Abstract
Anticipating how biodiversity will respond to climate change is challenged by the fact that climate variables affect individuals in competition with others, but interest lies at the scale of species and landscapes. By omitting the individual scale, models cannot accommodate the processes that determine future biodiversity. We demonstrate how individual-scale inference can be applied to the problem of anticipating vulnerability of species to climate. The approach places climate vulnerability in the context of competition for light and soil moisture. Sensitivities to climate and competition interactions aggregated from the individual tree scale provide estimates of which species are vulnerable to which variables in different habitats. Vulnerability is explored in terms of specific demographic responses (growth, fecundity and survival) and in terms of the synthetic response (the combination of demographic rates), termed climate tracking. These indices quantify risks for individuals in the context of their competitive environments. However, by aggregating in specific ways (over individuals, years, and other input variables), we provide ways to summarize and rank species in terms of their risks from climate change.
Keywords: biodiversity, climate change, forest dynamics, hierarchical models, model selection, risk analysis
1. Introduction
Ecologists struggle to anticipate how a 2–5°C rise in global temperatures and novel combinations of resources, precipitation, temperature and seasonality will change forest diversity [1–3]. Studies of trees concentrate on one or a few species, a single age class (e.g. seedlings), a single environmental variable (e.g. CO2, temperature or moisture), few response variables (growth or survival) and/or individuals small enough to be housed in chambers with suitable replication [4–7]. The price of this experimental control is a highly conditional view of how diverse forest communities will respond, missing the rich interactions between individuals of different species and sizes competing for resources locally, and the changing combination of growing season length, summer droughts and spatial variation in soils [8–10]. Recognition that such interactions are responsible for the diversity of natural ecosystems [11–17] casts doubt on the extrapolations that come from single factor experiments on seedlings of one or a few species. On the other hand, modelling studies that omit the individual scale arguably miss the processes that will control responses to climate. The most important question is not directly addressed by current research: how will the diversity of forests that depends on heterogeneity and interactions respond to a novel combination of climate variables? Here, we adopt individual-based inference, using the long-term health of individuals exposed to natural and experimental variation in risk factors. We introduce methods to infer and synthesize vulnerability to risk factors at the population scale based on studies conducted at the individual scale. These quantitative techniques provide ways to decide which variables are important and to summarize population-scale risk informed by inference at the individual plant scale.
Quantifying the biodiversity risks of climate change is frustrated by the fact that individuals, not species, respond to climate. Health of individual trees is typically studied from diameter growth, because it can be easily monitored or tracked retrospectively from ring-width data. However, population vulnerability depends on overall health, which entails not only growth, but also fecundity and survival [18]. Responses to climate will depend on competition for limiting resources, particularly local light and moisture availability [19–22] in ways that are not yet sufficiently understood for predictive modelling [7,23]. The univariate responses tracked by most studies (growth or survival) miss shifts in allocation [24]. The interactions between climate and competition that affect allocation occur within individuals. Fitted parameters that apply to the species as a whole marginalize over individuals exposed to a broad range of interactions that can hide or even reverse the conditional relationships that ultimately determine responses [25].
Here, we develop inference for climate-change vulnerability, shifting from the traditional aggregate approach to the scale at which climate actually impacts populations—individuals competing for resources. We transfer inference to the more relevant scale of individuals subjected to unique combinations of the environment and displaying multiple indicators of individual health, not all of which can be captured by population-level parameters. Our application makes use of an unusually large dataset at the individual-scale spanning many combinations of input variables, including climate and competition, and a state-space framework to accommodate uncertainty in the underlying processes and observations [13,26]. The range of inputs needed to infer their effects alone and in combination is achieved by exploiting natural variation with superimposed experimental manipulation [27–29].
The unusual size of the dataset allows us to confront a challenging model selection problem. Although we limit consideration to a few variables, widely recognized as critical, the complexity can be daunting. Seasonal winter and spring temperatures (w), summer drought (m), local moisture status (M), tree size (D), previous growth (d) and availability of light (C) affect the demographic rates of 40 dominant species over 20 years. We entertain up to 1062 main effects and two-way interactions, well beyond the scope of replicated controlled experiments in forests with dozens of species and not identifiable in geographically distributed datasets. Model fitting and selection tools allow consideration of many potential variables to identify those of most consequence [30]. Sensitivity analysis is implemented in a way that permits direct comparisons of the effects of all input variables on each demographic rate of all species. Climate tracking entails a composite demographic response to determine a more synthetic understanding of an input's impact on overall health.
2. Methods
Our analysis makes use of multivariate responses of individuals to multiple inputs, including climate variables, individual attributes and the local light environment. It extends a modelling approach that is detailed in Clark et al. [13]. Here, we summarize the approach, including sensitivity to individual input variables, dynamic inverse prediction (DIP) of climate and prior specifications for interactions.
Our approach emphasizes prediction, used here as a metric for the responsiveness of individual health to climate and competition. Individual health is multivariate, represented by a vector of estimates yi,t for states of individual i at time t informed by observations zi,t and a process model with inputs xi,t−1 (e.g. resources and climate) and fitted parameters θ. The sensitivity ∂yi,t(k)/∂xi,t(q) of a latent state k to input q relates two scalar quantities. This quantity may not provide much guidance about risks that operate on the individual-year scale and motivates a predictive approach. Predicting observations 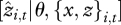 has long been used to check models (such as cross validation or predictive loss) [31]. Rather than ask how well the model predicts the data, the application here focuses on how well the individuals predict the environment xi,t, using the full response vector,
has long been used to check models (such as cross validation or predictive loss) [31]. Rather than ask how well the model predicts the data, the application here focuses on how well the individuals predict the environment xi,t, using the full response vector, 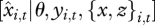 . The rationale here is that individual risk depends on combinations of inputs; vulnerability is most directly assessed from an individual's capacity to predict a risk factor based on its full response vector yi,t. We term this approach DIP. ‘Weak tracking’ represented by bias or large predictive variance means that an input has minor impact on an individual's health. The ensemble of predictions across individuals over time can provide insights concerning the context where different factors become important for individual health. The approach can be especially helpful when subjects are responding to inputs that operate at different scales, in this case, regional climate and local competition. Individuals can differ in terms of their predictive means (are predictions good on average or are they biased?) and predictive variances (are predictions confident or uncertain?). Because they are standardized, these indices have value for comparisons between input variables.
