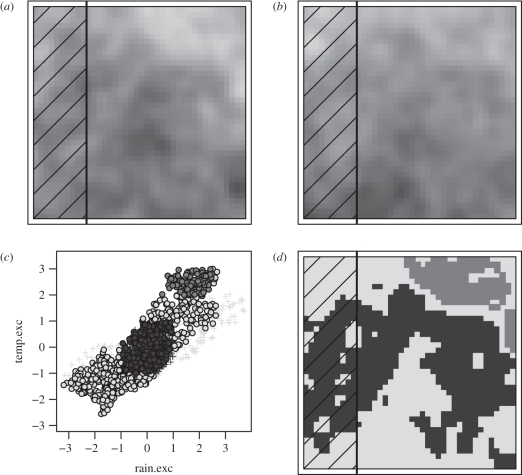
Figure 1.
A graphical demonstration that there is no necessary correlation between endemicity within a study area and ability to fit niche-based models. (a) Hypothetical temperature gradient across a square continent, with the eastern three-quarters (unmarked) being within the modelling area. (b) Hypothetical rainfall gradient across the same square continent, hashes again indicate portion of the continent outside the study area. (c) Niche-space within the continent—black rimmed circles fall within the surveyed area; plus symbols (‘+’) within the unsurveyed space. Fill indicates presence of two hypothetical species: dark or medium grey. (d) Geographical projections of the niches indicated in (c) for the two hypothetical species. Note that while panel (d) shows that the medium grey species' range falls completely within the surveyed area, it is impossible to identify upper niche limits for temperature for this species, while the dark grey species is not endemic to the study area, but all four niche limits can be identified from the data availability in panel (c).
Another source of modelling error in niche models involves identifying niche dimensions. Trivially, excluding an important covariate leads to a poor model and poor prediction. Equally, including redundant variables reduces the accuracy of parameter estimation, particularly, if unnecessary covariates correlate with useful variables [44]. Furthermore, addition of numerous redundant covariates increases the probability of over-fitting. This is of particular concern because many niche-based models use automated fitting methods with minimal or no selection of variables: it is not unusual for models to be built with ca 13 highly correlated covariates [45,46]. This problem is only one form of statistical model misspecification, all forms of which reduce prediction accuracy, while exaggerating precision. A further example is misspecification of the error term: many modelling methods either have no such term, or implicitly assume spatially independent errors when a model of spatially structured residual errors is appropriate (though leads to apparently less-precise predictions, as they do not overestimate precision [44]). Other often overlooked sources of uncertainty relate to the functional form of the niche model: to fit a plausible model requires considering the likely shape of the niche and, if necessary, constraining the statistical model to a biologically plausible form. It is, for example, unlikely that any real species has a fundamental niche that includes temperatures of 10–15°C, excludes 16–20°C but includes again 21–25°C, so fitting a unimodal model is better than one that is more flexible [19,47]—although additional flexibility may be preferable if modelling the realized niche [12]. Structural model misspecification also results in misidentification of covariates when a multi-model or covariate selection process is undertaken because the model likelihoods (and/or Akaike's information criterion (AIC)) will be wrong. When predicting, it is important to remember that fitted distributions are smoother than real distributions: they are more strongly autocorrelated than real distributions and generally have fewer isolated patches of presence. While inevitable (correct classification of presence and absence is weighted towards the main presence and absence areas, rather than correct classification of peripheral populations), this is important where small and isolated populations may be valuable. For example, calculating extinction risk requires knowing when the last population goes extinct: smooth modelled distributions with no outliers are more likely to predict total extinction than a real distribution with populations existing beyond the main climate envelope.
An important additional source of uncertainty in niche-based distribution models is in the assessment of model performance and goodness-of-fit [12,48,49]. Ultimately, all models must be assessed for performance and predictive ability, but this is not as straightforward as implied by many papers in this field. Typically, assessment is undertaken by comparison of observed with predicted distribution for current conditions. Apart from the problem of making genuinely independent predictions of distribution when these data (or at best a highly autocorrelated subset of test data) are used in model building [19], measures assessing pattern match are not without problems. Measures such as the area under the receiver-operating characteristics curve (AUC), or the true skill statistic are typical, but do not offer unbiased measures of pattern-matching ability [12,50,51] and are applied without penalization for model complexity, resulting in risks of over-fitting, which greatly reduces predictive ability [48,52]. A problem not generally noted is that increasing the study area by including additional unsuitable areas always results in an increased AUC or similar score, though the model is neither better nor worse than before: these scores are not a true goodness-of-fit statistic and do not explain how well a model predicts distribution, rather they estimate how well a prediction fits a particular dataset; a subtle, but important distinction. Furthermore, measures that assess overall significance of a model are not necessarily suitable for determining how much variation is explained by the model: statistical significance means neither biological significance nor strong predictive power. Moreover, models estimating the fundamental, rather than realized niche, are far harder to assess using simple pattern-matching: a geographical projection of the fundamental niche may be substantially larger than that of the realized niche, so a perfect model may have a relatively poor match to the current observed distribution, which reflects the realized niche. This source of uncertainty in ability to assess model fit applies to all classes of SDMs.
Prediction uncertainty of niche-based distribution models also occurs from the covariates used to predict future distribution. Such projections rely on the output of climate models and can be extremely uncertain, particularly for precipitation-related variables [53]. Moreover, not all variables are available as output of climate models, and projections of future values of covariates may be lacking. The availability of these future data can restrict initial covariate choice, despite the knowledge that factors for which future predictions are unavailable may be important [54]; i.e. models may be deliberately misspecified to reflect data availability, rather than for ecological reasons. Such sources of error are compounded when the uncertainty of climate predictions is ignored. Another source of prediction uncertainty comes from ‘novel climates’: future climates may have no analogues within the study area and accordingly the species response to this new environment cannot be known [55].
Many causes of uncertainty can be addressed by statistical methods that, while not eliminating uncertainty, allow correct assessment of precision. Usually, correctly modelling precision reduces apparent certainty in the output of SDMs because many exiting niche-based models give overconfident predictions with false precision. Suitable tools to improve uncertainty estimation are available for all stages of the modelling process. Many errors associated with distribution data can be incorporated within occupancy models to provide estimates of niche unbiased by observer ability [56,57]. Similarly, hierarchical models can explicitly incorporate uncertainty in covariates (both in input variables and in predicted future values where available), through resampling of plausible covariate values and model refitting [58]. The novelty of future climates may be a source of irreducible uncertainty for model extrapolations.
