Abstract
Rapamycin is an antibiotic inhibiting eukaryotic cell growth and proliferation by acting on target of rapamycin (TOR) kinase. Mammalian TOR (mTOR) is thought to work through 2 independent complexes to regulate cell size and cell replication, and these 2 complexes show differential sensitivity to rapamycin. Here we combine functional genetics and pharmacological treatments to analyze rapamycin-sensitive mTOR substrates that are involved in cell proliferation and tissue regeneration after partial hepatectomy in mice. After hepatectomy, hepatocytes proliferated rapidly, correlating with increased S6 kinase phosphorylation, while treatment with rapamycin derivatives impaired regeneration and blocked S6 kinase activation. In addition, genetic deletion of S6 kinase 1 (S6K1) caused a delay in S phase entry in hepatocytes after hepatectomy. The proliferative defect of S6K1-deficient hepatocytes was cell autonomous, as it was also observed in primary cultures and hepatic overexpression of S6K1-rescued proliferation. We found that S6K1 controlled steady-state levels of cyclin D1 (Ccnd1) mRNA in liver, and cyclin D1 expression was required to promote hepatocyte cell cycle. Notably, in vivo overexpression of cyclin D1 was sufficient to restore the proliferative capacity of S6K-null livers. The identification of an S6K1-dependent mechanism participating in cell proliferation in vivo may be relevant for cancer cells displaying high mTOR complex 1 activity and cyclin D1 accumulation.
Introduction
TOR is a protein kinase that was initially discovered as the intracellular target of rapamycin, an antibiotic macrolide produced by bacteria (1). mTOR is essential for life and regulates organismal growth, development, proliferation, and viability in response to nutrient availability. This is outlined by the severe phenotype of Mtor–/– mice that die during early embryonic development (2, 3). The antiproliferative efficacy of mTOR inhibitors is currently being evaluated in multiple clinical trials to treat human malignancies, and the use of rapamycin derivatives has already been approved for immune suppression after organ transplantation, to avoid vessel restenosis after angioplasty, in renal cell carcinomas, mantle T cell lymphomas, and tuberous sclerosis (4, 5). This therapeutic potential prompts further studies on the molecular mechanisms transducing growth and proliferative signals downstream of mTOR.
mTOR is found in the cell in 2 distinct protein complexes with specific binding partners, including raptor in mTOR complex 1 (mTORC1) and rictor in mTORC2 (1). Among the mTORC1 substrates are S6 kinases 1 and 2 (S6K1 and S6K2) and eIF4E-binding proteins 1, 2, and 3 (4E-BP1, 4E-BP2, and 4E-BP3), while mTORC2 phosphorylates the hydrophobic motif of Akt (Akt1–Akt3), SGK1, and PKCα. Rapamycin, in a gain-of-function complex with the FKBP12 protein, binds mTOR at the FRB domain close to the catalytic domain. Importantly, rapamycin has a striking specificity of action on mTOR. Rapamycin exclusively binds mTORC1, possibly because the FRB domain of mTOR is sterically hindered in mTORC2. However long-term treatment with rapamycin can also affect mTORC2 activity in a positive or negative manner depending on the cell types (6). The effect of rapamycin on mTORC2 activity is the resultant of contrasting actions on mTOR complex formation and on the negative feedback inhibition of mTORC1. In addition, rapamycin potency on mTORC1 substrates is also variable, as it leads to a complete inhibition of S6K phosphorylation, while the effect on 4EBP is milder and transient (7, 8). Hence, the therapeutic targets and efficacy of rapamycin are difficult to predict.
The multifaceted aspects of allosteric inhibition by rapamycin have led to the development of active site mTOR inhibitors (asTORis) that compete with ATP at the catalytic domain (7, 8). These general inhibitors block both mTORC1 and mTORC2 signaling. Importantly, they appear more potent than rapamycin in inhibiting proliferation of cultured cells (7, 8) and tumorigenesis in animal models of cancer (9, 10). Surprisingly, their antiproliferative action can also be observed in mTORC2-deficient cells, suggesting a dominant role of mTORC1 on cell cycle progression (7, 8). These findings are consistent with the early embryonic lethality of raptor-deficient mice, a phenotype equivalent to that of the Mtor–/– and more severe than the Rictor–/– embryos (11). Recently, 4EBPs have been demonstrated to be important players in cell cycle control by mTORC1. The proliferation of 4e-bp1–/–4E-BP2–/– mouse embryonic fibroblasts (MEFs) is resistant to asTORi (12). Conversely, deletion of another mTORC1 target, S6K1, mimics the effect of rapamycin and asTORi on cell size (12, 13). These findings have led to the model that mammalian cells have evolved a rapamycin-insensitive mTORC1/4EBP pathway for cell cycle control and a rapamycin-sensitive mTORC1/S6K1 pathway for cell size control. However, the proliferation of 4e-bp1–/–4E-BP2–/– MEFs remains sensitive to rapamycin, indicating additional mechanism(s) for cell cycle regulation downstream of mTORC1.
Liver regeneration after two-thirds partial hepatectomy (PH) is a well-characterized system to evaluate in vivo kinetics of cell cycle progression (14). After this surgical procedure, hepatocytes, which are differentiated and quiescent cells, reenter the cell cycle in a highly synchronized manner and restore the initial hepatocyte mass after 1 to 2 rounds of replication. Between 36 to 42 hours after two-thirds PH, most hepatocytes are in the S phase of the cell cycle, after having entered the cell cycle by transitioning from the G0 into the G1 phase, passed the restriction point in late G1, and initiated DNA replication. Since rapamycin treatment causes a significant delay in S phase entry of hepatocytes after hepatectomy (15–17), we set out here to study rapamycin-sensitive mechanisms that intervene during liver regeneration. We compare pharmacologic treatments and genetic invalidation of S6K and Akt family members. We reveal that S6K1 regulates cyclin D1 (Ccnd1) mRNA and protein levels and promotes hepatocyte proliferation. Thus S6K1 is not only required for cell size control by mTOR but may also provide a proliferative advantage in specific cell types and conditions.
Results
S6K1 deletion impairs liver regeneration in a cell-autonomous manner.
