Abstract
Intratumor heterogeneity is a major clinical problem because tumor cell subtypes display variable sensitivity to therapeutics and may play different roles in progression. We previously characterized 2 cell populations in human breast tumors with distinct properties: CD44+CD24– cells that have stem cell-like characteristics, and CD44–CD24+ cells that resemble more differentiated breast cancer cells. Here we identified 15 genes required for cell growth or proliferation in CD44+CD24– human breast cancer cells in a large-scale loss-of-function screen and found that inhibition of several of these (IL6, PTGIS, HAS1, CXCL3, and PFKFB3) reduced Stat3 activation. We found that the IL-6/JAK2/Stat3 pathway was preferentially active in CD44+CD24– breast cancer cells compared with other tumor cell types, and inhibition of JAK2 decreased their number and blocked growth of xenografts. Our results highlight the differences between distinct breast cancer cell types and identify targets such as JAK2 and Stat3 that may lead to more specific and effective breast cancer therapies.
Introduction
Breast tumors display a high degree of intratumor heterogeneity that drives the tumor evolution responsible for therapeutic resistance, recurrence, and tumor progression (1, 2). Cancer cells with stem cell–like properties in particular have been proposed to play a critical role in metastatic progression and resistance to commonly used cancer treatment (3–5). These cells can be identified by various functional assays (e.g., tumor spheres and xenograft assays) and using specific cell-surface markers. For example, the CD44+CD24– breast cancer cell population has been shown to be enriched for tumor-initiating (6) and chemotherapy-resistant (7, 8) cells.
We previously characterized the molecular profiles and functional properties of CD44+CD24– stem cell–like and CD44–CD24+ more differentiated luminal breast cancer cells and identified genes consistently differentially expressed between them (9, 10). CD44+CD24– cells highly express genes involved in invasion and angiogenesis and display activated TGF-β–, Hh-, and PLAU-signaling pathways, whereas markers and pathways of luminal epithelial differentiation are more abundant in CD44–CD24+ cells. The presence of these 2 cell populations within individual tumors and their dependence on different signaling pathways for growth and survival pose a challenge for the effective therapeutic eradication of breast tumors, especially when specific pathway-targeted approaches are used. As a proof of principle, we demonstrated that, in a pleural effusion sample, only CD44+CD24– cells respond to a TGFBR kinase inhibitor (10).
Based on immunohistochemical analyses of a large cohort of invasive and in situ breast carcinomas for markers of CD44+CD24– and CD44–CD24+ breast cancer cells, we also previously found that significant diversity both among and within tumors exists for these cell types. Overall, CD44+CD24– cells are more frequent in basal-like breast cancer, whereas luminal tumors are enriched in CD44–CD24+ cells (11, 12). Thus, therapies eliminating CD44+CD24– cells may represent a new approach for the clinical management of basal-like breast cancer, currently the only major breast tumor subtype without effective targeted treatment strategies and with poor prognosis (13). Our subsequent analysis of genetic alterations present in CD44+CD24– and CD44–CD24+ breast cancer cells at the single-cell level using immuno-FISH revealed extensive genetic diversity both within and between the 2 cell populations (14). Thus, even though these 2 cell populations may have consistently distinct gene expression profiles, they may not be genetically homogeneous, a characteristic that is likely to influence their sensitivity to therapeutic strategies targeting signaling pathways specifically activated in them.
In order to discover new ways to more effectively eliminate CD44+CD24– and CD44–CD24+ breast cancer cells, here we conducted a large-scale shRNA screen to identify genes and pathways on which each cell type or both cell types depend. The results of our screen demonstrate that several signaling pathways are preferentially active in CD44+CD24– breast cancer cells in primary human tumors and that by inhibiting these it is possible to obtain effective therapeutic response accompanied by the elimination of these cells.
Results
shRNA screen for genes required in CD44+CD24– and CD44–CD24+ breast cancer cells.
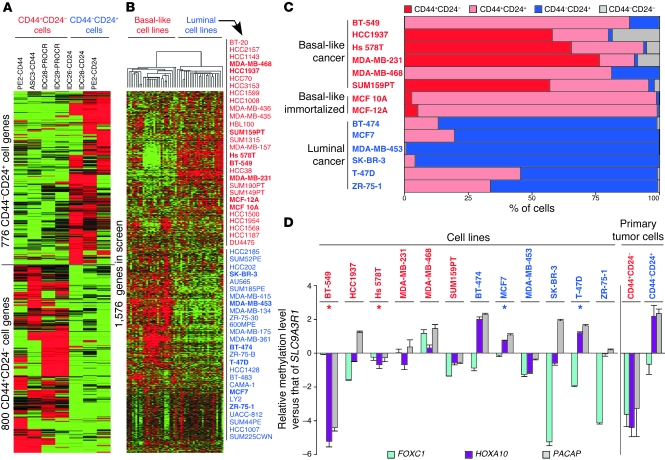
We hypothesized that genes differentially expressed between CD44+CD24– and CD44–CD24+ breast cancer cells might be enriched for signaling pathways on which these cells specifically depend. Thus, we identified 1,576 candidate genes to target based on their differential expression between these 2 cell populations in primary human breast tumor samples (10) and their representation in the TRC lentivirally delivered shRNA library (ref. 15, Figure 1A, and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI44745DS1). Due to the need to reliably and consistently grow large numbers of cells that maintain their properties, we chose to conduct the screen in a panel of breast cancer cell lines resembling CD44+CD24– and CD44–CD24+ breast cancer cells instead of using primary cultures (Supplemental Table 2). Suitable cell lines were selected based on their gene expression, cell-surface marker, and DNA methylation profiles (Figure 1, B–D, and Supplemental Figure 1). Specifically, we identified several basal-like (BT-549, HCC1937, Hs 578T, MDA-MB-231, MDA-MB-468, and SUM159PT) and luminal (BT-474, MCF7, MDA-MB-453, SK-BR-3, T-47D, and ZR-75-1) breast cancer cell lines that differentially expressed the 1,576 genes distinguishing CD44+CD24– and CD44–CD24+ breast cancer cells included in the screen (Figure 1B), including CD44 and CD24 (Figure 1C), and showed differential methylation for FOXC1, HOXA10, and PACAP relative to SLC9A3R1 (Figure 1D), genes we previously identified as hypo- and hypermethylated, respectively, in CD44+CD24– compared with in CD44–CD24+ cells (9). We also included MCF 10A and MCF-12A basal-like nontumorigenic, immortalized mammary epithelial cell lines as controls to be able to select for malignancy-associated targets. We used a decrease in viable cell numbers following infection with lentiviral shRNAs as the read-out in order to allow the identification of hits that are required either for cell survival or for proliferation.