. The rationale here is that individual risk depends on combinations of inputs; vulnerability is most directly assessed from an individual's capacity to predict a risk factor based on its full response vector yi,t. We term this approach DIP. ‘Weak tracking’ represented by bias or large predictive variance means that an input has minor impact on an individual's health. The ensemble of predictions across individuals over time can provide insights concerning the context where different factors become important for individual health. The approach can be especially helpful when subjects are responding to inputs that operate at different scales, in this case, regional climate and local competition. Individuals can differ in terms of their predictive means (are predictions good on average or are they biased?) and predictive variances (are predictions confident or uncertain?). Because they are standardized, these indices have value for comparisons between input variables.
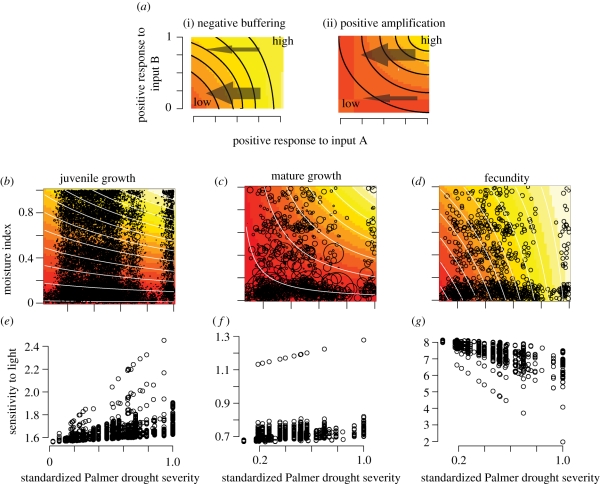
Bayesian implementation of DIP in models that include interactions motivates a specific type of prior dependence that we introduce as part of this analysis. Over the range of inputs in this study, effects of temperature, moisture and light availability are known to be positive. The interactions are not known and can mean negative buffering or positive amplification (figure 1). Negative interactions have convex contours (figure 1a(i)) meaning that the effect of increasing one input is greatest when the second input is low, i.e. one input buffers impacts of a second. For example, Frelich & Reich [9] hypothesize that moist locations will provide refuges as aridity increases. This represents a negative interaction between moisture change over time (increasing aridity) and spatial variation in hydrological status—reduced moisture supply during drought has greatest impact on sites where moisture is already low (figure 1b, top left). This would occur if individuals on wet sites are effectively buffered from drought, responding less than those on dry sites. The alternative positive interaction (figure 1b) could result if leaf area and transpiration demand (i.e. competition) increase to meet the greater moisture supply on wet sites, possibly making them more vulnerable to drought. Alternatively, a positive interaction means that increasing one input amplifies the response to a second input, and contours are concave (figure 1a(ii)). We show how to develop a prior specification for the full effects of inputs, where knowledge goes no further than the sign of the full effect, while placing no prior bias on the sign of the interaction. The DIP implementation makes use of this prior specification.
Figure 1.
(a) Examples of (i) negative and (ii) positive interactions when input variables are scaled to (0,1) (equation 2.10). Contours and shading indicate magnitude of the response. Strong interactions (thick arrows) cross many contours, weak interactions (thin arrows) cross few contours. Negative interactions indicate a large response to one input when the other is low, and vice versa. (b–d) Fraxinus interactions are positive for adult and juvenile growth and negative for fecundity. The distribution of data (size of circles proportional to magnitude of response) helps evaluate parameter space where observations are insufficient. (e–g) Sensitivity to light for Fraxinus depends on the summer drought experienced by each tree in each year (dots).
(a). Response variables
Response variables in the vector yij,t are demographic rates, including growth, fecundity and survival probability [13]. Response variables are informed by observations from tree censuses, tree increment cores, remote sensing and seed traps, detailed in Clark et al. [13]. Tree-year observations taken during censuses include tree diameter, survival status, crown class and reproductive status, conducted at 2 to 4 year intervals. Additional observations of growth are obtained from increment cores. Remote sensing is used to quantify exposed canopy area (ECA). Seed-year observations come from seed traps, collected two to five times annually.
(b). Input variables
Input variables xij,t include climate and competition (table 1). Manipulated forest canopies provide a full range of exposed canopy cover values lij,t, which range from 0 (completely shaded by neighbours) to greater than 100 m2. Plot selection provides a range of climate variation. The Palmer drought severity index (PDSI) mj,t is used for June through September for site j in year t. It expresses the departure of a given year from the long-term moisture availability for the site. Spatial variation in moisture availability Mij is the product of annual average precipitation (mm) at site j and the topographic convergence index [32] for the location of tree ij. Mij varies among the 12 stands owing to precipitation and within sites owing to topography. Thus, Mij represents spatial variation, and mj,t represents temporal variation, how a given growing season departs from the site average. Temperature is likewise decomposed into a site and a year effect. Spring temperature controls bud break, leaf and fruit set, and can have a large impact on tree carbon balance [33–36]. We use the annual temperature for January through March for site j in year t. The site effect is taken to be the average winter/spring temperature Wj, and the year effect is the annual departure from that average, wj,t.
Table 1.
Hypothesized direct effects and interactions by demographic response variable. (NI, non-informative.)
| input covariate | reference | summary rationale | prior distribution |
|
|---|---|---|---|---|
| growth response ln(dij,t−1) | fecundity potential ln(fij,t−1) | |||
| minimal model | ||||
| intercepts | species | no prior knowledge | NI | NI |
| canopy area ln(lij,t−1) | tree-year | light is a limiting resource for which plants compete | Aqr > 0 | Aqr > 0 |
| additional main effects | ||||
| diameter ln(Dij,t−1) | tree-year | fecundity potential can increase allometrically | NI | Aqr > 0.5 |
| large diameter effect ln2(Dij,t−1) | tree-year | physiological function may decline, but not improve with old age | Aqr < 0 | Aqr < 0 |
| previous year growth ln(dij,t−1) | tree-year | fecundity may depend on previous growth, beyond effects explained by past climate inputs | Aqr = 0 | NI |
| winter temperature deviation wj,t−1 | site-year | years with warm winters, long growing seasons increase carbon gain | Aqr > 0 | Aqr > 0 |
| summer (Jun, Jul, Aug and Sept) drought deviation mj,t | site-year | drought years decrease carbon gain | Aqr > 0 | Aqr > 0 |
| average winter (Jan, Feb and Mar) temperature Wj | site | sites with warm winters, long growing seasons increase carbon gain | Aqr > 0 | Aqr > 0 |
| average moisture index Mij | tree | moist sites support carbon gain | Aqr > 0 | Aqr > 0 |
| interactions | ||||
| light by winter temperature lij,t−1 × wj,t−1 | tree-year | NI | NI | |
| light by summer drought (PDSI)lij,t−1 × mj,t | tree-year | |||
| light by ave winter temperature lij,t−1 × Wj | site-year | |||
| light by ave moisture lij,t−1 × Mij | tree-year | |||
| winter temperature by summer drought (PDSI) wj,t−1 × mj,t | site-year | |||
| summer drought (PDSI) by ave moisture mj,t ×Mij | tree-year | |||
The ranges of input variables for this study are relevant for twenty-first century climate-change predictions. They span southeastern Piedmont to northern hardwoods in spatial variation and possess the low correlation needed for effective inference. Temperature variation spans the 2–5°C change predicted for twenty-first century climate change, both in space and over time. Within the 20 year study period, variation in summer PDSI spans the interval (−4,4), i.e. from severe drought to some of the wettest years since the 1930s. All pairwise correlations between input variables were less than 0.2 in absolute value.