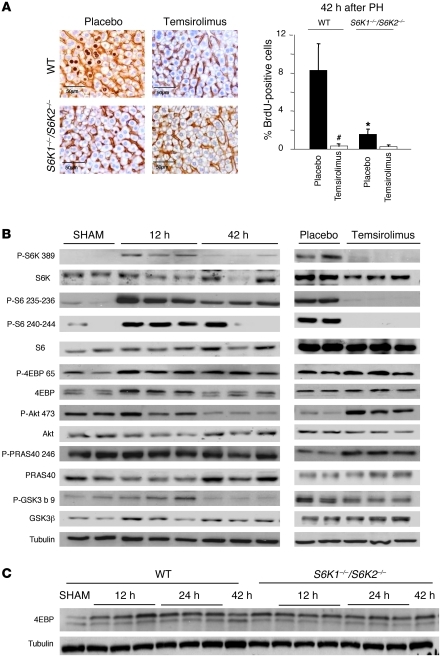
To in vivo dissect the contribution of the mTOR pathway in cell proliferation and tissue growth, two-thirds hepatectomy was performed in adult mice. Cell proliferation was assessed after a 2-hour BrdU pulse, while cell size was visualized by staining hepatocyte cell membrane with β-catenin antibodies (Figure 1A). Consistent with previous studies (18), S phase entry in WT mice peaked 42 hours after hepatectomy. Daily treatment with the rapamycin derivative temsirolimus at a 5 mg/kg dose inhibited cell cycle progression. Since rapamycin derivatives may differentially affect the phosphorylation of mTOR substrates (6–8), we compared the effects of temsirolimus treatment on 2 mTORC1 substrates, S6Ks and 4E-BPs, and on the mTORC2 substrate, Akt. As shown in Figure 1B and Supplemental Figure 1 (supplemental material available online with this article; doi: 10.1172/JCI44203DS1), phosphorylation of S6K and ribosomal protein S6 (rpS6) was upregulated during the G1 and S phases of the cell cycle, as illustrated, respectively, by the 12 and 42 hour time points after hepatectomy. 4E-BP1 phosphorylation was similarly controlled, as assessed by immunoblot analysis with anti–phospho-Ser 65 antibodies and by electromobility shift. During S phase, 42 hours after hepatectomy, S6K activity and 4E-BP1 phosphorylation started to decrease, though they were still higher than that in sham-operated control mice. Conversely, the Akt pathway was not significantly activated at these time points, as indicated by the phosphorylation state of Ser 473–Akt and of the downstream targets GSK3β and PRAS40. Thus liver regeneration is mainly accompanied by mTORC1, but not mTORC2, activation.
Figure 1. mTORC1 regulation of liver regeneration after partial hepatectomy.
(A) BrdU-positive hepatocytes of the indicated genotypes. Mice were injected intraperitoneally with 5 mg/kg of the rapamycin derivate temsirolimus or with placebo 2 hours before PH and then daily until sacrifice (42 hours after PH). Data are expressed as the percentage of BrdU-positive cells ± SEM for 4 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice; #P < 0.05 versus placebo mice. (B and C) Immunoblot analysis of protein extracts from liver of the indicated genotype from sham-operated mice (sham) or at different times after PH. When indicated, mice were injected intraperitoneally with 5 mg/kg of the rapamycin derivate temsirolimus or with placebo 2 hours before PH and then daily until sacrifice (24 hours after PH). Proteins were revealed using the indicated antibodies. P-S6K 389, phosphorylated S6K1 (Thr 389); P-S6 235-236, phosphorylated rpS6 (Ser 235/236); P-S6 240-244, phosphorylated rpS6 (Ser 240/244); P-4EBP 65, phosphorylated 4EBP (Ser 65); P-Akt 473, phosphorylated Akt (Ser 473); P-PRAS40 246, phosphorylated PRAS40 (Thr 246); P-GSK3b 9, phosphorylated GSK3β (Ser 9).
Strikingly, temsirolimus treatment completely abolished S6K activity, while it did not affect 4E-BP1 phosphorylation (Figure 1B). In addition, temsirolimus increased Akt and PRAS40 phosphorylation, possibly as a consequence of relieving the negative feedback inhibition of mTORC1 on mTORC2 activity. Since the inhibitory effects of rapamycin derivatives on cell cycle progression correlated with a specific downregulation of S6K activity, we addressed the functional role of this mTORC1 branch using S6K1–/–S6K2–/– mice. The percentage of hepatocytes in S phase 42 hours after hepatectomy was reduced by 77% in S6K-deficient mice as compared with that in WT controls (Figure 1A), indicating that S6K deletion partially mimics the effects of temsirolimus. S6K1–/–S6K2–/– hepatocyte proliferation was further blunted by temsirolimus treatment, although the tendency did not reach statistical significance, suggesting that additional rapamycin-sensitive mTOR targets may participate in this event. The phosphorylation of 4E-BP was not reduced in S6K1–/–S6K2–/– livers but slightly increased in S6K-null sham operated animals and at 42 hours after PH, consistent with the possibility that both mTORC1 substrates compete for raptor binding (Figure 1C). These data imply that the proliferative defects due to S6K deletion are specific and not a consequence of a general shutdown of mTORC1 signaling.
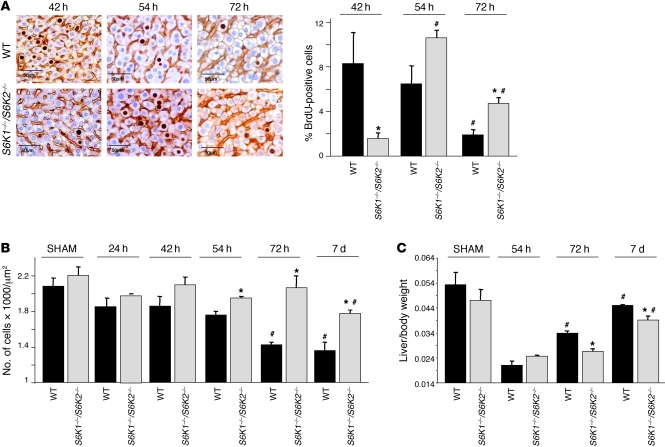
The S phase entry was delayed by 12 hours in S6K1–/–S6K2–/– livers, as assessed by BrdU staining (Figure 2A), indicating that S6K deletion lengthens the G0/G1 phase of hepatocyte cell cycle after hepatectomy. These results were surprising, as S6K expression did not impact cell proliferation in a variety of experimental models but rather specifically controlled cell growth (increase in cell size) (12, 13). To evaluate hepatocyte cell size during liver regeneration as a function of the S6K genotype, we stained hepatocyte membrane with β-catenin antibodies and counted the number of cells per surface. The cell density did not differ in sham-operated WT and S6K1–/–S6K2–/– livers (Figure 2B). However, 72 hours after hepatectomy, WT cell density decreased by 30%, reflecting an increased cell size that was maintained 7 days after surgery. This growth response was blunted at both time points analyzed after hepatectomy in S6K1–/–S6K2–/– livers. Consistently, the liver to whole body weight ratio was similar in sham-operated mice of both genotypes, though mass recovery after hepatectomy was less efficient in S6K1–/–S6K2–/– livers at 72 hours and 7 days after hepatectomy (Figure 2C). Taken together our findings demonstrate that S6K deletion slows down liver regeneration due to a combined defect in cell cycle progression and cell growth.
Figure 2. Deletion of both S6K1 and S6K2 induces a delay in liver regeneration.
(A) BrdU-positive hepatocytes of the indicated genotypes at different times after PH. Data are expressed as the percentage of BrdU-positive cells ± SEM for 4 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice; #P < 0.05 versus previous time for the same genotype. (B) Hepatocyte cell density analyzed at different times after PH in the indicated genotypes. Data are mean ± SEM for 4 mice per genotype. *P < 0.05 versus WT mice; #P < 0.05 versus previous time for the same genotype. (C) Liver to body weight ratio analyzed at different times after PH in the indicated genotypes. Data are mean ± SEM for 4 mice per genotype. *P < 0.05 versus WT mice; #P < 0.05 versus previous time for the same genotype.