Figure 1. Characteristics of genes and cell lines chosen for shRNA screen.
(A) Clustering of SAGE data for the genes in the screen. Genes were selected based on their differential expression between the groups of samples of CD44+CD24– and CD44–CD24+ (primary tumor) cells shown. Red and green (in A and B) indicate high and low gene expression, respectively. PE, pleural effusion; ASC, ascites; IDC, invasive ductal carcinoma. (B) Clustering of previously published microarray data (63) for 54 breast cell lines using the genes in the screen. Clusters of basal-like and luminal cell lines were formed. The 14 cell lines highlighted in bold were chosen for the screen. (C) Expression of CD44 and CD24 in the cell lines used in the screen. Results of flow cytometry analysis of CD44 and CD24 levels are shown. Flow cytometry profiles and gating are shown in Supplemental Figure 1. (D) qMSP data for the cell lines included in the screen for genes differentially methylated between CD44+CD24– and CD44–CD24+ breast cancer cells (as shown at right). Each bar represents the natural log of the ratio of the qMSP reading of either FOXC1, HOXA10, or PACAP (official gene symbol, MGC29506) to the qMSP reading of SLC9A3R1. Error bars show SD of triplicates (for cell lines) or 3 ER+PR+HER2– tumor samples (for primary tumor cells). Asterisks indicate the cell lines that were chosen for phase 1 of the screen based on these results and the ease of their growing in culture.
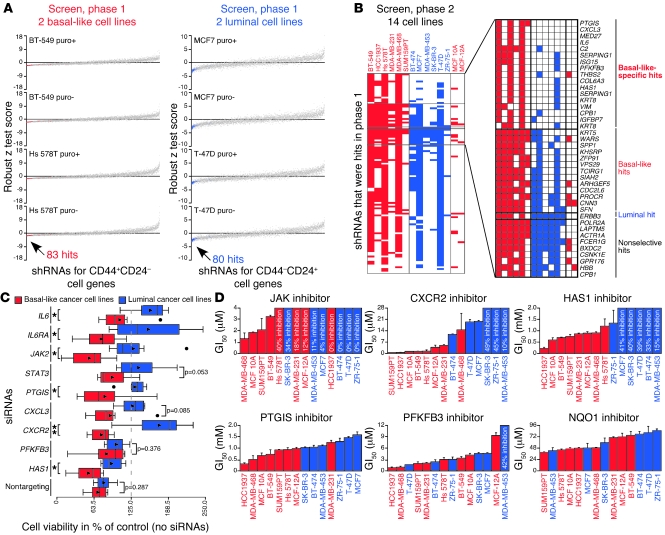
We performed the shRNA screen in 2 phases. In phase 1, we tested TRC shRNAs targeting genes more highly expressed in CD44+CD24– and CD44–CD24+ cells in BT-549 and Hs 578T or in MCF7 and T-47D cells, respectively, 2 cell lines highly resembling each corresponding cell type and easy to cultivate in vitro. Based on their effects on viability in each pair of cell lines (Figure 2A and Supplemental Table 3), we identified 83 hits targeting 67 genes in the basal-like and 80 hits targeting 65 genes in the luminal cell lines. In phase 2, we tested these shRNAs in all 14 cell lines and classified them as basal-like–specific, basal-like, luminal, and nonselective hits based on the cancer cell lines in which they scored based on percentage of control values (Figure 2B and Supplemental Tables 4 and 5). A detailed description of how phase 1 and 2 shRNA screen hits were defined is in the Supplemental Data. Briefly, in regard to the final classification of hits, “basal-like–specific” hits had to score numerous times almost only in basal-like cell lines, “basal-like” and “luminal” hits had to score in a large number of basal-like cell lines and fewer luminal cell lines or vice versa, and “nonselective” hits had to score in a large number of both basal-like and luminal cell lines. Besides their basal-like or luminal cell characteristics, each of the cell lines was derived from a different tumor and had unique genetic and epigenetic profiles. Thus, likely due to these differences between cell lines, not all hits that scored in the 4 cell lines used for the primary screen were classified as hits in the secondary screen conducted in 14 cell lines. Notably, the nonselective hits targeted genes known to be essential in cancer cells such as POLR2A and CSNK1E (16), and most hits did not affect the MCF 10A and MCF-12A cell lines that were used as controls. Also, none of the 5 types of negative control shRNAs included in the screen were hits in either phase 1 or 2. Thus, overall, the shRNA screen appeared to have worked successfully.
Figure 2. Identification and validation of genes required in basal-like breast cancer cells.