(c). Model development
The model that assimilates demographic observations to infer variation in diameter growth (cm yr−1), fecundity (seeds yr−1) and mortality risk (annual probability) for tree i at site j in year t is detailed in Clark et al. [13]. There is a nonlinear, multivariate state-space submodel for each individual, responding to its changing diameter, lagged growth rate (an AR(1) term), light availability, climate, random-individual effects and model error. There is observation error associated with each type of observation. We do not repeat from [13,18] details on datasets, prior specification, algorithm development and diagnostics. Here, we briefly summarize the model and extend it to evaluate the concept of DIP for climate vulnerability.
The process model tracks changing states of individuals each year, as they grow, reach reproductive maturity, produce seed and die. Maturation is a partially hidden Markov process, where an individual ij potentially changes from the immature Fij,t to mature Fij,t+1 state as it increases in size, depending on access to resources [13]. Seed trap data and individual observations of maturation status are the basis for inference on maturation status.
The DIP approach developed here focuses on the portion of the full model that directly relates to growth and fecundity and their relationships with tree size, resources, climate and interactions (table 1). The state-space model for the bivariate growth–fecundity response for a mature individual (Fij,t = 1),
| 2.1 |
includes fecundity potential fij,t (seeds per tree) and the diameter growth increment dij,t (cm), which determines change in diameter Dij,t = Dij,t−1 + dij,t−1. The submodel is
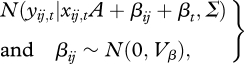 |
2.2 |
where xij,t is a 1 by Q vector of predictors (main effects and interactions), A is a Q by 2 matrix of fitted parameters, βij is the random effect associated with individual ij, Σ is a 2 by 2 covariance matrix for process error and Vβ is the 2 by 2 covariance matrix for random effects. The two columns in A match the two responses in equation (2.1). The Q rows in A match the predictor variables. For immature individuals, Fij,t = 0, yij,t is a scalar quantity for growth, and equation (2.2) is univariate. Note that a given predictor q ∈ {1, … ,Q} enters the model in three ways, for adult growth and fecundity (equation 2.1) and for juvenile growth.
Survival is conditional on growth rate, which is diagnostic of plant vigour. We additionally include tree diameter, to allow for senescence in especially large individuals in a non-parametric survival model of Clark et al. [37] and Vieilledent et al. [38]. Mortality risk declines with tree growth rate, which tends to be lowest in the small trees in the shaded understory and for large trees approaching senescence [39,40]. Each individual in each year has a pair of risks (μhd, μhD) associated with growth rate (monotonically decreasing) and diameter (monotonically increasing), with survival probability from year t − 1 to year t
| 2.3 |
The prior for the model m and its parameters θm is factored as
The set of models {m} and priors p(m) are discussed in §2d. Prior distributions for parameters p(θm|m) are developed in Clark et al. [13], with the exception of interactions, which are discussed in §2d. Posterior simulation is Metropolis-within-Gibbs, with adaptive sampling for Metropolis steps [13].
(d). Prior dependence
Prior belief on input effects is informative in sign, but not magnitude (table 1). Light, moisture and winter temperature have positive effects on plant vigour, but we cannot say how large. A traditional Gaussian prior on elements of the matrix A would not allow us to impose this prior belief in a transparent way. With prior knowledge limited to an input's sign, a Gaussian prior on a parameter matrix A in equation (2.2) would require adjusting the prior mean vector and covariance structure to ensure that ‘most of the posterior’ mass is positive or negative. With this approach, we cannot readily separate the magnitude of an input's effect from its weight, while at the same time restricting ‘most of the posterior’ to be positive or negative. To ensure that the magnitude of the estimate is determined by the data, rather than prior, we specify a flat prior, truncated at zero, and use model selection to determine whether or not the input should be included (see §2e). Transparency comes from the clear contribution of prior (truncation at zero) and data [13]. The priors for all elements of A are uniform with limits defined either by prior information, or they are set to large values [41]. The actual correlations between responses and inputs can take on positive or negative values, depending on how data are distributed and the many sources of variation and interactions. The priors we specify allow for such correlations, they simply assure that the full effects of inputs are consistent with prior knowledge.
Interaction terms complicate the implementation of prior belief, because product terms can reverse the relationship of a response to an input, depending on the value of a second input contributing to the interaction. Consider the relationship
where yij,t(r) is the rth response in vector y, and
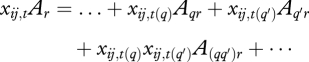 |
includes terms for direct effects of inputs q and q′ and their interaction (qq′). Further assume that a positive relationship is specified between input q and response r, i.e. Aqr > 0. This prior belief is only assured if xq′ = 0. But most variables are not measured on scales where 0 has any particular significance. The prior belief actually applies to the derivative
The interaction coefficient A(qq′)r can be positive or negative, but it should not be so negative that it violates prior belief that dyr/dxq > 0. When prior knowledge is limited to the sign of the effects of an input variable q on response r, e.g. positive dyr/dxq > 0 (table 1), it is not enough to specify a truncated prior on the main effect. Depending on sign and magnitude of the input xq, other inputs with which it interacts, and the strengths of the responses to those inputs, the full effect of input q could have either sign. We factor the prior for main effects and interactions
To preserve the prior belief that both q and q′ have positive effects, we specify a main-effects prior
| 2.4 |
and prior dependence on the interaction term,
| 2.5 |
This prior dependence is imposed where input variables are transformed to a (0,1) scale (§2e). This prior does not constrain the interaction to be positive or negative, but simply preserves the prior belief on the full effect.