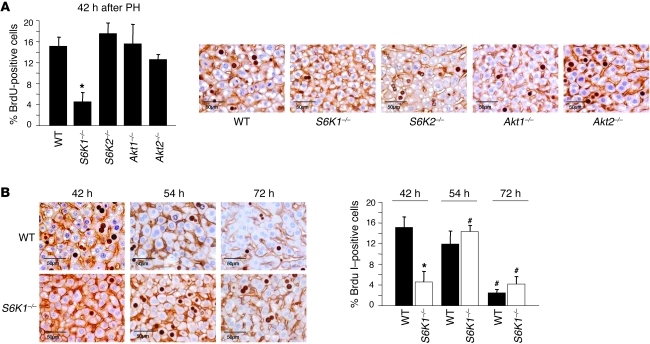
Since S6K1 and S6K2 are homologous genes that encode for protein kinases with overlapping yet distinct functions, we addressed what gene was required for hepatocyte cell cycle progression. The effects of single S6K mutants on S phase entry were also compared with Akt1- and Akt2-null mutants. As shown in Figure 3A, S6K1 deletion was sufficient to decrease the number of BrdU-labeled hepatocytes by 78% 42 hours after hepatectomy, whereas loss-of-function mutations of S6K2, Akt1, or Akt2 did not impair cell cycle progression. Strikingly, the kinetics of S phase entry in S6K1–/– livers was superimposable on the one of S6K1–/–S6K2–/– livers (Figure 2A and Figure 3B). However, the cell size of the S6K1-deficient liver was not significantly different to that of the WT control (data not shown), suggesting that both S6K1 and S6K2 concur to the growth regulation in liver. In conclusion, we demonstrated that the single deletions of Akt1 and Akt2 did not impact liver regeneration, consistent with the lack of mTORC2 activation. At this stage, we cannot exclude possible compensation between Akt1 and Akt2 during liver regeneration, as the double-mutant mice are not viable. Conversely, our data point to the important role of the S6 kinases on liver mass recovery after PH, with S6K1 having a specific role on hepatocyte proliferation.
Figure 3. Delay in liver regeneration correlates with the specific deletion of S6K1.
(A) BrdU-positive hepatocytes of the indicated genotypes 42 hours after PH. Data are expressed as the percentage of BrdU-positive cells ± SEM for 4 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice. (B) BrdU-positive hepatocytes of the indicated genotypes at different times after PH. Data are expressed as the percentage of BrdU-positive cells ± SEM for 5 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice; #P < 0.05 versus previous time for the same genotype.
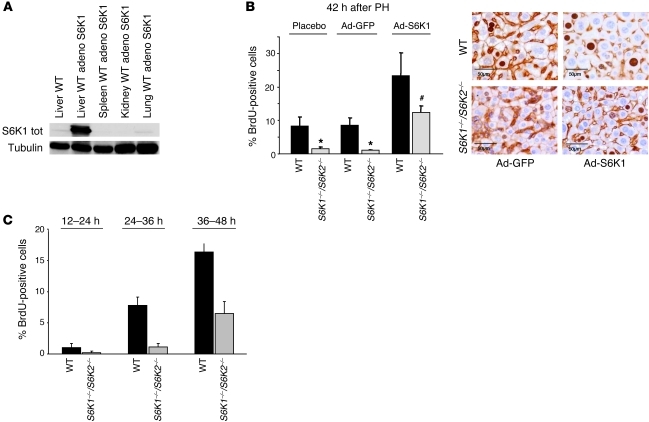
To evaluate whether transient expression of S6K1 in liver could rescue the proliferation defect of S6K1–/–S6K2–/– hepatocytes, adenoviruses carrying S6K1 cDNA were administered by retro-orbital intravenous injection 1 week before hepatectomy. This led to an overexpression of S6K1 in liver, while other tissues, including spleen, kidney, and lung, were poorly transduced (Figure 4A). Hepatic overexpression of S6K1 was sufficient to stimulate cell proliferation in WT livers as compared with GFP transduction (Figure 4B). More important, adenoviral S6K1 was also able to rescue the delay in the S phase of S6K1–/–S6K2–/– hepatocytes. To further address whether the proliferative defect of S6K-deficient hepatocytes is cell autonomous, primary hepatocyte cultures were prepared from WT and S6K1–/–S6K2–/– mice. As shown in Figure 4C, we observed a delayed entry in S phase in S6K1–/–S6K2–/– hepatocytes after stimulation with a defined mixture of growth factors and nutrients. In parallel cultures, cell viability was equivalent in the 2 genotypes, ruling out an effect of S6K deletion on apoptosis in these conditions (Supplemental Figure 2). Taken together, our data suggest that S6K expression in hepatocytes provides a cell-autonomous advantage promoting cell cycle progression.
Figure 4. Expression of S6K1 in liver rescues the proliferation defect.
(A) Immunoblot analysis of protein extracts from liver, spleen, kidney, and lung of the WT genotype using anti-S6K1 and anti-tubulin antibodies. Mice were treated or not with adenovirus S6K1 1 week before PH and sacrificed 42 hours after PH. S6K1 tot, S6K1 total. (B) BrdU-positive hepatocytes of the indicated genotypes were treated with adenovirus expressing GFP or S6K1 1 week before PH and sacrificed 42 hours after PH. Data are expressed as the percentage of BrdU-positive cells ± SEM for at least 4 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice; #P < 0.05 versus mice treated with GFP adenovirus. (C) BrdU-positive cultured hepatocytes of the indicated genotype at different times after plating. Data are expressed as the percentage of BrdU-positive cells ± SEM for duplicate plates per genotype. A representative experiment is shown. Data were reproduced in 3 independent cultures.
Delay in cell cycle progression of S6K1-deficient hepatocytes in spite of normal protein anabolic responses.
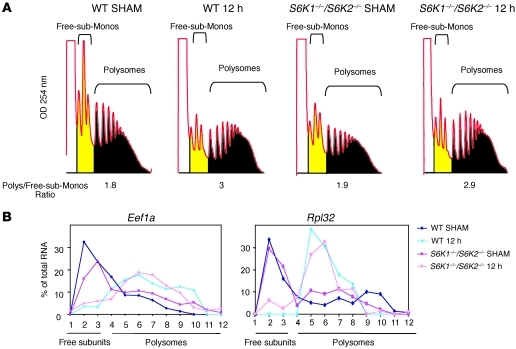
To understand how S6K1 regulates hepatocyte cell cycle, we considered key cellular responses for G1 phase progression. Hepatocytes undergo a massive stimulation of protein synthesis during early G1 phase (19). Since S6K1 is known to interact with the translational machinery (20), first we addressed whether S6K deletion affected global translational upregulation by assessing polysome formation in WT and S6K1–/–S6K2–/– livers 12 hours after hepatectomy. As shown in Figure 5A, the ratio between polysomes, monosomes, and free subunits was higher in hepatectomized livers as compared with that in sham-operated controls, indicating increased translation initiation. However, no difference in polysome profile was observed as a function of the S6K genotype. One major class of mRNAs, whose expression is controlled at the level of translation during the early phase of liver regeneration, are the 5′ terminal oligopyrimidine tract (5′TOP) mRNAs encoding ribosomal proteins and other translation factors (19). This regulation is under the control of mTOR and is required for ribosome biogenesis during growth responses (21). However, the recruitment of the 5′TOP-containing Eef1a and Rpl32 mRNAs onto polysomes 12 hours after hepatectomy was not affected by S6K deletion (Figure 5B), confirming previous observations in other cell types (21). Taken together, these findings suggest that the delay in cell cycle progression of S6K-deficient livers is not accompanied by a general deregulation of protein anabolic responses required for tissue growth.