(A) Robust z test scores ([average viability – plate median average viability]/[plate median absolute deviation of average viability]) from shRNA screen, phase 1, for shRNAs with infection efficiencies greater than 0.25 and robust z test score SD below the 99th percentile for both basal-like and luminal cell lines with and without puromycin (puro). Data representing basal-like and luminal hits, selected based on their 4 basal-like or luminal robust z test scores as described in the text, are red and blue, respectively. (B) Scoring of shRNAs with infection efficiencies greater than 0.25 for all cell lines and classification of hits in shRNA screen, phase 2. Shading indicates shRNAs scored based on percentage of control values ([100 × average viability]/[median average viability of plate controls]). Genes corresponding to hits are listed. (C) Box plots showing viability of breast cell lines treated with siRNAs in triplicate for 3 days. Triangles mark averages. Circles mark outliers (which were included in P value calculations). *P < 0.05, t test; **P < 0.01, t test. (D) GI50 values for inhibitors in breast cell lines. The maximum percentage of growth inhibition observed is shown when cells were not affected enough for GI50 calculation. Error bars show SD of triplicates. All percentage growth inhibition data used to prepare these graphs are depicted in Supplemental Figure 2C.
Validation of shRNA screen results.
We further investigated the 15 genes targeted by the basal-like–specific hits, since they represent promising therapeutic targets for CD44+CD24– breast cancer cells. We began our follow-up studies by validating the shRNA screen results. First, we treated the screened cancer cell lines with siRNAs specific for genes targeted by hits (IL6, PTGIS, CXCL3, PFKFB3, and HAS1), genes encoding proteins in their signaling pathways (STAT3; JAK2, encoding a kinase downstream of IL-6 that phosphorylates Stat3; IL6RA, encoding part of the IL-6 receptor; and CXCR2, encoding the CXCL3 receptor), or no genes (as control). All siRNAs effectively decreased the viability of basal-like compared with luminal cell lines, with nontargeting and PFKFB3 siRNAs only weakly showing this trend (Figure 2C and Supplemental Figure 2, A and B), lending support to the shRNA screen results. Second, we treated the screened cell lines with small molecule inhibitors for proteins encoded by genes validated by the siRNA experiments (PTGIS, CXCR2, PFKFB3, and HAS1), a commercial JAK inhibitor called JAK inhibitor I, which inhibits JAK2 (in addition to JAK1, JAK3, and TYK2), an inhibitor of NQO1, an enzyme encoded by a gene included in both phases of the screen but not ultimately classified as a hit (as a control), or vehicle alone for 3 days and calculated GI50 (concentration inhibiting cell growth 50%) values. In cell lines where the GI50 values of inhibitors were higher than all tested treatment concentrations, we recorded the maximum observed inhibition levels relative to vehicle-treated controls. Corroborating the shRNA screen and siRNA results, all inhibitors except the control NQO1 and PFKFB3 inhibitors effectively and preferentially decreased the viability of basal-like cell lines relative to vehicle-treated controls, at least partially by inducing apoptosis, which was also induced by the IL6 shRNA that was a hit in the shRNA screen (Figure 2D and Supplemental Figure 2, A–C).
Importance of the IL-6/JAK2/Stat3 pathway in basal-like breast cancer cells and in vivo.
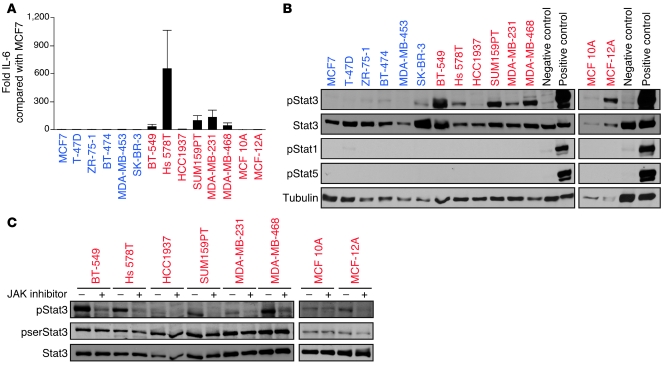
Due to the high level of specificity of the IL6 screening hit and JAK inhibitor in decreasing the viability of basal-like breast cancer cells (a characteristic not shared by any of the other inhibitors we tested, which each also decreased the growth of at least some luminal breast cancer cells by at least 50% with the concentrations used), we further examined the IL-6/JAK2/Stat3 pathway in this cell type. We confirmed that the IL6 screening hit effectively and specifically targeted IL6 by showing that it decreased IL6 mRNA and protein levels (Supplemental Figure 3, A and B), that its effect on cell viability could be rescued by the addition of recombinant human IL-6 (Supplemental Figure 3C), and that it decreased Stat3 activation, specifically the levels of phosphotyrosine-705–Stat3 (pStat3) (Supplemental Figure 3D). We also found that IL-6 was primarily secreted by basal-like breast cancer cell lines (Figure 3A), and we saw that IL-6 secretion was accompanied by high pStat3 levels (Figure 3B), suggesting the presence of an autocrine growth regulatory loop. This loop appears to be interrupted by JAK inhibitor, as demonstrated by the reduction of pStat3 levels upon such treatment (Figure 3C). The importance of other downstream targets activated by JAK2 (e.g., ERK/MAPK and NF-κB) in basal-like breast cancer cells cannot be excluded. However, the strong association among IL-6 secretion, basal-like phenotype, and pStat3 levels implied that Stat3 is the JAK2 target most relevant in these cells. Furthermore, inhibitors of ERK/MAPK and NF-κB signaling do not demonstrate the same degree of basal-like breast cancer cell specificity as we observed for the JAK inhibitor.