The final consideration concerns whether or not an input variable should be in the model at all. Including prior distributions truncated at zero does not mean that an input variable must have an effect, because only important main effects are retained. The prior implemented here only states that, if a main effect is important (see §2e), then its sign is prescribed. Section 2e describes how models are selected.
(e). Variable selection
Although we limit consideration to variables known to affect tree health, the number of potential main effects and interactions is large. All variables included in table 1 affect demographic rates; variable selection is implemented not to test a null hypothesis, but rather to identify which variables are most important and to quantify the direction and the strength of interactions. The minimal model includes only intercepts and light lij,t−1 (table 1), because all species are limited by light in shaded understories. The eight main effects in table 1 contribute up to 28 = 256 combinations. An interaction can be considered only if both main effects are included in the model. We have biological reasons for focusing on a subset of potential interactions, those most likely to have importance for climate-change vulnerability and its interaction with competition (table 1) for a total of 1062 variable combinations. The model space is large, but manageable with computational methods implemented here.
Models are evaluated based on the posterior model probability, derived from the model prior p(m) and marginal likelihood p(y|x,m),
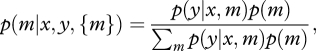 |
2.6 |
[42,43]. Equation (2.6) is the basis for many model choice criteria, including posterior odds and Bayes factors [44] and model averaging [45]. Following Scott & Berger [30], we apply fully Bayesian model choice, which automatically adjusts for multiple comparisons. The model prior is
| 2.7 |
[46], where km is the number of variables in model m (excluding intercept and canopy area in the minimal model) of M total. When p is treated as an unknown, this approach provides a multiplicity correction [30]. The marginal likelihood from equation (2.6) is
| 2.8 |
With prior beta(p|1,1), the posterior model probability is
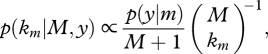 |
2.9 |
Large models are penalized in two ways. The first penalty comes from the fact that each variable adds a dimension requiring integration in the marginal likelihood. This penalty is roughly multiplicative and especially hard on interactions, which bear this burden thrice, once for each main effect, then again for the interaction term. This model size penalty is not a multiplicity correction, because it does not depend on the number of variables considered M [30]. The second penalty concerns multiplicity and enters through the prior (equation 2.7). In our application, the marginal likelihood is approximated using Chib [48]. The Metropolis-within-Gibbs algorithm described in Clark et al. [13] includes comparison of both the minimal model and a model proposed from a uniform distribution using log posterior probabilities. Low posterior probability is a cause for deletion of a model. Model selection is progressive, with especially poor models eliminated first and surviving models requiring more proposals before rejection.
(f). Population-scale sensitivity
Sensitivity coefficients quantify the full (main and interaction) effects of inputs in a way that permits comparison across inputs, demographic rates and species. Input variables are placed on a (0,1) scale
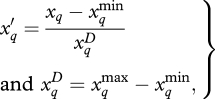 |
2.10 |
to allow prior specification on interaction terms as discussed in §2e. Although centring and scaling inputs by one or two standard deviations have advantages [49], they preclude the prior dependence we require on interaction terms. Extending the approach of Clark et al. [18] to include interactions, the sensitivity coefficient for response r = d or f (diameter growth or fecundity) to input q is
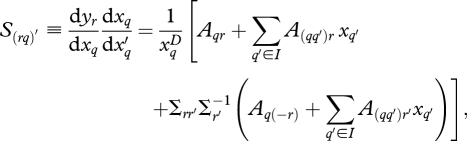 |
2.11 |
where subscripts ij,t are omitted from x and y to reduce clutter. The subscript (r) refers to the rth response column in A (growth or fecundity), and (−r) refers to the other response column. Σrr′ is the covariance between r and r′, and Σr′ is the variance on r′ in the matrix Σ (equation 2.2). Although parameters in A apply to species, interactions introduce individual differences in response, and can do so every year, depending on xq′. Thus, every individual has a different sensitivity to xq each year. In the absence of interactions, all individuals of a species show the same proportionate response,
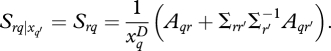 |
2.12 |
Thus, interactions bring individual variation in response.
The sensitivity of mortality rate to input q, conditional on tree diameter D, is
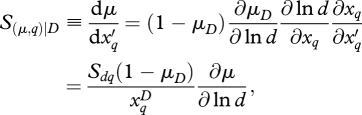 |
2.13 |
which can be evaluated for each tree ij in year t. Unconditionally, we have
By marginalizing over diameter in this way, the sensitivity is conditional on the diameter distribution of the population, but not on the diameter of the individual.
(g). Dynamic inverse prediction
DIP is derived directly from the fitted model. We separate the inputs (rows of A) into a group q that we wish to predict, based on the full-fitted model (including rows of A not included in q, designated (−q)). Equation (2.2) can be written in terms of these separate inputs
The first term in the mean vector includes only regression terms not involving q, neither as a main effect nor within interactions. The second term includes the main effect of q and interactions, where
xq′ is vector of variables that interact with q, and Aqq′ are the rows of A corresponding to these interactions. Conditionally, the predictive mean for the input variable xq is,
| 2.14 |
where
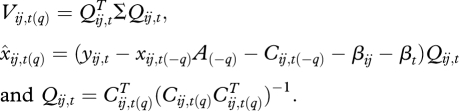 |
The predictive distribution entails marginalization, in this case, with Gibbs sampling and Metropolis.