Figure 5. Delay of cell cycle progression in S6K1 mutants in spite of normal protein anabolic responses.
(A) Liver polysomal profile of the indicated genotypes in sham-operated mice or 12 hours after PH. The area under the curve is indicated in yellow or black, and the ratio of the polysome/free-subunits plus monosomes (Free-sub-Monos) is shown. (B) Curves represent the Northern blot quantification of Eef1a or Rpl32 mRNA in liver of the indicated genotypes of sham-operated mice or mice 12 hours after PH. The relative amount of Eef1a or Rpl32 mRNA in each fraction is expressed as the percentage of the total in the polysome gradient.
Cyclin D regulation by S6K1.
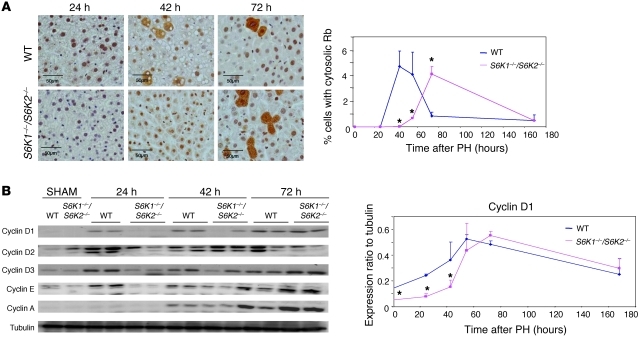
We next evaluated components of the cell cycle machinery involved in the progression through G1 phase. Inactivation and nuclear exclusion of the retinoblastoma protein, a late G1 phase event, was delayed in S6K1–/–S6K2–/– livers as compared with that in WT livers (Figure 6A). We therefore analyzed earlier events, such as the increase of cyclin D and E expression that was already detected 12 hours after hepatectomy (Figure 6B). As shown in Figure 6B, the kinetics of cyclin D1, cyclin D2, cyclin D3, and cyclin E expression were delayed in S6K1–/–S6K2–/– livers. These findings suggest that S6K1 controls cell cycle either by directly affecting phase G1 cyclin expression or by modifying the antecedents.
Figure 6. S6K1 deletion affects components of cell cycle machinery.
(A) Cytosolic Rb-positive hepatocytes of the indicated genotypes at different times after PH. Data are expressed as the percentage of cytosolic Rb-positive cells ± SEM for 4 mice per genotype. Representative images of Rb-immunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice. (B) Immunoblot analysis of protein extracts from the liver of the indicated genotypes in sham-operated mice or at different times after PH. Proteins were revealed using the indicated antibodies. The ratio of the densitometric assay is shown. Data are mean ± SEM. *P < 0.05 versus WT mice.
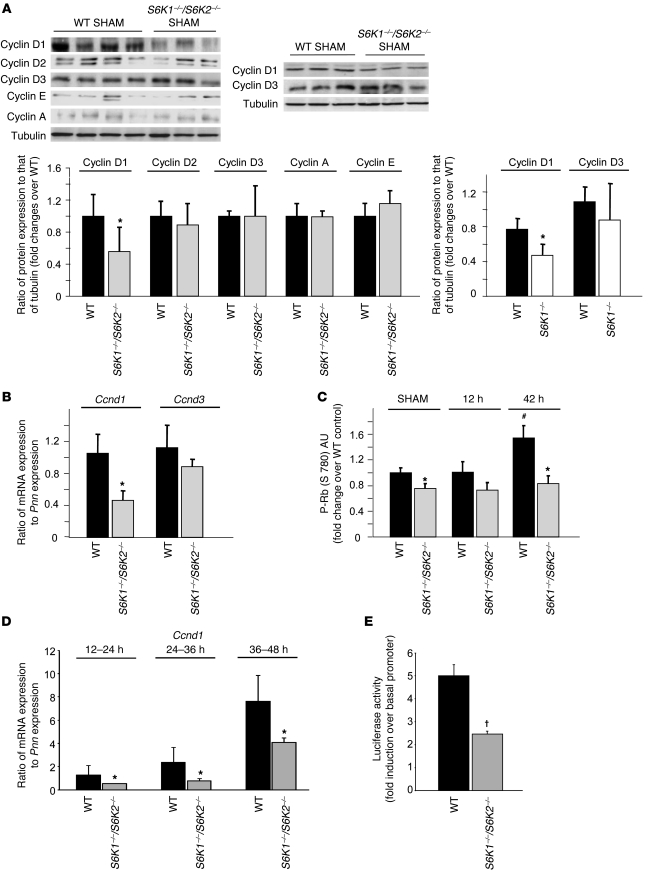
Cyclins were also expressed in adult livers in resting conditions (Figure 7A). Interestingly, cyclin D1 protein levels were lower in sham-operated S6K1–/–S6K2–/– livers as compared with those in WT livers, while cyclin D2, cyclin D3, cyclin E, and cyclin A levels were similar. The reduced cyclin D1 levels were due to S6K1 deletion (Figure 7A). The difference stemmed from a specific downregulation of Ccnd1 mRNA in S6K-deficient livers (Figure 7B), suggesting that S6K1 regulates Ccnd1 mRNA transcription and/or stability. In addition, the phosphorylation of Rb on Ser 780, a functional read-out of cyclinD/cdk4 activity, was also lower in sham-operated S6K1–/–S6K2–/– livers as compared with that in WT livers (Figure 7C). Taken together, our findings reveal a defect in the basal amount of Ccnd1 mRNA and protein, as well as associated kinase activities, that may account for the delayed cell cycle progression of S6K-deficient liver cells after hepatectomy.
Figure 7. Cyclin D1 regulation by S6K1.
(A) Immunoblot analysis of protein extracts from liver of the indicated genotypes in sham-operated mice. Proteins were revealed using the indicated antibodies. The ratio of the densitometric assay is shown. Data are mean ± SEM. *P < 0.05 versus WT mice. (B) RTqPCR analysis from liver of the indicated genotypes in sham-operated mice. Data are mean ± SEM. *P < 0.05 versus WT mice. (C) Immunoassay to detect Rb phosphorylation (P-Rb) on Ser 780 (S 780). Phospho-Rb protein in extracts from liver of the indicated genotypes in sham-operated mice or at different times after PH was revealed using an immunoassay. Data are mean ± SEM and expressed as fold change over sham-operated WT. *P < 0.05 versus WT mice; #P < 0.05 versus previous time for the same genotype. (D) RTqPCR analysis from primary hepatocyte cultures of the indicated genotypes at different time points after plating. Data are mean ± SEM. *P < 0.05 versus WT controls. (E) Hepatocytes were cotransfected with plasmids encoding firefly luciferase under a basal promoter (PA3-Luc) or under the 770-bp cyclin D1 promoter (770 bp cyclinD1-Luc) in combination with a plasmid encoding renilla luciferase under a constitutive promoter (pRL-TK). The ratio between firefly and renilla luciferase activity was measured 24 hours after plating. Data are mean ± SEM from 2 independent cultures. Data are expressed as fold increase of the cyclin D1 promoter activity over the basal promoter in the indicated genotypes. Data were confirmed at another time point (36 hours after plating). †P < 0.05 versus WT controls, using a Mann-Whitney test.