Figure 3. Preferential activation of the IL-6/JAK2/Stat3 pathway in basal-like breast cancer cells.
(A) ELISA results. Error bars show SD of triplicates. (B) Immunoblots. Positive and negative controls were T-47D cells treated or not treated, respectively, with prolactin and oncostatin M. Stat3, pStat1, pStat5, and tubulin were used as controls. (C) Immunoblots. Two-hour treatment with 2 μM JAK inhibitor reduces pStat3 in basal-like breast cells containing it. pserStat3 and Stat3 were used as controls.
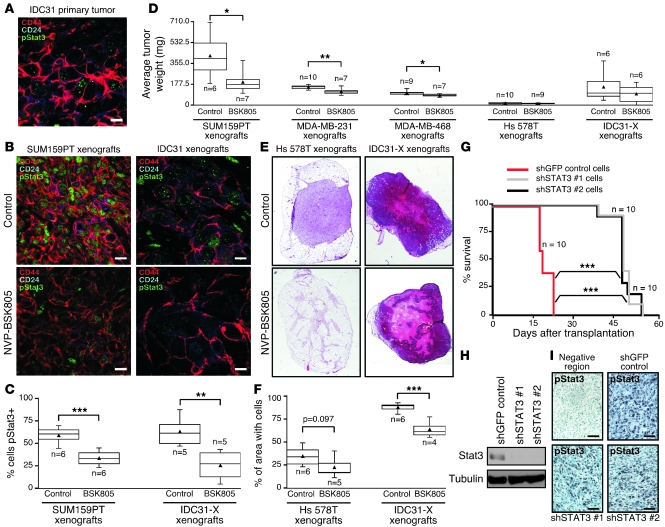
To determine whether JAK2 activity is required for breast tumor growth, we performed xenograft studies using the SUM159PT, MDA-MB-468, MDA-MB-231, and Hs 578T basal-like breast cancer cell lines (selected based on their tumorigenicity in mice and in vitro response to JAK inhibitors) and patient-derived primary human breast tumors in NOD/scid and NOD/scid/Il2rγ–/– mice. One of the primary tumors (IDC31) contained pStat3+ CD44+CD24– breast cancer cells, whereas 2 others (PE18 and IDC50) were mainly composed of pStat3– CD24+ cells (Figure 4A, Supplemental Figure 4A, and data not shown), a pattern that was reproduced in the xenografts derived from them (Figure 4B and Supplemental Figure 4B). We used the compound NVP-BSK805, a JAK2 inhibitor developed by Novartis (17) that, like the commercial JAK inhibitor we used earlier, is highly effective against basal-like breast cancer cells (Supplemental Figure 4, C–E), for these in vivo experiments since the commercial JAK inhibitor is not suitable for this type of study. Xenografts were allowed to grow to palpable size before starting treatment in order to test the efficacy of the inhibitor on established tumors. This treatment regimen would more closely resemble clinical treatment situations than would starting treatment before tumors are palpable. We observed a decrease in the number of pStat3+ cells in xenografts with NVP-BSK805 treatment (Figure 4, B and C), and NVP-BSK805 treatment significantly reduced tumor weights in most xenografts (Figure 4D). Notably, tumors that did not display statistically significant reduction in tumor weights (xenografts from Hs 578T cell line and IDC31 primary tumor) nonetheless responded with significant reduction in their cellularity (Figure 4, E and F), corroborating the trend toward smaller tumors with the inhibitor. Importantly, the effect of NVP-BSK805 treatment was only evident in xenografts derived from pStat3+ (mostly CD44+CD24–) (Figure 4, B–F) but not pStat3– (mostly CD24+) (Supplemental Figure 4, B, F, and G) primary tumors. In addition to the reduction of tumor weights and cellularity, we also observed reduced leukocyte infiltration and angiogenesis in mice treated with NVP-BSK805, which may reflect the inhibition of pStat3 in these cells or that of tumor-promoting paracrine epithelial-stromal and stromal-stromal cell interactions (Supplemental Figure 4H).
Figure 4. Importance of the IL-6/JAK2/Stat3 pathway in tumor growth.
(A) Representative immunofluorescence staining patterns for CD44, CD24, and pStat3 in ER–PR–HER2+ inflammatory breast carcinoma (IDC31). Scale bar: 10 microns. (B) Representative immunofluorescence staining patterns for CD44, CD24, and pStat3 in SUM159PT and IDC31 xenografts. Scale bars: 10 microns. (C) Box plots showing percentage of pStat3+ cells in SUM159PT and IDC31 mouse xenograft-derived (IDC31-X) xenografts by immunofluorescence and immunohistochemistry, respectively (counting 2–6 fields per sample). (D) Box plots showing xenograft tumor weights 34, 28, 28, 40, or 70 days after injecting SUM159PT, MDA-MB-231, MDA-MB-468, Hs 578T, or IDC31-X cells, respectively, into 2 fat pads of n mice. Mice were administered daily NVP-BSK805 (2 mg/mouse) or vehicle only (control) for 14, 16, 16, 24, or 24 days, respectively (after tumors reached palpable size), beginning 21, 13, 13, 17, or 47 days after injection, respectively. (E) H&E-stained Hs 578T and IDC31-X xenografts. (F) Box plots showing the percentage of area with cells in Hs 578T and IDC31-X xenografts calculated from whole tumor sections with H&E staining. (G) Kaplan-Meier curves of SUM159PT xenografts expressing STAT3 shRNAs (shSTAT3 #1 and #2) in n mice. (H) Immunoblots with cells used for xenografts in G. Tubulin was used as a loading control. (I) pStat3 immunohistochemistry staining for xenografts in G. Scale bars: 50 microns. Triangles in C, D, and F mark averages. *P < 0.05; **P < 0.01; ***P < 0.001, t test (C, D, and F). ***P < 0.001, log-rank test (comparing each STAT3 shRNA group to the shGFP control group) (G).