Note that there is a prediction for xq from every individual in every year. It is possible that individuals track an input differently at different locations, at different times, and over different ranges of input variables. Summaries are available by aggregating over individuals, years, or both. Aggregation over years can help identify healthy and unhealthy individuals, with mean goodness of prediction and variance
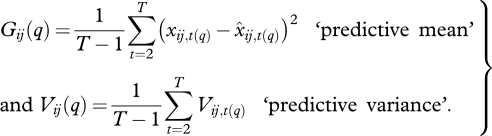 |
2.15 |
Large G(q) occurs if individual tree years are biased from true x(q). Large V(q) occurs if predictions are uncertain. The population-level prediction for the year can help to identify the types of years that pose large vulnerabilities, with mean fit and predictive variance determined as in equation (2.15), but with the summation taken over individuals, rather than years, and divided by number of individuals, rather than years. Likewise, the overall population prediction taken over all years summarizes variables that generally contribute risk, and sums over individuals and years. At each of these scales, we can define a mean square prediction error 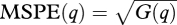 and predictive loss p(q) = G(q) + V(q), low values being indicative of accurate and confident prediction. Large MSPE occurs if there is bias in mean estimates. Large predictive variance occurs if there is large uncertainty.
and predictive loss p(q) = G(q) + V(q), low values being indicative of accurate and confident prediction. Large MSPE occurs if there is bias in mean estimates. Large predictive variance occurs if there is large uncertainty.
3. Results
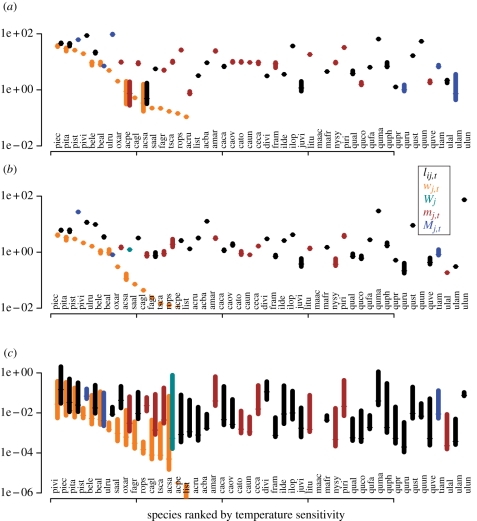
Products of this analysis include sensitivities and DIP of climate and competition, individually each year and in aggregate. At the species-level, fecundity was most sensitive to inputs, particularly summer drought (mj,t) and light (lij,t) availability (figure 2). Light was much more important for growth, overwhelmingly so for juveniles (not shown). Credible intervals of survival sensitivities are large (figure 4c), because there is less information—each individual contributes growth information every year and fecundity information every year that it is mature, but mortality affects only some individuals and, at most, once per individual.
Figure 2.
Sensitivities differ for each species to each climate variable in each of three (a) fecundity, (b) mature growth and (c) mature survival demographic rates. Species are ordered by sensitivity to winter temperature (wj,t). A second sensitivity is also shown for each species, the one that is largest. Variables are listed in table 1.
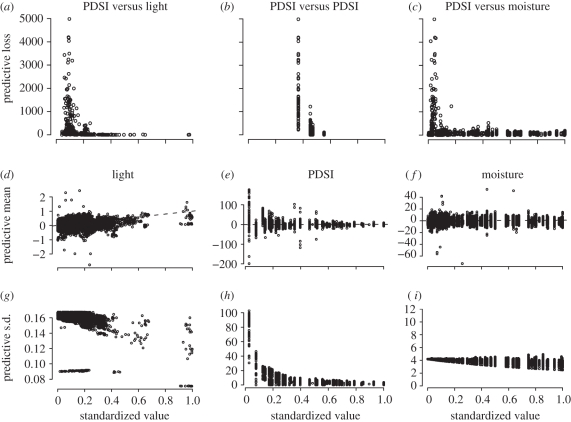
Figure 4.
Predictive loss for PDSI at each individual tree-year for Fraxinus P(q) = G(q) + V(q) decrease with (a) increasing light, (b) PDSI and (c) local moisture. PDSI is tracked more closely on average (low G(q)) and with greater confidence (low V(q)) when resources are high. (d–f) predictive means 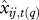 and (g–i) standard deviations
and (g–i) standard deviations 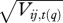 plotted against actual inputs xij,t(q) for all Fraxinus tree-years. The 1 : 1 line for predictive means indicates agreement.
plotted against actual inputs xij,t(q) for all Fraxinus tree-years. The 1 : 1 line for predictive means indicates agreement.
For a large number of species, a single model emerged as most probable. Sample sizes are large, so it is not surprising that the variables long known to affect tree health explain dynamic growth and fecundity. Despite the penalty against interactions, they emerged as important for a large number of predictors and responses. All six interactions considered were selected for multiple species. Moreover, with few exceptions, interactions were positive. To illustrate the approach, we use a single species, Fraxinus, as an example.
The individual scale is evident in interactions, when the response to one input depends on values of the other inputs to which an individual is exposed. Although all parameters in A are ‘population scale’, interactions impart an ‘i’ subscript to the sensitivity depending on the level of the other variables to which the individual is exposed in year t (equation (2.11)). The model finding most support for Fraxinus includes as inputs the previous year's growth rate, summer drought, local moisture index and three interactions. Several of these interactions are shown with the distribution of data in figure 1b–d. The population fit, shown as contours in figure 1b–d, aggregates over wide individual variation in sensitivity. Large variation and interactions (e.g. light and PDSI have a positive interaction on growth, negative on fecundity) result from the fact that each individual experiences different conditions that affect the sensitivity each year. The positive interactions for growth indicate that drought effects are amplified at high light (lower panels) and on wet sites (centre panels). Negative interactions for fecundity indicate that drought has the largest effects at low light and on dry sites.
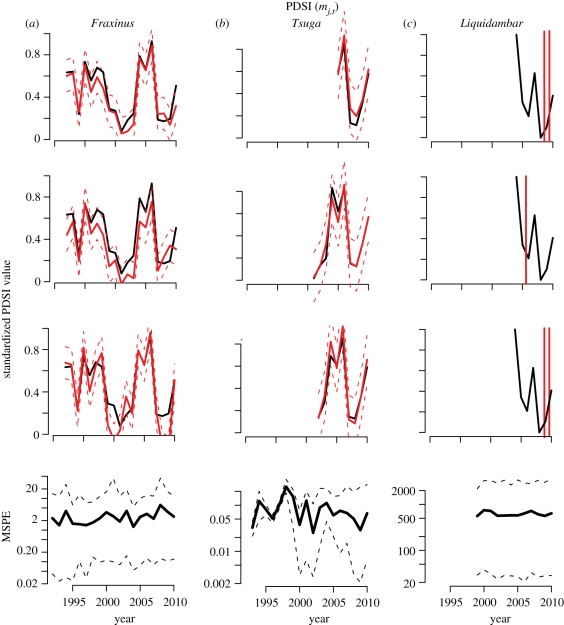
Although the population-scale interaction coefficients are valuable, there is more to be learned from DIP, i.e. individual/year-scale predictions by the full response yij,t. Each individual tracks each climate variable differently. For example, many Fraxinus individuals track summer drought closely. For some individuals in figure 3, summer PDSI tracking is least precise in drought years; responsiveness of growth and fecundity declines during drought. However, there is no indication that PDSI is tracked closely in particular years on a population-wide basis (lower left panel of figure 3). In contrast to Fraxinus, all individuals of Tsuga track PDSI closely (figure 3b), whereas no individuals of Liquidambar closely track PDSI (figure 3c). At the population level, tracking deteriorates for Tsuga with increasing mortality after losses owing to the hemlock woolly adelgid (bottom panel). The huge spread within and among Liquidambar trees contrasts with the two other examples here (bottom panels–note different scales).