Additionally, appropriate spatial error models can be incorporated during model building [12,19,41]. It is more challenging to ascertain how to model interactions between species given the complexity seen in even simple food webs and the analytical challenge presented by multi-species spatial data. Yet, methods such as Bayesian network tools are developing that allow complex interaction networks to be inferred from distribution data while simultaneously estimating the influence of covariates [59,60]. Ultimately, new methods will develop that incorporate this full range of statistical advances, allowing credible estimation of uncertainty.
4. Demographic models
In this category, we identify two main types of model. First, there are classic metapopulation models explaining local patch occupancy as a dynamic process of birth, death, immigration and emigration models linked by dispersal [61]. Such models can be parametrized by correlating demographic components with weather and then used for future prediction using climate projections [62]. Second, models have been developed that again correlate demographic components with climate or weather, often identifying intermediate linkage via food availability but are not patch-based [63–66]. In reality, models of both types fall on a continuum from the purely statistical to those that include climate links to physiology and/or intermediate processes such as food availability that tend towards process-based models.
Metapopulation models of the stochastic patch occupancy type would be excellent tools for modelling fragmented species distributions (species interactions aside). They can accommodate patch structure, fine-scale habitat fragmentation and landscape architecture, population density and Allee effects, local adaptation and evolution, local environmental conditions, and even the behaviour of individuals [67]. Metapopulation models can predict future distributions using two approaches. Firstly, future environments may modify the availability of suitable habitat patches, with some patches becoming unsuitable, and others becoming suitable as the environment changes [62,68,69]. Alternatively, demographic parameters within patches may be adjusted according to climate-related processes measured in the field or laboratory. For example, temperature-related hatching success is widespread among many insect species to which metapopulation models have been applied [70]. Clearly, the patch loss and demographic shifts are not mutually exclusive and both can be incorporated into the same predictive model. Uncertainty in these models is explicitly recognized in their stochastic nature, provided variation in demographic components is accurately measured.
While useful when applied to particular species at the landscape scale (but see [71,72]), in reality these models require intensive fieldwork to parametrize and are typically practical only for small proportions of a species range [71], thereby limiting their usefulness to meet the challenge of global environmental impacts on biodiversity. Furthermore, metapopulation models are criticized for their difficulty in dealing with landscapes other than simply suitable patches within a matrix of unsuitable (though maybe permeable) habitat, and also have little to offer when biotic interactions (beyond direct host–plant relationships) are important in determining distribution limits [67,71].
The second type of model within the class which we call demographic models is less well-defined. At base, these models use information on breeding and survival, correlate these with weather variables and from this generate a simple demographic model [64]. When this statistical model is embedded in a geographical grid with cells with different climate conditions, estimates of the intrinsic rate of population change become spatially explicit, generating an SDM. Complexity (and further realism) can be added by identifying the mechanism linking weather and demographic components. Typically, this involves the linkage between weather and food availability, which, in turn, is linked to breeding success. For example, Both et al. [66] describe how climate change is likely to alter the distribution of pied flycatchers (Ficedula hypoleuca) through generating a temporal mismatch between timing of the bird's breeding and the peak spring availability of their caterpillar prey, resulting in reduced breeding success in areas where early spring has rapidly advanced. Similar studies of other birds also identify range-margin populations under pressure from climate change [65]. So far, there are few examples of this model type and particularly their projection into geographical distributions. To date, those that have use only mean estimates from estimated relationships [65] and therefore underestimate the uncertainties in predictions, yet there is no reason why future predictions may not be generated stochastically to quantify uncertainty appropriately.
In general, demographic models are appealing as SDMs, as they are explicit about the demographic hypotheses involved and offer transparent estimation of uncertainty. Unfortunately, geographical predictions from these methods are accurate only if the demographic model correctly identifies the important factors determining population limitation. As these correlative methods implicitly (but often incorrectly) assume that the demographic impacts of weather and climate are identical, model misspecification errors may be large. For example, consider that many upland birds are restricted in geographical range to places with cool, wet climates [73]. While these species may struggle in a warm, dry climate, many of them show higher breeding success and better survival in warm, dry years than in the typical conditions they usually experience [74–76]. Clearly, demographic models would predict a more widespread population than their preferred upland habitats, suggesting that while warm, dry weather may be good, a warm, dry climate would not. Instead, the distribution of these species relates to climate via potentially complex interactions with other species which, if this class of model is to be correctly specified, must be simultaneously modelled.
Predictions of future distribution from metapopulation models are clouded by uncertainties in determining patch suitability for currently unoccupied patches. This has been attempted using statistical models of habitat quality (with some success [77]), but the uncertainties in determining unoccupied patch suitability are rarely quantified. As with niche-based models, there is also a problem extrapolating to future conditions when expected conditions exceed the variability sampled in current data: wherever relationships between climate and demography are based on observation, extrapolations are subject to large but unmeasurable uncertainties.
Looking forward, it seems likely that the fundamental components of demographic models will not change greatly, but there will continue to be refinement of individual models as data on additional mechanisms accrue. We expect that these models will be fitted to more species: usable data are routinely collected in existing monitoring schemes for many more species than so far modelled. As these models are developed further, the same uncertainties about measures of model fit that affect niche-based model predictions will become important.
5. Process-based models
This third and final class of SDM takes an approach opposite to that of niche-based models. Instead of building a statistical model of niche from geographical distribution, process-based models instead determine niche physiology and then reconstruct geographical distribution from this. This is a relatively new field, with comparatively few working models. Examples of the genre include models for trees that identify niche using aspects of phenology (e.g. PHENOFIT [78]), which assumes that a species can survive only if all components of its life cycle are completed within a calendar year: producing leaves while avoiding frost, growing flowers, setting seed, etc. Similar approaches for animals may be possible, but the best examples focus on determining whether animals meet energetic demands [79]. Kearney et al. [80] describe a model of the greater glider (Petauroides volans) using assessments of energetic and evaporative costs converted to units of milk. They identify the physiological niche where sufficient milk can be produced to successfully raise young and project this to geographical space. Similarly, Kearney et al.'s [81] model of the cane toad (Bufo marinus) measures the impacts of temperature on movement, reproduction and survival, then projects this to geographical distribution. At an even more fundamental level, there are models of plants that use the chemistry of photosynthesis and physiology of water-stress to model tree responses to global change (indeed, some argue that only models based directly on physiology should truly be described as process-based models, all other approaches being descriptions of the relationship between climate and demography). Functioning at this level means such models can incorporate processes such as responses to changing CO2 levels [82].
Process-based models often require numerous parameters (e.g. timing of phenological events, or sensitivity of activities to climate), each of which carries uncertainty. Correctly assessing uncertainties in process-based models involves identification of sources of variation for each parameter, including individual variation and population differences caused by local adaptation. It is simple to incorporate this variation into output through resampling of observed variation and re-running the model, though to date most models discard information on variation and instead use averages [78] or at best undertake sensitivity analyses [81]. More complex models have more parameters and suffer more from parameter estimation uncertainty, as error propagation results in increasingly imprecise estimates as complexity increases. Indeed, creating geographical predictions from the output of the most complex process-based models is problematic as, for example, tree growth rates (raw output from some physiological models) must be translated into population dynamics [82].