To further strengthen the link between JAK2 and Stat3 in basal-like breast cancer cells, we also transplanted mice with SUM159PT cells in which STAT3 had been knocked down using lentivirally delivered shRNAs from the TRC library. Tumors derived from cells with Stat3 knockdown displayed substantial delay in their outgrowth (Figure 4, G and H). Notably, all of the mice in this experiment eventually suffered tumor-related morbidity due to their outgrowth of pStat3+ xenografts (Figure 4I), suggesting strong selective pressure to restore pStat3 activity. Despite these results demonstrating strong requirements for both pStat3 and JAK2 for tumorigenicity, we cannot exclude the possibility that some effects of the JAK2 inhibitor are independent of IL-6 and Stat3. For example, recent data implicate JAK2 in the regulation of histone modification patterns (18, 19). However, based on our results and building upon the findings of others (20–23), the IL-6/JAK2/Stat3 pathway appears to actively and preferentially promote the growth and survival of basal-like breast cancer cells.
Causes and effects of Stat3 activation and its clinical significance.
Because not all basal-like breast cancer cells depended on IL6 despite their high pStat3 and JAK inhibitor sensitivity, and since we found no evidence for the mutational activation of this pathway in these cells (data not shown), we explored potential links between the JAK2/Stat3 pathway and the genes targeted by the 15 basal-like–specific hits by creating a signature network with them using MetaCore (24). Interestingly, this network includes Stat3 as a key downstream transcriptional effector, emphasizing its critical role in CD44+CD24– basal-like breast cancer cells (Figure 5). To test the validity of this network in breast cancer, we treated 3 pStat3+ basal-like breast cancer cell lines with the 4 basal-like–specific hit inhibitors that we used in earlier experiments in this study (and the commercial JAK inhibitor, NQO1 inhibitor, and vehicle alone as controls) and analyzed pStat3 levels at an early time point (6 hours) after treatment, prior to signs of apoptosis and cell cycle arrest. The PTGIS and HAS1 inhibitors decreased pStat3 relative to total Stat3 in the 3 basal-like cell lines (Figure 6, A and B), and, along with the CXCR2 and PFKFB3 inhibitors, decreased Stat3 transcriptional activity in Hs 578T cells in a statistically significant manner (Figure 6C). Observation of a more significant inhibition of Stat3 transcriptional activity than decrease in pStat3 levels is likely due to the higher sensitivity of the transcriptional activity assay relative to immunoblot analysis. The striking lack of effect of the NQO1 inhibitor on pStat3 activity seems to indicate that NQO1 is not important in the IL-6/JAK2/Stat3 pathway in basal-like breast cells, a finding consistent with the absence of a strong preference of the inhibitor on basal-like breast cell viability (Figure 2D). In fact, an NQO1-targeting shRNA was a luminal hit in phase 1 of the shRNA screen, so perhaps this gene affects some pathway more specific to luminal breast cancer cells.
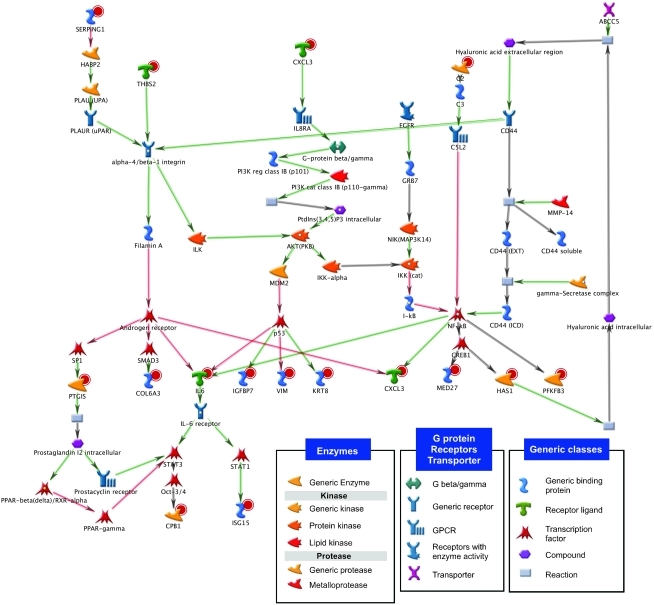
Figure 5. Network of genes targeted by the basal-like–specific screening hits.
The MetaCore software platform was used to construct the diagram shown. The 15 genes targeted by the basal-like–specific hits from the shRNA screen and links among them based on published findings are included in the network. Genes targeted by screening hits are marked with red concentric circles. Red and green arrows indicate inhibitory and activating interactions, respectively. Gray arrows indicate interactions between substrate and reaction or reaction and product or indicate unspecified effects.
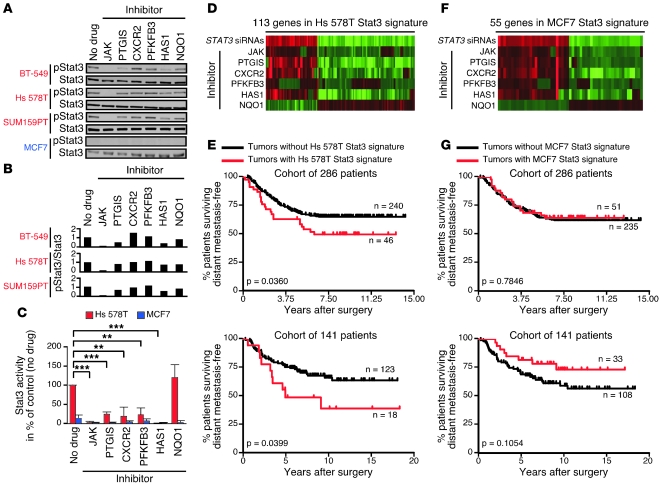
Figure 6. Regulation of the JAK2/Stat3 pathway in basal-like breast cancer cells and its clinical significance in primary human breast tumors.