Figure 3.
Predictive means and 95% predictive intervals (red lines) for individuals selected at random for (a) Fraxinus, (b) Tsuga and (c) Liquidambar in response to summer drought. The true values are in black. The lower panels show the median and 95% of the individual deviations 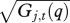 , i.e. they summarize the population.
, i.e. they summarize the population.
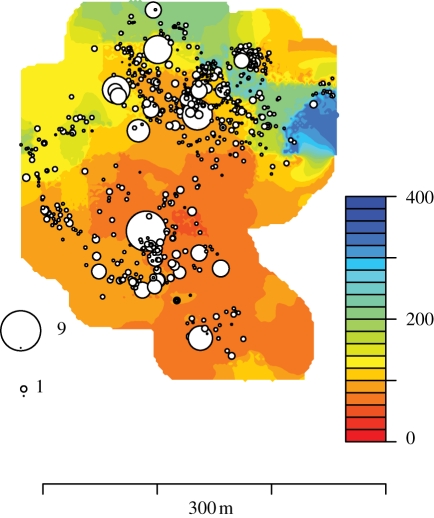
Combining indices for predictive mean and variance indicates that PDSI is tracked more closely on average (low G(q)) and with greater confidence (low V(q)) when resources are high (figure 4a–c). Separating the two components of predictive loss shows that predictive means improve (figure 4d–f) and are more certain (figure 4g–i) for all inputs when they are at high values. Predictive distributions can be mapped to provide a spatial view of responsiveness. For Fraxinus, predictions are best (tracking is tight) in the dry habitats of the site shown in figure 5.
Figure 5.
Fraxinus showing 1/MSPE for summer PDSI at a site on the Piedmont plateau. Large circles indicate close tracking of PDSI on dry sites at this site.
The patterns for Fraxinus do not generalize to other species; we found a range of patterns. The responses in figures 2 and 3 can be compared across species and input variables. They combine responses of growth and fecundity. When sensitivities and DIP are aggregated by species (equation 2.15), we found clear differences in terms of how the full response vectors of individuals respond to each input. For example, all individuals of Pinus taeda and Pinus strobus track winter temperatures, whereas Fagus grandifolia and Liquidambar styraciflua individuals show variable responses. Of still greater interest is the fact that several species track closely more than one climate variable; aggregate tracking of both winter temperature and summer drought is tight for P. taeda. Pinus strobus closely tracks both winter temperature variation and spatial variation in soil moisture. By contrast, L. styraciflua and Robinia pseudoacacia do not track closely any input. Species showing a broad range of tracking levels for spatial variation in soil moisture suggest the importance of interactions, where individuals vary in their tracking levels, depending on the other variables to which they are exposed.
4. Discussion
DIP of vulnerability to climate change (figures 3–5) complements current approaches that include dynamic simulation and climate envelopes. Individuals, not species, respond to climate change, but current efforts to predict biodiversity response are based on aggregated parameters for a species. Disaggregation to the individual scale shows profound differences between species in terms of the distribution of responses among individuals, with vulnerability depending on the local competitive setting (figures 3–5). The high dimensionality of these differences is synthesized here by aggregating individual responses, to capture the multivariate and conditional responses to many inputs. The disaggregation by input and response at the tree-year scale allows for analysis at the scale where climate matters—individuals competing for resources in the context of changing weather. The reaggregation of sensitivities and DIP by year, site and individual provides a basis for anticipating which species will respond in what way to each combination of climate variables, depending on the local competitive context (figure 2).
Resource and climate tracking are the multivariate demographic response to an input variable evaluated as the predictive distribution of that variable. If an input variable has impact on individual health then the individual is responding to variation in that input, over time, and potentially in multiple ways. If so, the fitted model for the individual should produce a predictive distribution for that variable that closely tracks its variation in space and time. Conversely, an input that has no consequence for that individual's health will not be closely tracked, and thus cannot be predicted from the individual demographic responses. We are currently applying this method to determine the vulnerabilities of species to climate change.
Acknowledgements
This research was supported by NSF grants DDDAS054845, CDI0940671, and the Coweeta LTER.