As with niche-based models, a further source of error in process-based models is the challenge of identifying the important niche axes. Although niche-based models use pattern-matching to identify likely niche components, process-based models rely on an understanding of the species' ecology to identify important niche axes. Where direct impacts of temperature or water-stress are important, experiments can measure tolerance and identify limiting factors [78,81]. Often, however, the interaction between environment and organism is complicated and experiments are impossible or unethical: it is hard to justify using temperature chambers to identify stressful environmental conditions on many animal species, especially rare ones. In such cases, tolerance must be measured differently [80] with increasing errors. Moreover, identification of climate impacts on life-history components is challenging, requiring detailed knowledge of the ecology of the target species: once options are identified, the only appropriate test is to run the model and see how well predicted matches observed distribution. This comparison is even more challenging than for niche-based models because the prediction is based on the fundamental niche (though if dispersal is incorporated within the model, the prediction can be expected to be closer to the realized niche), while the geographical distribution is formed by the realized niche: over-prediction is virtually guaranteed.
An important advantage of process-based models is that true prediction into conditions beyond observed conditions (e.g. into areas with novel climates) is possible [82], and can incorporate appropriate uncertainty. Moreover, genetic evolution can be included within process-based models, enabling this potentially important mechanism to be incorporated [83]. Indeed, models containing competitive interactions with other species are possible, though have not yet been attempted. Without such multi-species modelling, however, the same problems with lack of interactions remain for process-based models as other distribution modelling approaches.
Distribution models based on physiology can be further abstracted to generic trait types and not for individual species. Such models predict not species, but collections of plant functional types defining vegetation classes, such as biomes. They are termed dynamic global vegetation models (DGVMs; [84,85]) and not usually considered as SDMs, but their conceptual and mathematical similarity to single-species process-based models makes it appealing to consider their use alongside more standard distribution models. In contrast to single-species models, DGVMs explicitly model vegetation assemblages and the boundaries between major community types from basic plant physiology, using plant functional types to define communities [47]. Most DGVMs are run only for single or small sets of parameter values, providing sensitivity and not uncertainty estimates, but increased computer power will soon enable uncertainty assessments [86]. Other refinements, such as the incorporation of additional traits, will follow [47]. Assessment of DGVM accuracy, like that of typical SDMs, lies in the match between predicted current distribution and that of observed distribution [84,85]. As with all models, this comparison is subject to uncertainty in the observed distributions (usually assessed from satellite imagery), but also in the definition of plant communities: many biomes do not have clear edges, but rather grade from one to another through intermediate states, and boundary choice is arbitrary and introduces uncertainty. Unlike single-SDMs, however, DGVMs are not subject to uncertainty from interactions, as the competition between biomes and associations between species are explicitly included in the models. There are, however, uncertainties in this for those systems where factors other than climate drive biome limits, such as grassland–forest interfaces mediated through herbivory and fire [87], only the latter of which is sometimes incorporated into DGVMs. Furthermore, current DGVMs are subject to uncertainties from the lack of explicit modelling of dispersal [88]. Despite these uncertainties, however, DGVMs are remarkably accurate in their overall prediction of global vegetation communities [85]. It seems that sacrificing information on species identity makes it possible to improve predictions about collections of species as vegetation types. This may be because community or local assemblage membership rules operating via species interactions combine with environmental filters acting on species traits to make the average properties more predictable than specific presence or absence of individual species.
Uncertainty in process-based models can easily be incorporated into predictions. However, uncertainties from the modelling process are not the only sources of uncertainty, as predictions are generated from an assumed model of the niche. Using a process-based model, one cannot quantify the uncertainty that occurs owing to missing niche components. For example, if Kearney et al. [81] had not measured mobility and assumed only reproduction and survival were temperature-sensitive they could have built a process-based model including full uncertainty from the variability in temperature and demography—but their model would have been less accurate than the uncertainty thus derived supposes. Unfortunately, this uncertainty is irreducible and the only test of the model is to compare observed with modelled distribution, with all the problems this involves. So far, few process-based models have been published and it remains unknown how difficult it is to identify niche parameters—studies published to date have been successful, predicting current distributions similar to observed patterns [79]. However, it is unlikely that process-based models which do not predict current distribution well will be publishable, when one cannot determine if it is the model that is misspecified, or if realized and fundamental niche are simply too different [89].
Thanks to their bottom-up approach and the ability to predict distributions independent of the data used to generate them, process-based models have an important contribution to make in this field. Models of individual species will become increasingly well-defined, as information accrues about niche dimensions and physiological responses to temperature and water-stress are better understood. There is little doubt that models will be developed for more species and new software is becoming available to assist the process. We suggest, however, that the requirement for detailed, species-specific information (often involving laboratory experiments) means that process-based models will never be available for the number and diversity of species that are required, for example to make global assessments of species sensitivity to climate change. Instead, we suggest that an abstraction is likely, similar to that of DGVMs, where species identity is ignored and trait-based analyses develop that allow sensitivity to be assessed based on readily available trait information.
6. Discussion
From this overview, it is clear that multiple sources of uncertainty apply to all model types. Data uncertainties in environmental covariates affect output from all models, and should always be incorporated in model predictions but this is rarely done [90]. Uncertainties in species distribution data present particular challenges to niche-based models, but also need considering when validating predictions from all model types using observed data—another issue often overlooked. Prediction uncertainty of future climate from climate models needs to be considered in predictions from all model types; e.g. rather than using median outputs for each climate model variable, individual realizations should be used. Parameter uncertainty should be estimated and recorded, whether using a niche-based model, or a model of fundamental niche based on laboratory-measured physiological tolerance. And all examples are subject to uncertain influences of biotic interactions. Other sources of uncertainty are specific to model class: niche-based models are subject to uncertainty from having no mechanism to include dispersal and source–sink processes, whereas mechanistic niches modelled in process-based models never reflect true complexity of biological systems. Although most sources of uncertainty have been previously identified and occasionally measured [15], we know of no example where all known sources of uncertainty have been measured. As many sources of uncertainty have multiplicative effects, it is inevitable that uncertainty in SDMs is generally underestimated. Indeed, once appropriate measures of prediction uncertainty are available, we expect that for many species our best models are completely uninformative.