To gain further insights on the regulation of cyclin D1 expression by S6K, we established primary cultures from WT and S6K-null genotypes. Importantly, the decrease of Ccnd1 mRNA levels was reliably observed in S6K-deficient hepatocytes at different time points during the transition from G0/G1 to S phases (Figure 7D). To address whether S6K activity regulates the amount of Ccnd1 mRNA at the transcriptional or posttranscriptional levels, a plasmid encoding a luciferase reporter gene under the control of a 770-bp region of the cyclin D1 promoter was ectopically expressed in hepatocyte cultures. This promoter region contains a number of functional response elements that have been partly characterized in previous studies (22, 23). As shown in Figure 7E, the activity of the cyclin D1 promoter was reduced by half in S6K-deficient hepatocytes. These findings indicate that the transcriptional control of the cyclin D1 promoter by S6K plays a major role in the observed effects on Ccnd1 mRNA and protein levels.
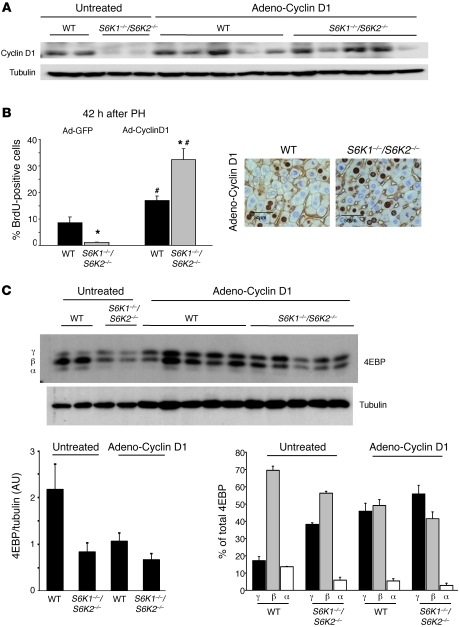
To establish a causal link among S6K activities, cyclin D1 expression, and liver proliferation after PH, adenoviral-mediated cyclin D1 expression was induced in livers before the PH (Figure 8A). Consistent with previous data (16, 24), cyclin D1 overexpression was sufficient to promote hepatocyte proliferation in adult livers that did not undergo PH, as assessed by immunostaining for the proliferating cell nuclear antigen (Supplemental Figure 3). Interestingly, the effect of ectopic cyclin D1 on proliferation was equivalent in WT and S6K-deficient genotypes. Next, the proliferative responses in cyclin D1–overexpressing cells were analyzed 42 hours after PH. As shown in Figure 8B, adenoviral-mediated cyclin D1 promoted S phase entry of S6K1–/–S6K2–/– livers at an even higher rate, as compared with that of WT controls. The hypersensitivity of S6K mutant livers to cyclin D1 expression suggests that these cells may have upregulated parallel pathways in an attempt to circumvent the proliferative defect. We therefore analyzed expression and phosphorylation of 4EBP1, an mTORC1 substrate that participates in the cell cycle control (12). In conclusion, restoring the levels of cyclin D1 is sufficient to rescue the proliferative defects due to S6K inactivation during liver regeneration.
Figure 8. Cyclin D1 expression rescues the proliferation defeat due to S6K inactivation during liver regeneration.
(A) Immunoblot analysis of protein extracts from liver before PH using anti–cyclin D1 and anti-tubulin antibodies. Mice were treated or not with adenovirus cyclin D1 2 days before tissue resection. (B) BrdU-positive hepatocytes of the indicated genotypes treated with adenovirus expressing GFP or cyclin D1 2 days before PH and sacrificed 42 hours after PH. Data are expressed as the percentage of BrdU-positive cells ± SEM for 4 mice per genotype. Representative images of BrdU and β-catenin coimmunostaining are shown. Scale bar: 50 μm. *P < 0.05 versus WT mice; #P < 0.05 versus mice treated with GFP adenovirus. (C) Immunoblot analysis of protein extracts from liver of the indicated genotypes after PH and cyclin D1 transduction (adeno-cyclin D1) or sham operation (untreated). Proteins were revealed using the indicated antibodies. The ratio of the densitometric assay for 4EBP1/tubulin and the percentage of α (hypophosphorylated), β, γ (hyperphosphorylated) forms of 4EBP1 are shown. Data are mean ± SEM.
Discussion
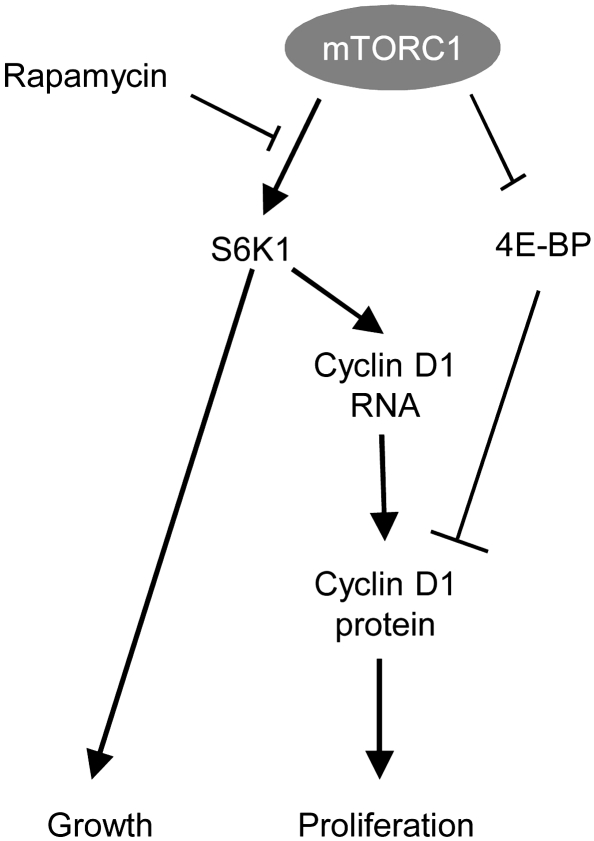
The kinetics of mouse liver regeneration after two-thirds PH allows a precise evaluation of in vivo hepatocyte growth and proliferation and the assessment of whether these anabolic responses are coordinated (14). The allosteric mTOR inhibitor rapamycin is known to significantly delay S phase entry and liver regeneration, indicating an important functional role for mTOR signaling (15–17). We show here that treatment with rapamycin derivatives severely blunts phosphorylation of the mTORC1 substrates S6K1 and S6K2, while having no effect on the 4E-BP family members of mTORC1 substrates and upregulating the activity of the mTORC2 substrates Akt1–Akt3 (Figure 1). We describe a rapamycin-sensitive pathway that is mediated by S6K1 activity and participates in the regulation of growth and cell cycle progression (Figures 2 and 3). Finally, we point to the impaired cyclin D1 expression in S6K1-deficient hepatocytes in resting conditions as a contributing factor for the proliferative defect (Figures 6–8).