(A) Immunoblots of pStat3 in basal-like breast cancer cell lines (and MCF7, in which pStat3 is undetectable by immunoblot) after 6-hour inhibitor treatments. Concentrations used were 2 μM JAK, 1 mM PTGIS, 1.5 μM CXCR2, 10 μM PFKFB3, 1 mM HAS1, and 50 μM NQO1 inhibitor. Stat3 was used as a loading control. (B) Quantitation of basal-like immunoblots in A. pStat3/Stat3 values represent ratios to control (no drug). (C) Luciferase assay results using Hs 578T and MCF7 cells treated with inhibitors for 2 days. Error bars show SD of triplicates. **P < 0.01; ***P < 0.001, t test. (D and F) Fold changes in tag counts for genes in our Hs 578T and MCF7 Stat3 signatures in SAGE-Seq libraries prepared from Hs 578T or MCF7 cells treated for 2 days with STAT3 siRNAs (versus nontargeting siRNAs) or inhibitors (versus no drug). Red and green indicate high and low fold changes, respectively. Each gene in the signatures had |fold change| > 2 with STAT3 siRNAs and in the same direction with at least 4 inhibitors (not NQO1). (E and G) Significant association of the presence of the Hs 578T Stat3 signature with shorter distant metastasis-free survival in 2 cohorts of breast cancer patients and lack of such an association for the MCF7 Stat3 signature. Kaplan-Meier curves (for n patients with and without each signature) and their corresponding log-rank test P values are shown.
To further investigate the link between Stat3 transcriptional activity and other signaling pathways, we determined the gene expression profiles of Hs 578T basal-like and MCF7 luminal breast cancer cells treated with the PTGIS, HAS1, CXCR2, PFKFB3, commercial JAK, and NQO1 inhibitors, inhibitor treatment vehicle alone, STAT3 siRNAs, or nontargeting siRNAs (Supplemental Figure 5A). For each cell line, among genes significantly regulated by STAT3 siRNAs (|fold change| > 2 and |z test score| > 2.33), we identified those similarly affected (same direction fold change) by at least 4 inhibitors but not by the NQO1 inhibitor (Figure 6, D and F, and Supplemental Tables 6 and 7). We called the 2 sets of genes identified in this way our Hs 578T and MCF7 Stat3 signatures. The signature definition process was designed to select genes regulated specifically by activated Stat3 (by requiring genes to be regulated by STAT3 siRNAs and also by several of the inhibitors that regulate pStat3 levels but not by the NQO1 inhibitor, which does not affect pStat3 levels). In Hs 578T cells, 968 genes were significantly regulated by STAT3 siRNAs, and 113 of these were identified as the Stat3 signature; in MCF7 cells, 556 genes were significantly regulated by STAT3 siRNAs, and 55 of these were identified as the Stat3 signature. Interestingly, the Hs 578T but not the MCF7 Stat3 signature was enriched in genes involved in development (Supplemental Figure 5B), such as IRX3, HOXA10, and FGF1. We repeated the expression profiling experiment in SUM159PT basal-like breast cancer cells and found some overlap between the genes regulated by STAT3 siRNAs in this cell line and in Hs 578T cells but very little overlap between the genes identified as a SUM159PT Stat3 signature using the same method used to identify the Hs 578T and MCF7 signatures and the Hs 578T Stat3 signature (data not shown), a finding consistent with prior studies demonstrating strong cell-type specificity of STAT3 transcriptional responses (25). Interestingly, we did find less overlap between the genes significantly regulated by STAT3 siRNAs and the Stat3 gene signatures identified in MCF7 and in either Hs 578T or SUM159PT.
To investigate the potential clinical relevance of our Hs 578T Stat3 gene signature and to determine whether, similar to our CD44+CD24– cell gene signature (10), it identifies breast cancer patients with poor clinical outcome, we compared its presence in 2 independent sets of public gene expression data with corresponding clinical outcome information (26–28). In each data set, tumors were considered to have the Stat3 signature if they had average expression values for all genes in the signature downregulated by STAT3 siRNAs above the 60th percentile and average expression values for all genes in the signature upregulated by STAT3 siRNAs below the 40th percentile. We found that the activation of the Stat3 pathway, represented as expression of our Hs 578T Stat3 gene signature, in primary lymph node–negative invasive breast tumors was associated with shorter distant metastasis-free survival at a statistically significant rate (Figure 6E and Supplemental Figure 5C). While this signature was not associated with estrogen receptor (ER) status in a statistically significant way, we observed a trend toward shorter distant metastasis-free survival in the presence of the signature among the groups of ER+ tumors only from each data set (but not among both groups of ER– tumors only, probably due to small sample size) (Supplemental Figure 5D and Supplemental Table 8), indicating that the Hs 578T Stat3 signature is likely clinically relevant in ER+ tumors. We also found that expression of our MCF7 Stat3 signature in the same sets of primary tumors is not associated with shorter distant metastasis-free survival (Figure 6G and Supplemental Figure 5C). The expression of the set of genes significantly regulated by STAT3 siRNAs in Hs 578T cells in primary tumors (defined as average expression values for all genes downregulated by STAT3 siRNAs above the 60th percentile and average expression values for all genes upregulated by STAT3 siRNAs below the 40th percentile) was not associated with shorter distant metastasis-free survival in the 2 public gene expression data sets used (data not shown). These findings are consistent with the preferential activation of Stat3 in stem cell–like CD44+CD24– breast cancer cells in primary tumors, as we previously associated the presence of more of these cells with increased risk of distant metastasis in the same patient cohorts (9, 10). Furthermore, the convergence to Stat3 of multiple other signaling pathways on which these cells depend indicates that the activation of Stat3 is centrally important for the maintenance of CD44+CD24– stem cell–like breast cancer cells.