We have further identified gaps where research can reduce the uncertainties associated with current SDMs. Primary among these is an appropriate measure of model fit. Analysts have relied heavily on internal cross-validation and AUC measures of predictive performance, but this approach can misidentify over-fitted models as well-fitting and strongly predictive. Until we know with confidence how well a model performs on independent datasets, we remain fundamentally uncertain about predictions of all SDMs. Such a measure must identify how well predictions fit observed data (dealing appropriately with the uncertainty in the observations themselves) while penalizing for model complexity, and should remain unaffected by both autocorrelation in the observed pattern [51] and prevalence [50]. Ideally, models should be tested against independent data, such as in an introduced range [91,92], through use of historic [93] or palaeontological [47] datasets to retrodict distribution, and through the use of simulated data where the ability of the model to recover known processes is a measure of performance.
Measures appropriate to many uncertainty sources are already available, both through mechanisms to incorporate uncertainty into the modelling framework (e.g. hierarchical models allow estimates and errors to propagate through various submodels [57]) and through those designed to increase accuracy. In the latter class fall ensemble forecasts which average across many alternative models [94]. However, while averaging is useful for increasing precision, it does not affect the accuracy and current implementations of ensemble modelling do not preserve measures of uncertainty through the averaging process. Nevertheless, comparisons of multiple models are useful for reducing uncertainty and examples exist where comparisons have been made between different classes of distribution model, allowing differences and similarities to be identified. Notably, Kearney et al. [80] compared niche-based models and a process-based model of the distribution of the invasive cane toad in Australia. They showed that while some niche-based models give different outputs, one method in particular (maximum entropy [95]) gave a similar predicted distribution to the process-based model. By contrast, Keenan et al. [82] compared niche-based models and process-based models for three tree species and found that if impacts of increasing CO2 are ignored, then both niche- and process-based models give qualitatively similar predictions, but if CO2 is incorporated into the process-based model, the predictions are reversed: the effect of CO2 is overwhelmingly important, but cannot be identified by niche-based modelling methods. Other comparisons report similarly disparate results, and in cases of disagreement, we suggest the more fundamental insights coming from a good process-based model should be preferred to those of a pattern-based approach. These comparisons are important because they allow uncertainties from different sources to be identified, and the potential magnitude of mistakes to be estimated.
While comparisons are useful, they are not the only way multiple model types can be combined: different model types can be joined statistically, in similar ways different types of climate models have been linked to generate localized predictions [96]. For example, niche-based models and demographic- or process-based models could be integrated across spatial scales in a hierarchical framework [97], or more simply, DGVM output could feed into SDMs to better predict species reliant on particular biomes. These methods could further be linked to network recovery tools to estimate and incorporate the impacts of biotic interactions. Implementing such ‘second-generation’ SDMs will require further statistical research developing methods to identify biotic interactions and to develop specific hierarchical models relevant to this problem. The bases for such models are already available [59,60], and progress in these areas will likely be rapid: any attempt to move models from a simple pattern-based approach to a more fundamental understanding of ecological processes is to be welcomed.
Returning to the use of SDMs to inform management decisions, the final area for further development is in the incorporation of uncertainty into decision-making frameworks. Decision theory is a well-developed field, but has only recently appeared in the ecological management literature [98–100]. Perhaps ironically, for many of the species of most interest to conservation (often those with small ranges), modelling uncertainty will always be large. Indeed, not only may uncertainty be great, but also some sources are unquantifiable, such that we are uncertain about the degree of uncertainty in many distribution models. We argue that it is just as important to quantify uncertainty in model predictions as to make the predictions themselves, yet the importance of prediction uncertainty is rarely emphasized. By correctly identifying the existence and sources of uncertainty, even if in the worst case predictions are impossible, advice and management actions will be better informed than if based on false certainty.
References
- 1.Gaston K. J., Blackburn T. M. 1999. A critique for macroecology. Oikos 84, 353–368 10.2307/3546417 (doi:10.2307/3546417) [DOI] [Google Scholar]
- 2.Kearney M., Porter W. P., Williams C., Ritchie S., Hoffmann A. A. 2009. Integrating biophysical models and evolutionary theory to predict climatic impacts on species' ranges: the dengue mosquito Aedes aegypti in Australia. Funct. Ecol. 23, 528–538 10.1111/j.1365-2435.2008.01538.x (doi:10.1111/j.1365-2435.2008.01538.x) [DOI] [Google Scholar]
- 3.Rouget M., Richardson D. M., Milton S. J., Polakow D. 2001. Predicting invasion dynamics of four alien Pinus species in a highly fragmented semi-arid shrubland in South Africa. Plant Ecol. 152, 79–92 10.1023/A:1011412427075 (doi:10.1023/A:1011412427075) [DOI] [Google Scholar]