The recovery of liver function after hepatectomy is promoted by a variety of signals that lead to the prompt re-establishment of liver mass by upregulating cell growth and/or proliferation. Rare are the mutations that completely abrogate liver regeneration, as the redundancy of signal transduction elements may mask functional defects (25). For instance, the transcription factor STAT3 has been described as an important element promoting liver proliferation after hepatectomy downstream IL-6 signaling, though liver STAT3-deficient mice compensate for the hepatocyte cell cycle defect with an increased cell size and display no defect in mass recovery (26, 27). Thus, the mutations causing a significant delay in liver regeneration are considered to hit essential pathways for liver function. Relevant to our study, the phosphoinositide-dependent protein kinase 1 (PDK1), which is required for the activities of Akt, S6K, and other AGC kinases, strongly impacts liver regeneration, as 70% of liver-specific PDK1-knockout mice die after PH. The 30% of viable mice display impaired liver mass recovery, mainly due to cell size defects (28). In the same study, treatment of mice with sodium salicylate, which inhibits S6K signaling, leads to a delay in S phase entry after PH. Here we show that S6K deletion impaired both liver proliferation and cell growth, determining a considerable delay in mass recovery (Figures 1 and 2). It has previously been shown that protein composition in the diet influences liver recovery (29). In particular, it was shown that plasma levels of branched-chain amino acids decrease after hepatectomy, indicating an increased intracellular utilization, and that supplementation with branched-chain amino acids and glutamine ameliorates liver recovery. Interestingly these amino acids stimulate mTORC1/S6K activity while suppressing mTORC2/Akt (30, 31) (C. Chauvin, V. Koka, and M. Pende, unpublished observations). Thus, the preferential activation of the mTORC1/S6K1 pathway over the mTORC2/Akt branch is likely to mediate the nutritional input from amino acids during liver regeneration.
Previous studies have identified S6K as a cell size regulator. Drosophila S6K mutant flies have normal cell number in adult wing and eye tissues but selectively display a cell size defect (32). However, Drosophila larvae are delayed, and it is possible that a proliferation defect is masked by the additional developmental period. In mice, the cell size defect is observed in a number of tissues, including skeletal muscle, fat, and pancreatic islets, in resting conditions (13, 33, 34), and in liver during regeneration (Figure 1). In addition, S6K deletion in MEF and myoblast cultures mimics the effects of rapamycin and asTORi on cell size (12, 13). Conversely, mutant cells in culture do not display impaired proliferation when maintained with maximal supplies of mitogens and nutrients, and their cell cycle remains sensitive to the inhibitory action of rapamycin and asTORi (12, 13, 21). Although these findings have been interpreted as evidence for a selective control by S6K on cell size, but not cell number, downstream of mTOR, no precise kinetic analysis of cell proliferation has been performed in vivo. Our studies after liver hepatectomy and in hepatocyte primary cultures reveal that S6K1 activity also provides a proliferative advantage in vivo and in vitro to the hepatocytes. This effect is likely to be largely cell autonomous, as it is observed in cultured cells in defined medium, and hepatic overexpression of S6K1 rescues the defect of mutant livers in vivo. Thus, in hepatocytes, the lack of S6K1 signaling impairs the proliferative response, a defect that is not entirely compensated by other rapamycin-sensitive mTORC1 targets in this specific cell type.
S6K1 deletion does not cause a general delay in anabolic responses during liver regeneration, which might be expected if S6K1 were acting on humoral factors or the priming phase of regeneration or the general fitness of the mice. Although S6K1 was shown to interact with the translational machinery (20), the upregulation of translation initiation, an early event during regeneration, occurred normally in S6K1-deficient livers, as assessed by polysome formation 12 hours after hepatectomy (Figure 5). Similarly, the recruitment onto polysomes of 5′TOP mRNAs coding for ribosomal proteins and translation factors is not delayed by S6K1. Thus 2 typical growth responses downstream of mTOR do not depend on S6K1 activity. In contrast, the induction of early G1 phase cyclin levels, namely cyclin D1, cyclin D2, cyclin D3, and cyclin E, is already altered during the first day after hepatectomy in S6K1–/– hepatocytes (Figure 6). Taken together these surprising findings reveal changes in cell cycle regulators, depending on S6K1 activity regardless of unaltered translational responses.
Although additional mechanisms can be involved, it is likely that the downregulation of cyclin D1 expression greatly contributes to the impaired regeneration of S6K1-null livers based on the following considerations. First, the reduction of Ccnd1 mRNA and protein levels is already observed in resting conditions, suggesting a direct target of S6K1 activity and not a consequence of the delay in cell cycle progression (Figure 7). Second, cyclin D1 is known to promote hepatocyte cell cycle progression, as demonstrated by ectopic overexpression and genetic invalidation experiments (24, 35). Importantly, cyclin D1 overexpression rescues the inhibitory effect of S6K deletion on S phase entry after PH (Figure 8), providing a causal link among mTOR/S6K pathway, cyclin D1 expression, and cell cycle progression. Third, cyclin D1 is an early target after transient inhibition of mTOR activity by pharmacological agents, as shown in multiple systems (36). Interestingly, cyclin D1 expression can be regulated by the mTOR pathway at the level of transcription and translation. While 4E-BPs have been demonstrated to control translational regulation of Ccnd1 mRNA (12, 37), our studies define a distinct output from mTOR that regulates cyclin D1 promoter activity and mRNA levels dependent on S6K1 activity (Figure 9). It is tempting to speculate on whether the regulation of cyclin D1 expression may also explain other phenotypes of S6K1-deficient mice. Cyclin D in Drosophila and mammals is also involved in cell size control (24, 38, 39). In addition cyclin D1 also regulates mitochondrial biogenesis (40). Future studies should assess whether the atrophy of S6K1-deficient cells and their altered mitochondrial mass may be at least in part a consequence of cyclin D1 regulation (13, 34).
Figure 9. Model of growth and proliferation control by mTORC1.
Elucidating the molecular mechanisms of cell cycle control by the mTOR pathway is crucial for all the disease states in which treatment with mTOR inhibitors is expected to be beneficial. Recently, the use of asTORi has uncovered the importance of a rapamycin-insensitive branch of mTORC1 that involves 4E-BPs in cell cycle regulation (ref. 12 and Figure 9). However, distinct rapamycin-sensitive components exist, as outlined by the sensitivity of 4E-BP–deficient cell proliferation to the inhibitory action of rapamycin. It is possible that the relative contributions of these mTORC1 branches vary depending on cell types and pathophysiological context. For instance, the growth of kidney tumors in a mouse model of tuberous sclerosis is equally sensitive to a rapamycin derivative and a dual pan class I PI3K/mTOR catalytic inhibitor (41), suggesting that a rapamycin-sensitive pathway may mediate a major proliferative signal in these benign tumors. It would be interesting to test the functional relevance of the S6K1/cyclin D1 axis in this settings, though we are aware that other yet uncharacterized proliferative mechanisms may intervene, given the sensitivity of S6K-deficient cell cycle to rapamycin (13, 21).