References
- 1.Parmesan C., Yoh G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42 10.1038/nature01286 (doi:10.1038/nature01286) [DOI] [PubMed] [Google Scholar]
- 2.Karl T. R., Melillo J. M., Peterson T. C. (eds) 2009. Global climate change impacts in the United States. New York, NY: Cambridge University Press [Google Scholar]
- 3.Field C. B., Mortsch L. D., Brklacich M., Forbes D. L., Kovacs P., Patz J. A., Running S. W., Scott M. J. 2007. North America. In Climate change 2007: impacts, adaptation and vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (eds Parry M. L., Canziani O. F., Palutikof J. P., van der Linden P. J., Hanson C. E.), pp. 617–652 Cambridge, UK: Cambridge University Press [Google Scholar]
- 4.Körner C. 2000. Biosphere responses to CO2 enrichment. Ecol. Appl. 10, 1590–1619 [Google Scholar]
- 5.Rustad L. E., Campbel J. L., Marion G. M., Norby R. J., Mitchell M. J., Hartley A. E., Cornelissen J. H. C., Gurevitch J. 2001. A meta-analysis of the response of soil respiration, net nitrogen mineralization, and aboveground plant growth to experimental ecosystem warming. Oecologia 126, 543–562 10.1007/s004420000544 (doi:10.1007/s004420000544) [DOI] [PubMed] [Google Scholar]
- 6.Norby R. J., Luo Y. 2004. Evaluating ecosystem responses to rising atmospheric CO2 and global warming in a multi-factor world. New Phytol. 162, 281–294 10.1111/j.1469-8137.2004.01047.x (doi:10.1111/j.1469-8137.2004.01047.x) [DOI] [Google Scholar]
- 7.Way D. A., Oren R. 2010. Differential responses to changes in growth temperature between trees from different functional groups and biomes: a review and synthesis of data. Tree Physiol. 30, 669–688 10.1093/treephys/tpq015 (doi:10.1093/treephys/tpq015) [DOI] [PubMed] [Google Scholar]
- 8.McMahon S. M., Parker G. G., Miller D. R. 2010. Evidence for a recent increase in forest growth. Proc. Natl Acad. Sci. USA 107, 3611–3615 10.1073/pnas.0912376107 (doi:10.1073/pnas.0912376107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frelich L. E., Reich P. B. 2010. Will environmental changes reinforce the impact of global warming on the prairie-forest border of central North America? Front. Ecol. Environ. 8, 371–378 10.1890/080191 (doi:10.1890/080191) [DOI] [Google Scholar]
- 10.Mohan J., Cox R., Iverson L. 2009. Composition and carbon dynamics of forests in northeastern North America in a future, warmer world. Can. J. Forest Res. 39, 213–230 10.1139/X08-185 (doi:10.1139/X08-185) [DOI] [Google Scholar]
- 11.Hutchinson G. E. 1961. The paradox of the plankton. Am. Nat. 95, 137–145 [Google Scholar]
- 12.Ibáñez I., Clark J. S., Dietze M. 2008. Evaluating the sources of potential migrant species: implications under climate change. Ecol. Appl. 18, 1664–1678 10.1890/07-1594.1 (doi:10.1890/07-1594.1) [DOI] [PubMed] [Google Scholar]
- 13.Clark J. S., et al. 2010. High dimensional coexistence based on individual variation: a synthesis of evidence. Ecol. Monogr. 80, 569–608 10.1890/09-1541.1 (doi:10.1890/09-1541.1) [DOI] [Google Scholar]
- 14.Classen A. T., Norby R. J., Campany C. E., Sides K. E., Weltzin J. F. 2010. Climate change alters seedling emergence and establishment in an old-field ecosystem. PLoS ONE 5, e13476. 10.1371/journal.pone.0013476 (doi:10.1371/journal.pone.0013476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angert A., Biraud S., Bonfils C., Henning C. C., Buermann W., Pinzon J., Tucker C. J., Fung I. 2005. Drier summers cancel out the CO2 uptake enhancement induced by warmer springs. Proc. Natl Acad. Sci. USA 102, 10 823–10 827 10.1073/pnas.0501647102 (doi:10.1073/pnas.0501647102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernal M., Estiarte M., Peñuelas J. 2010. Drought advances spring growth phenology of the Mediterranean shrub Erica multiflora. Plant Biol. 13, 252–257 10.1111/j.1438-8677.2010.00358.x (doi:10.1111/j.1438-8677.2010.00358.x) [DOI] [PubMed] [Google Scholar]
- 17.Robson T. M., Rodríguez-Calcerrada J., Sánchez-Gómez D., Aranda I. 2009. Summer drought impedes beech seedling performance more in a sub-Mediterranean forest understory than in small gaps. Tree Physiol. 29, 249–259 10.1093/treephys/tpn023 (doi:10.1093/treephys/tpn023) [DOI] [PubMed] [Google Scholar]
- 18.Clark J. S., Bell D. M., Hersh M. H., Nichols L. 2011. Climate change vulnerability of forest biodiversity: climate and competition tracking of demographic rates. Global Change Biol. 17, 1834–1849 10.1111/j.1365-2486.2010.02380.x (doi:10.1111/j.1365-2486.2010.02380.x) [DOI] [Google Scholar]
- 19.Souza G. M. I., Ribeiro R. V., Sato A. M., Oliveira M. S. 2008. Diurnal and seasonal carbon balance of four tropical tree species differing in successional status. Braz. J. Biol. 68, 781–793 10.1590/S1519-69842008000400012 (doi:10.1590/S1519-69842008000400012) [DOI] [PubMed] [Google Scholar]
- 20.Valladares F., Zaragoza-Castells J., Sánchez-Gómez D., Matesanz S., Alonso B., Portsmuth A., Delgado A., Atkin O. K. 2008. Is shade beneficial for Mediterranean shrubs experiencing periods of extreme drought and late-winter frosts? Ann. Bot. 102, 923–933 10.1093/aob/mcn182 (doi:10.1093/aob/mcn182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valladares F., Pearcy R. W. 2002. Drought can be more critical in the shade than in the sun: a field study of carbon gain and photo-inhibition in a Californian shrub during a dry El Niño year. Plant Cell Environ. 25, 749–759 10.1046/j.1365-3040.2002.00856.x (doi:10.1046/j.1365-3040.2002.00856.x) [DOI] [Google Scholar]
- 22.Welp L. R., Randerson J. T., Liu H. P. 2007. The sensitivity of carbon fluxes to spring warming and summer drought depends on plant functional type in boreal forest ecosystems. Agric. Forest Meteorol. 147, 172–185 10.1016/j.agrformet.2007.07.010 (doi:10.1016/j.agrformet.2007.07.010) [DOI] [Google Scholar]