- 4.Loiselle B. A., Howell C. A., Graham C. H., Goerck J. M., Brooks T., Smith K. G., Williams P. H. 2003. Avoiding pitfalls of using species distribution models in conservation planning. Conserv. Biol. 17, 1591–1600 10.1111/j.1523-1739.2003.00233.x (doi:10.1111/j.1523-1739.2003.00233.x) [DOI] [Google Scholar]
- 5.Rodríguez J. P., Brotons L., Bustamante J., Seoane J. 2007. The application of predictive modelling of species distribution to biodiversity conservation. Div. Distrib. 13, 243–251 10.1111/j.1472-4642.2007.00356.x (doi:10.1111/j.1472-4642.2007.00356.x) [DOI] [Google Scholar]
- 6.Anderson B. J., et al. 2009. Using distribution models to test alternative hypotheses about a species' environmental limits and recovery prospects. Biol. Conserv. 142, 488–499 10.1016/j.biocon.2008.10.036 (doi:10.1016/j.biocon.2008.10.036) [DOI] [Google Scholar]
- 7.Parmesan C., Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 10.1038/nature01286 (doi:10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 8.Kremen C., et al. 2008. Aligning conservation priorities across taxa in Madagascar with high-resolution planning tools. Science 320, 222. 10.1126/science.1155193 (doi:10.1126/science.1155193) [DOI] [PubMed] [Google Scholar]
- 9.Vaughan I. P., Ormerod S. J. 2003. Improving the quality of distribution models for conservation by addressing shortcomings in the field collection of training data. Conserv. Biol. 17, 1601–1611 10.1111/j.1523-1739.2003.00359.x (doi:10.1111/j.1523-1739.2003.00359.x) [DOI] [Google Scholar]
- 10.Huntley B., Collingham Y. C., Willis S. G., Green R. E. 2008. Potential impacts of climatic change on European breeding birds. PLoS ONE 3, e1439. 10.1371/journal.pone.0001439 (doi:10.1371/journal.pone.0001439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julliard R., Jiguet F., Couvet D. 2004. Common birds facing global changes: what makes a species at risk? Glob. Change Biol. 10, 148–154 10.1111/j.1365-2486.2003.00723.x (doi:10.1111/j.1365-2486.2003.00723.x) [DOI] [Google Scholar]
- 12.Araújo M. B., Guisan A. 2006. Five (or so) challenges for species distribution modelling. J. Biogeogr. 33, 1677–1688 10.1111/j.1365-2699.2006.01584.x (doi:10.1111/j.1365-2699.2006.01584.x) [DOI] [Google Scholar]
- 13.Heikkinen R. K., Luoto M., Araujo M. B., Virkkala R., Thuiller W., Sykes M. T. 2006. Methods and uncertainties in bioclimatic envelope modelling under climate change. Prog. Phys. Geogr. 30, 751–777 10.1177/0309133306071957 (doi:10.1177/0309133306071957) [DOI] [Google Scholar]
- 14.Thuiller W. 2004. Patterns and uncertainties of species' range shifts under climate change. Glob. Change Biol. 10, 2020–2027 10.1111/j.1365-2486.2004.00859.x (doi:10.1111/j.1365-2486.2004.00859.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buisson L., Thuiller W., Casajus N., Lek S., Grenouillet G. 2010. Uncertainty in ensemble forecasting of species distribution. Glob. Change Biol. 16, 1145–1157 10.1111/j.1365-2486.2009.02000.x (doi:10.1111/j.1365-2486.2009.02000.x) [DOI] [Google Scholar]
- 16.Thomas C. D., et al. 2004. Extinction risk from climate change. Nature 427, 145–148 10.1038/nature02121 (doi:10.1038/nature02121) [DOI] [PubMed] [Google Scholar]
- 17.Berkhout F. 2010. Reconstructing boundaries and reason in the climate debate. Glob. Environ. Change 20, 565–569 10.1016/j.gloenvcha.2010.07.006 (doi:10.1016/j.gloenvcha.2010.07.006) [DOI] [Google Scholar]
- 18.Raiffa H., Schlaifer R. 1968. Applied statistical decision theory. Chichester, UK: Wiley [Google Scholar]
- 19.Austin M. P. 2002. Spatial prediction of species distribution: an interface between ecological theory and statistical modelling. Ecol. Model. 157, 101–118 10.1016/S0304-3800(02)00205-3 (doi:10.1016/S0304-3800(02)00205-3) [DOI] [Google Scholar]
- 20.Elith J., et al. 2006. Novel methods improve prediction of species' distributions from occurrence data. Ecography 29, 129–151 10.1111/j.2006.0906-7590.04596.x (doi:10.1111/j.2006.0906-7590.04596.x) [DOI] [Google Scholar]
- 21.Elith J., Leathwick J. R. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677–697 10.1146/annurev.ecolsys.110308.120159 (doi:10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 22.Guisan A., Thuiller W. 2005. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009 10.1111/j.1461-0248.2005.00792.x (doi:10.1111/j.1461-0248.2005.00792.x) [DOI] [PubMed] [Google Scholar]
- 23.Hernandez P. A., Graham C. H., Master L. L., Albert D. L. 2006. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 29, 773–785 10.1111/j.0906-7590.2006.04700.x (doi:10.1111/j.0906-7590.2006.04700.x) [DOI] [Google Scholar]
- 24.Pearson R. G., Raxworthy C. J., Nakamura M., Peterson A. T. 2007. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J. Biogeogr. 34, 102–117 10.1111/j.1365-2699.2006.01594.x (doi:10.1111/j.1365-2699.2006.01594.x) [DOI] [Google Scholar]
- 25.Pulliam H. R. 2000. On the relationship between niche and distribution. Ecol. Lett. 3, 349–361 10.1046/j.1461-0248.2000.00143.x (doi:10.1046/j.1461-0248.2000.00143.x) [DOI] [Google Scholar]
- 26.Alstroem P. E. R., Davidson P., Duckworth J. W., Eames J. C., Le T. T., Nguyen C., Olsson U., Robson C., Timmins R. 2010. Description of a new species of Phylloscopus warbler from Vietnam and Laos. Ibis 152, 145–168 10.1111/j.1474-919X.2009.00990.x (doi:10.1111/j.1474-919X.2009.00990.x) [DOI] [Google Scholar]
- 27.Ceballos G., Ehrlich P. R. 2009. Discoveries of new mammal species and their implications for conservation and ecosystem services. Proc. Natl Acad. Sci. USA 106, 3841. 10.1073/pnas.0812419106 (doi:10.1073/pnas.0812419106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eriksson O. 1996. Regional dynamics of plants: a review of evidence for remnant, source–sink and metapopulations. Oikos 77, 248–258 10.2307/3546063 (doi:10.2307/3546063) [DOI] [Google Scholar]
- 29.Kawecki T. J. 1995. Demography of source—sink populations and the evolution of ecological niches. Evol. Ecol. 9, 38–44 10.1007/BF01237695 (doi:10.1007/BF01237695) [DOI] [Google Scholar]