Methods
Animals and surgery.
Generation of S6K1;S6K2-deficient mice (C56BL/6-129/Ola) and S6K1-, S6K2-, Akt1-, and Akt2- deficient mice (C57BL/6) has been previously described (13, 21, 42, 43). Each genotype was compared with WT mice of the same genetic background. Mice were maintained at 22°C with a 12-hour-dark/12-hour-light cycle and had free access to food. All animal studies were approved by the Direction Départementale des Services Véterinaires, Prefecture de Police, Paris, France (authorization number 75-1313). All studies were done in male animals. Eight- to twelve-week-old mice were subjected to sham operation or two-thirds PH, as previously described (18), under general anaesthesia with inhaled isoflurane (n = 4 for each genotype and time point). Animals were killed at different times after surgery, as indicated in the figure legends. Sham animals had operations without any liver resection.
When indicated, mice were infected with 5 × 109 infectious particles of GFP, S6K1, or cyclin D1 adenovirus by retro-orbital intravenous injection. Surgery was performed 2 days after infection for cyclin D1 or 1 week after infection for GFP and S6K1.
Hepatocyte cell cultures.
Hepatocytes were isolated from mice by in situ perfusion and were seeded in complete medium, as described before (44). After cell spreading, the culture medium was deprived of fetal bovine serum and proliferation was induced with 50 ng/m of EGF (Sigma-Aldrich). Incorporation of the thymidine analog BrdU was used as an index of cell proliferation. After fixation, BrdU-positive cells were detected using the BrdU Detection Kit I (Roche). Apoptotic cells were detected using In Situ Cell Death Detection Kit, Fluorescein (Roche). Transfection of expression plasmids was performed in 6-well plates in the presence of Lipofectamine LTX with PLUS Reagent (InVitrogen) 4 hours after plating. Four μg expression plasmids and 5 μl Lipofectamine per well were applied in a final volume of 2 ml Opti-MEM. After 4 hours, the medium was renewed with complete medium. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Plasmid encoding a firefly luciferase under the control of 770-bp human cyclin D1 promoter was provided by Anil Rustgi (Department of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA) (22).
Immunohistochemistry and morphometric analysis.
Liver tissue was fixed overnight in phosphate-buffered 10% formalin, embedded in paraffin, and sectioned in 4 μm. Immunohistochemistry was performed with the following antibodies: anti-BrdU antibody (Roche), anti–β-catenin antibody (Calbiochem), and anti-Rb antibody (Abcam), and they were all counterstained with hematoxylin. Hepatocyte proliferation was determined by BrdU incorporation. Mice were injected intraperitoneally with 50 mg/kg of BrdU 2 hours prior to sacrifice. The results were expressed as the percentage of BrdU-positive cells. At least 3,000 hepatocytes were counted. Hepatocyte cell density was determined by anti–β-catenin immunohistochemistry. The results were expressed as the number of cells in a liver surface. At least 10 areas of 33,500 μmβ were analyzed.
Cytosolic presence of Rb was determined by Rb immunohistochemistry. The results were expressed as the percentage of cytosolic Rb-positive cells. At least 3,000 hepatocytes were counted.
Western blot.
A piece of frozen tissue was ground to powder under liquid N2 and lysed in 20 mM Tris-HCl (pH 8.0), 5% glycerol, 138 mM NaCl, 2.7 mM KCl, 1% NP-40, 20 mM NaF, 5 mM EDTA, 1 mM sodium orthovanadate, 5 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM DTT. To remove cell debris, homogenates were spun at 8,000 g for 10 minutes at 4°C. Protein extract from liver was resolved by SDS-PAGE before transfer onto PVDF membrane and incubation with anti-S6K1, anti–cyclin D1, anti–cyclin D2, anti–cyclin D3, anti–cyclin A, and anti–cyclin E (all from Santa Cruz Biotechnology Inc.); anti-phospho S6K1 (Thr 389), anti-4EBP, anti-phospho 4EBP (Ser 65), anti-Akt, anti-phospho Akt (Ser 473), anti-S6, anti-phospho rpS6 (Ser 235/236), anti-phospho rpS6 (Ser 240/244), anti-PRAS40, anti-phospho PRAS40 (Thr 246), anti-GSK3β, anti-phospho GSK3β (Ser 9), and anti–cylin D3 (all from Cell Signalling Technology); and anti-tubulin (Sigma-Aldrich).
Immunoassay.
Levels of phospho-Rb (Ser 780) in liver protein extracts were measured by ELISA using Human Rb (pS780) Immunoassay Kit (Invitrogen) according to the manufacturer’s instructions. Five μg and twenty μg of protein extracts were used.
Real-time quantitative PCR.
Total RNA was isolated from liver tissue using an RNeasy Mini Kit (QIAGEN), according to the manufacturer’s instructions, and single-strand cDNA was synthesized from 1 μg of total RNA with random hexamer primers and SuperScript II (Invitrogen). Real-time quantitative PCR (RTqPCR) was performed using a TaqMan instrument (Applied Biosystems), according to the manufacturer’s instructions, using a SYBR Green PCR Master Mix (Applied Biosystems). We determined the relative amounts of the mRNAs studied by means of the 2–ΔΔCT method, with pinin and WT mice as the invariant control for all studies. The murine primer sequences used were as follows: cyclin D1, sense 5′-GCGTACCCTGACACCAATCTC-3′, antisense 5′-CTCCTCTTCGCACTTCTGCTC-3′; cyclin D2, sense 5′-GAGTGGGAACTGGTAGTGTTG-3′, antisense 5′-CGCACAGAGCGATGAAGGT-3′; cyclin D3, sense 5′-CGAGCCTCCTACTTCCAGTG-3′, antisense 5′-GGACAGGTAGCGATCCAGGT-3′; and pinin, sense 5′-ACCTGGAAGGGGCAGTCAGTA-3′, antisense 5′-ATCATCGTCTTCTGGGTCGCT-3′. The results of RTqPCR are given in arbitrary units and expressed as fold changes in mRNA levels relative to those of WT controls.
Polysome fractionation and Northern blot analysis.
Sucrose density gradient centrifugation was used to separate the subpolysomal from the polysomal ribosome fractions. A piece of frozen liver was ground to powder under liquid N2 and lysed in 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 240 mM KCl, 250 mM sucrose, 2% Triton X-100, 5 mM dithiothreitol, 100 μg/ml cycloheximide, and 100 U/ml RNase inhibitor. To remove cell debris, homogenates were spun at 8,000 g for 5 minutes at 4°C. An aliquot of the supernatant was removed to measure protein concentration. Heparin was added to the supernatants at a final concentration of 1 mg/ml. The extracts were rapidly frozen into liquid nitrogen and stored at –80°C. 1.2 mg protein were layered on a 0.5–1.5 M linear sucrose gradient (20 mM Tris-HCl, pH 7.5, 80 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol) and centrifuged in a SW41 rotor at 160,000 g for 2 hours at 4°C. After centrifugation, the gradient was displaced upward through a flow cell recording absorbance at 260 nm with the use of the density gradient fractionation system (Isco) and fractionated in 12 fractions. RNA isolation and Northern blot analysis of each fraction were performed as described previously (45). Northern blots were hybridized with specific probes against Eef1a (5′-GCCGGAATCTACGTGTCCGATTACGACGATGTTGATGTGAGTCTTTTCCTTTCCCAT-3′) and Rpl32 (5′-TTCACATATCGGTCTGACTGGTGCCTGATGAACTTCTTGG-3′). Quantification was made using Storm ImageQuant software (GE Healthcare Life Science).