Specific activation of Stat3 in CD44+CD24– breast cancer cells in primary human tumors.
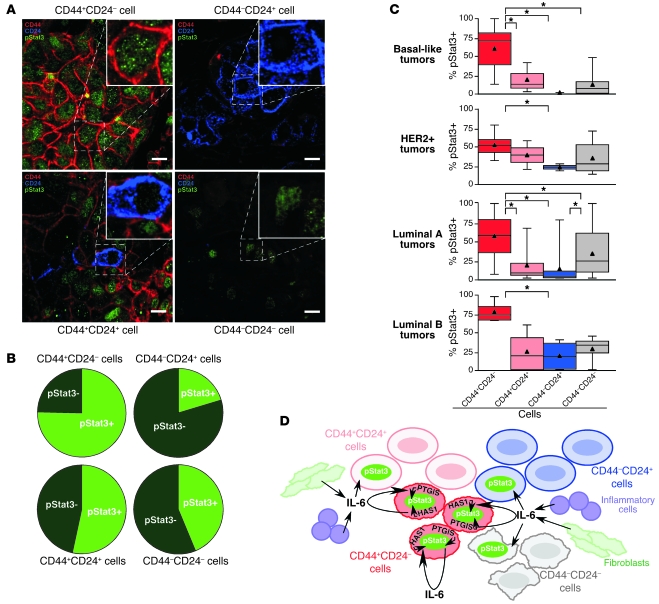
To investigate the specificity of Stat3 activation in primary human breast tumors in further detail, we performed triple immunofluorescence analysis of CD44, CD24, and pStat3 expression in 170 invasive ductal breast carcinomas (Figure 7A), most of which were on a tissue microarray. We have previously analyzed slides from the same tissue microarray for the expression of multiple CD44+CD24– and CD44–CD24+ cell–specific markers and also for cytokeratins (11); thus, we were able to differentiate the tumor epithelial and stromal cells with high confidence. CD24 is mainly expressed in tumor epithelial cells and a minor subset of leukocytes, whereas the expression of CD44 is more promiscuous and can also be detected in various stromal cells. Based on these analyses, we found that, statistically, CD44+CD24– breast cancer cells are significantly more likely than other breast cancer cell types to contain pStat3, whereas CD44–CD24+ breast cancer cells are the least frequently pStat3+ (Figure 7B). This was true within each breast tumor subtype examined (Figure 7C), even though the frequency of the 4 cell types defined based on CD44 and CD24 staining patterns varied according to subtype, with basal-like tumors containing the highest frequency of CD44+CD24– cells (Supplemental Figure 6).
Figure 7. Specific activation of Stat3 in CD44+CD24– cells in primary human breast tumors.
(A) Representative immunofluorescence staining patterns for CD44, CD24, and pStat3 in primary human breast tumor samples in each of the 4 indicated cell types. pStat3 is primarily in CD44+CD24– cells. Scale bars: 10 microns. (B) Frequency of pStat3 positivity in the indicated breast cancer cell types in 170 samples analyzed by triple immunofluorescence. (C) Box plots showing cells of 4 types positive for pStat3 based on triple immunofluorescence in 4 breast tumor subtypes. Triangles mark averages. *P < 0.001. (D) Model of Stat3 activation in breast cancer. CD44+CD24– stem cell–like cancer cells have constitutive pStat3 due to expression of genes such as IL6, PTGIS, and HAS1. Other cancer cells are sometimes pStat3+ due to uptake of IL-6 secreted by CD44+CD24– cancer cells, fibroblasts, and inflammatory cells.
Discussion
Breast cancer cells with a CD44+CD24– phenotype and stem cell–like features have been proposed to be resistant to cancer therapies (3–5, 7, 8), suggesting that their effective elimination may require the identification of signaling pathways on which they are dependent. Here we demonstrate that using unbiased screening strategies, it is possible to identify such genes and pathways and that therapeutic inhibition of these can be used for the effective elimination of these cells.
Technical considerations forced us to conduct the shRNA screen reported here on breast cancer cell lines resembling CD44+CD24– and CD44–CD24+ primary tumor cells rather than primary tumor cell cultures. However, the results were validated in primary human breast tumors as well as in xenografts derived from them. Although the genetic differences among the breast cancer cell lines may have influenced the shRNA screen results, as signaling pathways may function differently in different contexts, we previously showed that, even within primary human tumors, CD44+CD24– and CD44–CD24+ breast cancer cells are highly genetically heterogeneous despite having consistent gene expression patterns (14). Thus, our screen demonstrated that it is possible to identify signaling pathways active in CD44+CD24– breast cancer cells, the targeting of which can be used for the elimination of these cells despite their underlying genetic heterogeneity. Therefore, we believe that our findings are directly applicable to primary patient tumors.