- 30.Dennis R. L., Sparks T. H., Hardy P. B. 1999. Bias in butterfly distribution maps: the effects of sampling effort. J. Insect Conserv. 3, 33–42 10.1023/A:1009678422145 (doi:10.1023/A:1009678422145) [DOI] [Google Scholar]
- 31.Hortal J., Jiménez-Valverde A., Gómez J. F., Lobo J. M., Baselga A. 2008. Historical bias in biodiversity inventories affects the observed environmental niche of the species. Oikos 117, 847–858 10.1111/j.0030-1299.2008.16434.x (doi:10.1111/j.0030-1299.2008.16434.x) [DOI] [Google Scholar]
- 32.Prendergast J. R., Wood S. N., Lawton J. H., Eversham B. C. 1993. Correcting for variation in recording effort in analyses of diversity hotspots. Biodivers. Lett. 1, 39–53 10.2307/2999649 (doi:10.2307/2999649) [DOI] [Google Scholar]
- 33.Guisan A., Zimmermann N. E. 2000. Predictive habitat distribution models in ecology. Ecol. Model. 135, 147–186 10.1016/S0304-3800(00)00354-9 (doi:10.1016/S0304-3800(00)00354-9) [DOI] [Google Scholar]
- 34.Johnson G. L., Daly C., Taylor G. H., Hanson C. L. 2010. Spatial variability and interpolation of stochastic weather simulation model parameters. J. Appl. Meteorol. 39, 778–796 (doi:10.1175/1520-0450(2000)039<0778:SVAIOS>2.0.CO;2) [DOI] [Google Scholar]
- 35.Vetaas O. R. 2002. Realized and potential climate niches: a comparison of four Rhododendron tree species. J. Biogeogr. 29, 545–554 10.1046/j.1365-2699.2002.00694.x (doi:10.1046/j.1365-2699.2002.00694.x) [DOI] [Google Scholar]
- 36.Malanson G. P., Westman W. E., Yan Y. L. 1992. Realized versus fundamental niche functions in a model of chaparral response to climatic change. Ecol. Model. 64, 261–277 10.1016/0304-3800(92)90026-B (doi:10.1016/0304-3800(92)90026-B) [DOI] [Google Scholar]
- 37.Soberón J. 2007. Grinnellian and Eltonian niches and geographic distributions of species. Ecol. Lett. 10, 1115–1123 10.1111/j.1461-0248.2007.01107.x (doi:10.1111/j.1461-0248.2007.01107.x) [DOI] [PubMed] [Google Scholar]
- 38.Case T. J., Taper M. L. 2000. Interspecific competition, environmental gradients, gene flow, and the coevolution of species' borders. Am. Nat. 155, 583–605 10.1086/303351 (doi:10.1086/303351) [DOI] [PubMed] [Google Scholar]
- 39.Secondi J., Bretagnolle V., Compagnon C., Faivre B. 2003. Species-specific song convergence in a moving hybrid zone between two passerines. Biol. J. Linnean Soc. 80, 507–517 10.1046/j.1095-8312.2003.00248.x (doi:10.1046/j.1095-8312.2003.00248.x) [DOI] [Google Scholar]
- 40.Pearson R. G., Dawson T. P. 2004. Bioclimate envelope models: what they detect and what they hide—response to Hampe (2004). Glob. Ecol. Biogeogr. 13, 471–473 10.1111/j.1466-822X.2004.00112.x (doi:10.1111/j.1466-822X.2004.00112.x) [DOI] [Google Scholar]
- 41.Hampe A. 2004. Bioclimate envelope models: what they detect and what they hide. Glob. Ecol. Biogeogr. 13, 469–471 10.1111/j.1466-822X.2004.00090.x (doi:10.1111/j.1466-822X.2004.00090.x) [DOI] [Google Scholar]
- 42.Peterson A. T., et al. 2009. The climate envelope may not be empty. Proc. Natl Acad. Sci. USA 106, E47. 10.1073/pnas.0809722106 (doi:10.1073/pnas.0809722106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiménez-Valverde A., Barve N., Lira-Noriega A., Maher S. P., Nakazawa Y., Papeş M., Soberón J., Sukumaran J., Peterson A. P. 2011. Dominant climate influences on North American bird distributions. Glob. Ecol. Biogeogr. 20, 114–118 10.1111/j.1466-8238.2010.00574.x (doi:10.1111/j.1466-8238.2010.00574.x) [DOI] [Google Scholar]
- 44.Beale C. M., Lennon J. J., Yearsley J. M., Brewer M. J., Elston D. A. 2010. Regression analysis of spatial data. Ecol. Lett. 13, 246–264 10.1111/j.1461-0248.2009.01422.x (doi:10.1111/j.1461-0248.2009.01422.x) [DOI] [PubMed] [Google Scholar]
- 45.Beaumont L. J., Hughes L., Poulsen M. 2005. Predicting species distributions: use of climatic parameters in BIOCLIM and its impact on predictions of species' current and future distributions. Ecol. Model. 186, 251–270 10.1016/j.ecolmodel.2005.01.030 (doi:10.1016/j.ecolmodel.2005.01.030) [DOI] [Google Scholar]
- 46.Carpenter G., Gillison A. N., Winter J. 1993. DOMAIN: a flexible modelling procedure for mapping potential distributions of plants and animals. Biodivers. Conserv. 2, 667–680 10.1007/BF00051966 (doi:10.1007/BF00051966) [DOI] [Google Scholar]
- 47.McMahon S. M., et al. 2011. Improving assessment and modelling of climate change impacts on global terrestrial biodiversity. Trends Ecol. Evol. 26, 249–259 10.1016/j.tree.2011.02.012 (doi:10.1016/j.tree.2011.02.012) [DOI] [PubMed] [Google Scholar]
- 48.Vaughan I. P., Ormerod S. J. 2005. The continuing challenges of testing species distribution models. J. Appl. Ecol. 42, 720–730 10.1111/j.1365-2664.2005.01052.x (doi:10.1111/j.1365-2664.2005.01052.x) [DOI] [Google Scholar]
- 49.Guisan A., Lehmann A., Ferrier S., Austin M., Overton J., Aspinall R., Hastie T. 2006. Making better biogeographical predictions of species' distributions. J. Appl. Ecol. 43, 386–392 10.1111/j.1365-2664.2006.01164.x (doi:10.1111/j.1365-2664.2006.01164.x) [DOI] [Google Scholar]
- 50.Lobo J. M., Jiménez-Valverde A., Real R. 2008. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145–151 10.1111/j.1466-8238.2007.00358.x (doi:10.1111/j.1466-8238.2007.00358.x) [DOI] [Google Scholar]
- 51.Beale C. M., Lennon J. J., Gimona A. 2009. European bird distributions still show few climate associations. Proc. Natl Acad. Sci. USA 106, E41–E43 10.1073/pnas.0902229106 (doi:10.1073/pnas.0902229106) [DOI] [Google Scholar]
- 52.Stockwell D. R., Peterson A. T. 2002. Effects of sample size on accuracy of species distribution models. Ecol. Model. 148, 1–13 10.1016/S0304-3800(01)00388-X (doi:10.1016/S0304-3800(01)00388-X) [DOI] [Google Scholar]
- 53.Murphy J. M., Sexton D. M., Barnett D. N., Jones G. S., Webb M. J., Collins M., Stainforth D. A. 2004. Quantification of modelling uncertainties in a large ensemble of climate change simulations. Nature 430, 768–772 10.1038/nature02771 (doi:10.1038/nature02771) [DOI] [PubMed] [Google Scholar]