Statistics.
Analysis was performed by 2-tailed, unpaired Student’s t test unless otherwise indicated. P values of less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank the Novartis Foundation and the George Thomas laboratory for the use of S6K mutant mice, the Morris Birnbaum laboratory for the use of Akt mutant mice, the Anil Rustgi laboratory for providing the cyclin D1 promoter plasmid, and Pfizer for the gift of temsirolimus. We are grateful to the members of INSERM-U845 for support and to Lluis Fajas for helpful discussions and sharing reagents. We thank Sophie Berissi, Stephanie Boumard, and Alexandra Dubois for their technical help. This work was supported by grants from the European Research Council, from Fondation de la Recherche Medicale (DEQ20061107956), from Fondation Schlumberger pour l’Education et la Recherche, from the Association pour la Recherche sur le Cancer to M. Pende, and from the NIH (DK54921) to J.H. Albrecht. C. Espeillac received a fellowship from the MRT and FRM.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(7):2821–2832. doi:10.1172/JCI44203.
Claudia Mitchell’s present address is: Halo-Bio, RNAi Therapeutics Inc. Seattle, Washington, USA.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Murakami M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24(15):6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangloff YG, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24(21):9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houghton PJ, Huang S. mTOR as a target for cancer therapy. Curr Top Microbiol Immunol. 2004;279:339–359. doi: 10.1007/978-3-642-18930-2_20. [DOI] [PubMed] [Google Scholar]
- 5.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 6.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Thoreen CC, et al. An ATP-competitive mTOR inhibitor reveals rapamycin-insensitive functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janes MR, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16(2):205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17(3):249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328(5982):1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohanna M, et al. Atrophy of S6K1(–/–) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. . Nat Cell Biol. 2005;7(3):286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- 14.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 15.Jiang YP, Ballou LM, Lin RZ. Rapamycin-insensitive regulation of 4E-BP1 in regenerating rat liver. . J Biol Chem. 2001;276(14):10943–10951. doi: 10.1074/jbc.M007758200. [DOI] [PubMed] [Google Scholar]
- 16.Nelsen CJ, Rickheim DG, Tucker MM, Hansen LK, Albrecht JH. Evidence that cyclin D1 mediates both growth and proliferation downstream of TOR in hepatocytes. J Biol Chem. 2003;278(6):3656–3663. doi: 10.1074/jbc.M209374200. [DOI] [PubMed] [Google Scholar]
- 17.Buitrago-Molina LE, et al. Rapamycin delays tumor development in murine livers by inhibiting proliferation of hepatocytes with DNA damage. Hepatology. 2009;50(2):500–509. doi: 10.1002/hep.23014. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3(7):1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 19.Aloni R, Peleg D, Meyuhas O. Selective translational control and nonspecific posttranscriptional regulation of ribosomal protein gene expression during development and regeneration of rat liver. Mol Cell Biol. 1992;12(5):2203–2212. doi: 10.1128/mcb.12.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 21.Pende M, et al. S6K1(–/–)/S6K2(–/–) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. . Mol Cell Biol. 2004;24(8):3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan YX, Nakagawa H, Lee MH, Rustgi AK. Transforming growth factor-alpha enhances cyclin D1 transcription through the binding of early growth response protein to a cis-regulatory element in the cyclin D1 promoter. J Biol Chem. 1997;272(52):33181–33190. doi: 10.1074/jbc.272.52.33181. [DOI] [PubMed] [Google Scholar]
- 23.Vartanian R, et al. AP-1 regulates cyclin D1 and c-MYC transcription in an AKT-dependent manner in response to mTOR inhibition: role of AIP4/itch-mediated JUNB degradation. Mol Cancer Res. 2011;9(1):115–130. doi: 10.1158/1541-7786.MCR-10-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelsen CJ, Rickheim DG, Timchenko NA, Stanley MW, Albrecht JH. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res. 2001;61(23):8564–8568. [PubMed] [Google Scholar]
- 25.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176(1):2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Liang X, Kellendonk C, Poli V, Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem. 2002;277(32):28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 27.Haga S, et al. Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J Hepatol. 2005;43(5):799–807. doi: 10.1016/j.jhep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 28.Haga S, et al. The survival pathways phosphatidylinositol-3 kinase (PI3-K)/phosphoinositide-dependent protein kinase 1 (PDK1)/Akt modulate liver regeneration through hepatocyte size rather than proliferation. Hepatology. 2009;49(1):204–214. doi: 10.1002/hep.22583. [DOI] [PubMed] [Google Scholar]
- 29.Holecek M. Nutritional modulation of liver regeneration by carbohydrates, lipids, and amino acids: a review. Nutrition. 1999;15(10):784–788. doi: 10.1016/S0899-9007(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 30.Patti ME, Brambilla E, Luzi L, Landaker EJ, Kahn CR. Bidirectional modulation of insulin action by amino acids. J Clin Invest. 1998;101(7):1519–1529. doi: 10.1172/JCI1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greiwe JS, Kwon G, McDaniel ML, Semenkovich CF. Leucine and insulin activate p70 S6 kinase through different pathways in human skeletal muscle. Am J Physiol Endocrinol Metab. 2001;281(3):E466–E471. doi: 10.1152/ajpendo.2001.281.3.E466. [DOI] [PubMed] [Google Scholar]
- 32.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285(5436):2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 33.Pende M, et al. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature. 2000;408(6815):994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 34.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431(7005):200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 35.Ledda-Columbano GM, Pibiri M, Concas D, Cossu C, Tripodi M, Columbano A. Loss of cyclin D1 does not inhibit the proliferative response of mouse liver to mitogenic stimuli. Hepatology. 2002;36(5):1098–1105. doi: 10.1053/jhep.2002.36159. [DOI] [PubMed] [Google Scholar]
- 36.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene. 2008;27(8):1106–1113. doi: 10.1038/sj.onc.1210715. [DOI] [PubMed] [Google Scholar]
- 38.Frei C, Edgar BA. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev Cell. 2004;6(2):241–251. doi: 10.1016/S1534-5807(03)00409-X. [DOI] [PubMed] [Google Scholar]
- 39.Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol. 2005;289(6):C1457–C1465. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103(31):11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollizzi K, Malinowska-Kolodziej I, Stumm M, Lane H, Kwiatkowski D. Equivalent benefit of mTORC1 blockade and combined PI3K-mTOR blockade in a mouse model of tuberous sclerosis. Mol Cancer. 2009;8:38. doi: 10.1186/1476-4598-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho H, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta). Science. 2001;292(5522):1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 43.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276(42):38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 44.Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278(21):19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 45.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17(22):6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.