- 23.Bonan G. B. 2008. Forests and climate change: forcings, feedbacks, and the climate benefits of forests. Science 320, 1444–1449 10.1126/science.1155121 (doi:10.1126/science.1155121) [DOI] [PubMed] [Google Scholar]
- 24.Granier A., Bŕeda N., Longdoz B., Gross P., Ngao J. 2008. Ten years of fluxes and stand growth in a young beech forest at Hesse, North-eastern France. Ann. For. Sci. 64, 703 [Google Scholar]
- 25.Clark J. S., Bell D. M., Hersh M. H., Kwit M. C., Moran E., Salk C., Stine A., Valle D., Zhu K. 2011. Individual-scale variation, species-scale differences: inference needed to understand diversity. Ecol. Lett. 14, 1273–1287 10.1111/j.1461-0248.2011.01685.x (doi:10.1111/j.1461-0248.2011.01685.x) [DOI] [PubMed] [Google Scholar]
- 26.Clark J. S. 2010. Individuals and the variation required for high species diversity. Science 327, 1129–1132 10.1126/science.1183506 (doi:10.1126/science.1183506) [DOI] [PubMed] [Google Scholar]
- 27.Clark J. S., Gelfand A. E. 2006. A future for models and data in ecology. Trends Ecol. Evol. 21, 375–380 10.1016/j.tree.2006.03.016 (doi:10.1016/j.tree.2006.03.016) [DOI] [PubMed] [Google Scholar]
- 28.Cressie N., Calder C. A., Clark J. S., Ver Hoef J. M., Wikle C. K. 2009. Accounting for uncertainty in ecological analysis: the strengths and limitations of hierarchical statistical modeling. Ecol. Appl. 19, 553–570 10.1890/07-0744.1 (doi:10.1890/07-0744.1) [DOI] [PubMed] [Google Scholar]
- 29.Ogle K. 2009. Hierarchical Bayesian statistics: merging experimental and modeling approaches in ecology. Ecol. Appl. 19, 577–581 10.1890/08-0560.1 (doi:10.1890/08-0560.1) [DOI] [PubMed] [Google Scholar]
- 30.Scott J. G., Berger J. O. 2010. Bayes and empirical-Bayes multiplicity adjustment in the variable-selection problem. Ann. Stat. 38, 2587–2619 10.1214/10-AOS792 (doi:10.1214/10-AOS792) [DOI] [Google Scholar]
- 31.Gelfand A. E., Ghosh S. K. 1998. Model choice: a minimum posterior predictive loss approach. Biometrika 85, 1–11 [Google Scholar]
- 32.Bevin K., Kirkby M. 1979. A physically based, variable contributing area model of basin hydrology. Hydrol. Sci. Bull. 24, 43–69 10.1080/02626667909491834 (doi:10.1080/02626667909491834) [DOI] [Google Scholar]
- 33.Houle G. 1999. Mast seedling in Abies balsamea, Acer saccharum, and Betula alleghaniensis in an old growth, cold temperate forest of north-eastern North America. J. Ecol. 87, 413–422 10.1046/j.1365-2745.1999.00352.x (doi:10.1046/j.1365-2745.1999.00352.x) [DOI] [Google Scholar]
- 34.Masaki T., Oka T., Osumi K., Suzuki W. 2008. Geographical variation in climatic cues for mast seeding of Fagus crenata. Popul. Ecol. 50, 357–366 10.1007/s10144-008-0104-6 (doi:10.1007/s10144-008-0104-6) [DOI] [Google Scholar]
- 35.Kon H., Noda T. 2007. Experimental investigation on weather cues for mast seeding of Fagus crenata. Ecol. Res. 22, 802–806 10.1007/s11284-006-0320-5 (doi:10.1007/s11284-006-0320-5) [DOI] [Google Scholar]
- 36.Delpierre N., et al. 2008. Exceptional carbon uptake in European forests during the warm spring of 2007: a data–model analysis. Global Change Biol. 15, 1455–1474 10.1111/j.1365-2486.2008.01835.x (doi:10.1111/j.1365-2486.2008.01835.x) [DOI] [Google Scholar]
- 37.Clark J. S., Dietze M., Agarwal P., Chakraborty S., Ibanez I., LaDeau S., Wolosin M. 2007. Resolving the biodiversity debate. Ecol. Lett. 10, 647–662 10.1111/j.1461-0248.2007.01041.x (doi:10.1111/j.1461-0248.2007.01041.x) [DOI] [PubMed] [Google Scholar]
- 38.Vieilledent G., Courbaud B., Kunstler G., Dhôte J.-F., Clark J. S. 2009. Biases in the estimation of size dependent mortality models: advantages of a semi-parametric approach. Can. J. Forest Res. 39, 1430–1443 10.1139/X09-047 (doi:10.1139/X09-047) [DOI] [Google Scholar]
- 39.Batista W. B., Platt W. J. 2003. Tree population response to hurricane disturbance: syndromes in a southeastern United States old-growth forest. J. Ecol. 91, 197–212 [Google Scholar]
- 40.Uriarte M., Canham C. D., Thompson J., Zimmerman J. K. 2004. A maximum-likelihood, neighborhood analysis of tree growth and survival in a tropical forest. Ecol. Monogr. 71, 591–614 10.1890/03-4031 (doi:10.1890/03-4031) [DOI] [Google Scholar]
- 41.Mitchell T. J., Beauchamp J. J. 1988. Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83, 1023–1032 10.2307/2290129 (doi:10.2307/2290129) [DOI] [Google Scholar]
- 42.Zellner A. 1978. Jeffreys-Bayes posterior odds ratio and the Akaike information criterion for discriminating between models. Econom. Lett. 1, 337–342 10.1016/0165-1765(78)90008-3 (doi:10.1016/0165-1765(78)90008-3) [DOI] [Google Scholar]
- 43.Clyde M. A., Ghosh J., Littman M. L. 2011. Bayesian adaptive sampling for variable selection and model averaging. J. Comp. Graph. Stat. 20, 80–101 10.1198/jcgs.2010.09049 (doi:10.1198/jcgs.2010.09049) [DOI] [Google Scholar]
- 44.Clyde M., George E. I. 2004. Model uncertainty. Stat. Sci. 19, 81–94 10.1214/088342304000000035 (doi:10.1214/088342304000000035) [DOI] [Google Scholar]
- 45.Hoeting J. A., Madigan D., Raftery A. E., Volinsky C. T. 1999. Bayesian model averaging: a tutorial. Stat. Sci. 14, 382–417 10.1214/ss/1009212519 (doi:10.1214/ss/1009212519) [DOI] [Google Scholar]
- 46.George E. I., McCullough R. E. 1997. Approaches for Bayesian variable selection. Stat. Sin. 7, 339–373 [Google Scholar]
- 47.George E. I., McCullough R. E. 1993. Variable selection via Gibbs sampling. J. Am. Statist. Assoc. 88, 881–889 10.2307/2290777 (doi:10.2307/2290777) [DOI] [Google Scholar]
- 48.Chib S. 1995. Marginal likelihood from the Gibbs output. J. Am. Stat. Assoc. 90, 1313–1321 10.2307/2291521 (doi:10.2307/2291521) [DOI] [Google Scholar]
- 49.Gelman A. 2008. Scaling regression inputs by dividing by two standard deviations. Stat. Med. 27, 2865–2873 10.1002/sim.3107 (doi:10.1002/sim.3107) [DOI] [PubMed] [Google Scholar]