- 54.Heikkinen R. K., Luoto M., Virkkala R., Pearson R. G., Körber J. H. 2007. Biotic interactions improve prediction of boreal bird distributions at macro-scales. Glob. Ecol. Biogeogr. 16, 754–763 10.1111/j.1466-8238.2007.00345.x (doi:10.1111/j.1466-8238.2007.00345.x) [DOI] [Google Scholar]
- 55.Williams J. W., Jackson S. T., Kutzbach J. E. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738. 10.1073/pnas.0606292104 (doi:10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Royle J. A., Link W. A. 2006. Generalized site occupancy models allowing for false positive and false negative errors. Ecology 87, 835–841 10.1890/0012-9658(2006)87[835:GSOMAF]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[835:GSOMAF]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 57.Royle J. A., Kery M., Gautier R., Schmid H. 2007. Hierarchical spatial models of abundance and occurrence from imperfect survey data. Ecol. Monogr. 77, 465–481 10.1890/06-0912.1 (doi:10.1890/06-0912.1) [DOI] [Google Scholar]
- 58.Draper D. 1995. Assessment and propagation of model uncertainty. J. R. Stat. Soc. B 57, 45–97 [Google Scholar]
- 59.Faisal A., Dondelinger F., Husmeier D., Beale C. M. 2010. Inferring species interaction networks from species abundance data: a comparative evaluation of various statistical and machine learning methods. Ecol. Inform. 5, 451–464 10.1016/j.ecoinf.2010.06.005 (doi:10.1016/j.ecoinf.2010.06.005) [DOI] [Google Scholar]
- 60.Milns I., Beale C. M., Smith V. A. 2010. Revealing ecological networks using Bayesian network inference algorithms. Ecology 91, 1892–1899 10.1890/09-0731.1 (doi:10.1890/09-0731.1) [DOI] [PubMed] [Google Scholar]
- 61.Hanski I. 1999. Metapopulation ecology. Oxford, UK: Oxford University Press [Google Scholar]
- 62.Wilson R. J., Davies Z. G., Thomas C. D. 2010. Linking habitat use to range expansion rates in fragmented landscapes: a metapopulation approach. Ecography 33, 73–82 10.1111/j.1600-0587.2009.06038.x (doi:10.1111/j.1600-0587.2009.06038.x) [DOI] [Google Scholar]
- 63.Sillett T. S., Holmes R. T., Sherry T. W. 2000. Impacts of a global climate cycle on population dynamics of a migratory songbird. Science 288, 2040. 10.1126/science.288.5473.2040 (doi:10.1126/science.288.5473.2040) [DOI] [PubMed] [Google Scholar]
- 64.Pearce-Higgins J. W., Yalden D. W., Dougall T. W., Beale C. M. 2009. Does climate change explain the decline of a trans-Saharan Afro-Palaearctic migrant? Oecologia 159, 649–659 10.1007/s00442-008-1242-4 (doi:10.1007/s00442-008-1242-4) [DOI] [PubMed] [Google Scholar]
- 65.Pearce-Higgins J. W., Dennis P., Whittingham M. J., Yalden D. W. 2010. Impacts of climate on prey abundance account for fluctuations in a population of a northern wader at the southern edge of its range. Glob. Change Biol. 16, 12–23 10.1111/j.1365-2486.2009.01883.x (doi:10.1111/j.1365-2486.2009.01883.x) [DOI] [Google Scholar]
- 66.Both C., Bouwhuis S., Lessells C. M., Visser M. E. 2006. Climate change and population declines in a long-distance migratory bird. Nature 441, 81–83 10.1038/nature04539 (doi:10.1038/nature04539) [DOI] [PubMed] [Google Scholar]
- 67.Ovaskainen O., Hanski I. 2004. From individual behavior to metapopulation dynamics: unifying the patchy population and classic metapopulation models. Am. Nat. 164, 364–377 10.1086/423151 (doi:10.1086/423151) [DOI] [PubMed] [Google Scholar]
- 68.Griffiths R. A., Sewell D., McCrea R. S. 2010. Dynamics of a declining amphibian metapopulation: survival, dispersal and the impact of climate. Biol. Conserv. 143, 485–491 10.1016/j.biocon.2009.11.017 (doi:10.1016/j.biocon.2009.11.017) [DOI] [Google Scholar]
- 69.McLaughlin J. F., Hellmann J. J., Boggs C. L., Ehrlich P. R. 2002. Climate change hastens population extinctions. Proc. Natl Acad. Sci. USA 99, 6070. 10.1073/pnas.052131199 (doi:10.1073/pnas.052131199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berger D., Walters R., Gotthard K. 2008. What limits insect fecundity? Body size-and temperature-dependent egg maturation and oviposition in a butterfly. Funct. Ecol. 22, 523–529 10.1111/j.1365-2435.2008.01392.x (doi:10.1111/j.1365-2435.2008.01392.x) [DOI] [Google Scholar]
- 71.Baguette M. 2004. The classical metapopulation theory and the real, natural world: a critical appraisal. Basic Appl. Ecol. 5, 213–224 10.1016/j.baae.2004.03.001 (doi:10.1016/j.baae.2004.03.001) [DOI] [Google Scholar]
- 72.Hanski I. 2004. Metapopulation theory, its use and misuse. Basic Appl. Ecol. 5, 225–229 10.1016/j.baae.2004.03.002 (doi:10.1016/j.baae.2004.03.002) [DOI] [Google Scholar]
- 73.Stillman R., Brown A. F. 1998. Pattern in the distribution of Britain's upland breeding birds. J. Biogeogr. 25, 73–82 10.1046/j.1365-2699.1998.251169.x (doi:10.1046/j.1365-2699.1998.251169.x) [DOI] [Google Scholar]
- 74.Cattadori I. M., Haydon D. T., Hudson P. J. 2005. Parasites and climate synchronize red grouse populations. Nature 433, 737–741 10.1038/nature03276 (doi:10.1038/nature03276) [DOI] [PubMed] [Google Scholar]
- 75.Lindström Å., Agrell J. 1999. Global change and possible effects on the migration and reproduction of Arctic-breeding waders. Ecol. Bull. 47, 145–159 [Google Scholar]
- 76.Moss R., Oswald J., Baines D. 2001. Climate change and breeding success: decline of the capercaillie in Scotland. J. Anim. Ecol. 70, 47–61 10.1046/j.1365-2656.2001.00473.x (doi:10.1046/j.1365-2656.2001.00473.x) [DOI] [Google Scholar]
- 77.Willis S. G., Hill J. K., Thomas C. D., Roy D. B., Fox R., Blakeley D. S., Huntley B. 2009. Assisted colonization in a changing climate: a test-study using two UK butterflies. Conserv. Lett. 2, 46–52 10.1111/j.1755-263X.2008.00043.x (doi:10.1111/j.1755-263X.2008.00043.x) [DOI] [Google Scholar]
- 78.Morin X., Viner D., Chuine I. 2008. Tree species range shifts at a continental scale: new predictive insights from a process-based model. J. Ecol. 96, 784–794 10.1111/j.1365-2745.2008.01369.x (doi:10.1111/j.1365-2745.2008.01369.x) [DOI] [Google Scholar]
- 79.Kearney M., Porter W. 2009. Mechanistic niche modelling: combining physiological and spatial data to predict species' ranges. Ecol. Lett. 12, 334–350 10.1111/j.1461-0248.2008.01277.x (doi:10.1111/j.1461-0248.2008.01277.x) [DOI] [PubMed] [Google Scholar]
- 80.Kearney M. R., Wintle B. A., Porter W. P. 2010. Correlative and mechanistic models of species distribution provide congruent forecasts under climate change. Conserv. Lett. 3, 203–213 10.1111/j.1755-263X.2010.00097.x (doi:10.1111/j.1755-263X.2010.00097.x) [DOI] [Google Scholar]
- 81.Kearney M., Phillips B. L., Tracy C. R., Christian K. A., Betts G., Porter W. P. 2008. Modelling species distributions without using species distributions: the cane toad in Australia under current and future climates. Ecography 31, 423–434 10.1111/j.0906-7590.2008.05457.x (doi:10.1111/j.0906-7590.2008.05457.x) [DOI] [Google Scholar]
- 82.Keenan T., MariaSerra J., Lloret F., Ninyerola M., Sabate S. 2011. Predicting the future of forests in the Mediterranean under climate change, with niche-and process-based models: CO2 matters! Glob. Change Biol. 17, 565–579 10.1111/j.1365-2486.2010.02254.x (doi:10.1111/j.1365-2486.2010.02254.x) [DOI] [Google Scholar]
- 83.McInerny G. J., Turner J. R. G., Wong H. Y., Travis J. M. J., Benton T. G. 2009. How range shifts induced by climate change affect neutral evolution. Proc. R. Soc. B 276, 1527. 10.1098/rspb.2008.1567 (doi:10.1098/rspb.2008.1567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonan G. B., Levis S., Sitch S., Vertenstein M., Oleson K. W. 2003. A dynamic global vegetation model for use with climate models: concepts and description of simulated vegetation dynamics. Glob. Change Biol. 9, 1543–1566 10.1046/j.1365-2486.2003.00681.x (doi:10.1046/j.1365-2486.2003.00681.x) [DOI] [Google Scholar]
- 85.Sitch S., et al. 2003. Evaluation of ecosystem dynamics, plant geography and terrestrial carbon cycling in the LPJ dynamic global vegetation model. Glob. Change Biol. 9, 161–185 10.1046/j.1365-2486.2003.00569.x (doi:10.1046/j.1365-2486.2003.00569.x) [DOI] [Google Scholar]
- 86.Fisher R., McDowell N., Purves D., Moorcroft P., Sitch S., Cox P., Huntingford C., Meir P., Woodward F. I. 2010. Assessing uncertainties in a second-generation dynamic vegetation model caused by ecological scale limitations. New Phytol. 187, 666–681 10.1111/j.1469-8137.2010.03340.x (doi:10.1111/j.1469-8137.2010.03340.x) [DOI] [PubMed] [Google Scholar]
- 87.Belsky A. J. 1990. Tree/grass ratios in East African savannas: a comparison of existing models. J. Biogeogr. 17, 483–489 10.2307/2845380 (doi:10.2307/2845380) [DOI] [Google Scholar]
- 88.Neilson R. P., Pitelka L. F., Solomon A. M., Nathan R., Midgley G. F., Fragoso J. M., Lischke H., Thompson K. 2005. Forecasting regional to global plant migration in response to climate change. Bioscience 55, 749–759 10.1641/0006-3568(2005)055[0749:FRTGPM]2.0.CO;2 (doi:10.1641/0006-3568(2005)055[0749:FRTGPM]2.0.CO;2) [DOI] [Google Scholar]
- 89.Rosenthal R. 1979. The file drawer problem and tolerance for null results. Psychol. Bull. 86, 638–641 10.1037/0033-2909.86.3.638 (doi:10.1037/0033-2909.86.3.638) [DOI] [Google Scholar]
- 90.Rivington M., Matthews K. B., Bellocchi G., Buchan K. 2006. Evaluating uncertainty introduced to process-based simulation model estimates by alternative sources of meteorological data. Agric. Syst. 88, 451–471 10.1016/j.agsy.2005.07.004 (doi:10.1016/j.agsy.2005.07.004) [DOI] [Google Scholar]
- 91.Duncan R. P., Cassey P., Blackburn T. M. 2009. Do climate envelope models transfer? A manipulative test using dung beetle introductions. Proc. R. Soc. B 276, 1449–1457 10.1098/rspb.2008.1801 (doi:10.1098/rspb.2008.1801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Randin C. F., Dirnböck T., Dullinger S., Zimmermann N. E., Zappa M., Guisan A. 2006. Are niche-based species distribution models transferable in space? J. Biogeogr. 33, 1689–1703 10.1111/j.1365-2699.2006.01466.x (doi:10.1111/j.1365-2699.2006.01466.x) [DOI] [Google Scholar]
- 93.Tingley M. W., Monahan W. B., Beissinger S. R., Moritz C. 2009. Birds track their Grinnellian niche through a century of climate change. Proc. Natl Acad. Sci. USA 106, 19 637–19 643 10.1073/pnas.0901562106 (doi:10.1073/pnas.0901562106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Araújo M. B., New M. 2007. Ensemble forecasting of species distributions. Trends Ecol. Evol. 22, 42–47 10.1016/j.tree.2006.09.010 (doi:10.1016/j.tree.2006.09.010) [DOI] [PubMed] [Google Scholar]
- 95.Phillips S. J., Dudik M. 2008. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175 10.1111/j.0906-7590.2008.5203.x (doi:10.1111/j.0906-7590.2008.5203.x) [DOI] [Google Scholar]
- 96.Wood A. W., Leung L. R., Sridhar V., Lettenmaier D. P. 2004. Hydrologic implications of dynamical and statistical approaches to downscaling climate model outputs. Clim. Change 62, 189–216 10.1023/B:CLIM.0000013685.99609.9e (doi:10.1023/B:CLIM.0000013685.99609.9e) [DOI] [Google Scholar]
- 97.Anderson B. J., Akcakaya H. R., Araujo M. B., Fordham D. A., Martinez-Meyer E., Thuiller W., Brook B. W. 2009. Dynamics of range margins for metapopulations under climate change. Proc. R. Soc. B 276, 1415. 10.1098/rspb.2008.1681 (doi:10.1098/rspb.2008.1681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Possingham H. P. 1996. Decision theory and biodiversity management: how to manage a metapopulation. In Frontiers of population ecology (eds Floyd R. B., Sheppard A. W., de Barro P. J.), pp. 391–398 Melbourne, Australia: CSIRO Publishing [Google Scholar]
- 99.Possingham H. 2010. The business of biodiversity: applying decision theory principles to nature conservation. Fitzroy, Australia: Australian Conservation Foundation [Google Scholar]
- 100.Regan H. M., Ben-Haim Y., Langford B., Wilson W. G., Lundberg P., Andelman S. J., Burgman M. A. 2005. Robust decision-making under severe uncertainty for conservation management. Ecol. Appl. 15, 1471–1477 10.1890/03-5419 (doi:10.1890/03-5419) [DOI] [Google Scholar]