We mainly focused on the 15 genes targeted by the basal-like–specific hits in our follow-up studies since they represent promising therapeutic targets for CD44+CD24– breast cancer cells, which we observed at the highest frequency in basal-like breast tumors, a tumor subtype with no effective targeted therapy. Interestingly, many of these 15 genes encode secreted and extracellular matrix–related proteins, suggesting that CD44+CD24– cells may produce and depend on their own niche. In addition, many of them have been associated with stem cell maintenance, cancer cell survival, or poor breast cancer prognosis. In particular, and most relevant to the bulk of our follow-up studies, IL6 is implicated in the maintenance of stem cell–like cancer cells (29) and progenitor-enriched mammosphere formation (30), and its downstream effector Stat3 enforces the undifferentiated state in murine embryonic stem cells (31). Furthermore, high IL-6 levels have been associated with poor clinical outcome in breast cancer patients (32). Recent studies have demonstrated a critical role for IL-6 and Stat3 for the survival of intestinal epithelial cells in colitis-associated cancer (29, 33, 34), and a similar link between IL-6–mediated inflammation and cellular transformation was also observed in mammary epithelial cells (35).
Two other genes encoded by basal-like–specific hits besides IL6 that also appear to be of prime importance in CD44+CD24– breast cancer cells are PTGIS and HAS1. PTGIS, encoding prostacyclin synthase, is related to PTGES, a gene required for hematopoietic stem cell maintenance (36). Furthermore, the use of NSAIDs has been associated with decreased breast cancer risk (37) and with improved clinical outcome in breast cancer patients largely due to decreased risk of distant metastasis (38). NSAIDs inhibit the production of prostaglandins, which is catalyzed by PTGIS and PTGES. Thus, inhibition of the prostaglandin pathway may be beneficial in breast cancer treatment by reducing the number of CD44+CD24– stem cell–like cells. HAS1 encodes hyaluronan synthase 1, which catalyzes the production of hyaluronic acid, a ligand for CD44, and SPP1 (encoding osteopontin and targeted by a basal-like hit) regulates HAS1 and also binds CD44 (39), supporting the idea that CD44 promotes the viability of stem cell–like breast cancer cells in addition to marking them (40). Correlating with this, CD44 was shown to be required for the survival of leukemia cancer stem cells (41), and hyaluronic acid synthases have been linked to invasiveness and metastatic behavior in multiple cancer types (42). Surprisingly, CD44 shRNAs were included in our shRNA screen but did not show up as hits. There are many possible explanations for why a HAS1 shRNA was a hit and CD44 shRNAs were not. One possibility is that CD44 is indeed required for cell growth or proliferation but was not represented by a hit due to a technical reason such as inefficient targeting of the gene by the CD44 shRNAs used in the screen. Another is that HAS1 may be acting functionally independently of CD44 in its cell viability role, as there are other receptors for hyaluronic acid.
Additional genes encoded by basal-like–specific hits of interest include CXCL3, ISG15, PFKFB3, and IGFBP7. CXCR1, a homolog of the CXCL3 receptor gene CXCR2, is implicated in breast stem cell survival (43), and CXCL3 expression is higher in more aggressive breast tumors (44). Similarly, ISG15 is associated with poor prognosis in breast cancer patients (45). PFKFB3 encodes a glycolytic enzyme, and its higher expression and specificity for basal-like breast cancer cells correlate with our findings that basal mammary epithelial cells have higher glycolytic activity than luminal ones (46). Inhibition of PFKFB3 has been able to reduce tumor growth in preclinical models (47). Furthermore, PFKFB3 is one of the genes in the CD44+CD24– cell gene signature we previously linked to increased risk of distant metastasis and poor clinical outcome in breast cancer patients (10). IGFBP7 is a target of the TGF-β pathway (48), which we showed is specifically activated in CD44+CD24– breast cancer cells (10), and the phenotype of the Igfbp7–/– mouse suggests that this gene may be critical in the maintenance of mammary epithelial stem cells (A. Burger, personal communication). Thus, the identity of the basal-like–specific hits is consistent with CD44+CD24– cells displaying more stem cell–like features, as several of the signaling pathways targeted by these hits have been shown to be required for the survival of stem cells in breast or other organ types and are potential therapeutic targets.
Based on our follow-up studies, we demonstrated that the 15 basal-like–specific hits form a compact network with Stat3 as a key downstream transcriptional mediator. Inhibition of genes that encode proteins that regulate Stat3 in this network would be predicted to downregulate Stat3 activity. For instance, HAS1 is linked to Stat3 in Figure 5 via the production of hyaluronic acid, which then binds to hyaluronic acid receptors (e.g., CD44) and activates downstream signaling pathways leading to the activation of Stat3. Experimental validation of this network showed that inhibition of PTGIS, CXCR2, HAS1, and PFKFB3 decreases pStat3 levels and transcriptional activity. These findings correlate with prior reports describing a link between the enzymatic activities of PTGIS and HAS1 and Stat3 signaling (49, 50). Based on our comprehensive gene expression profiling of basal-like breast cancer cells treated with STAT3 siRNAs and the various inhibitors, we also identified a Stat3 gene signature commonly affected by them and demonstrated that this is associated with increased risk of distant metastasis in breast cancer patients. These results emphasize the central importance of Stat3 in CD44+CD24– stem cell–like breast cancer cells and the clinical relevance of this cell type. This Stat3 signature is not simply associated with or significant in ER- (basal-like) tumors, which is in line with our findings that tumors of all different types can contain a proportion of CD44+CD24– cells. We suppose that tumors containing more Stat3 activation, either due to the presence of many CD44+CD24– cells or to paracrine activation of other cell types by a few of these cells, are more aggressive.
The JAK2/Stat3 pathway has been intensely investigated in breast and other cancer types (51). Constitutive activation of Stat3 signaling due to mutations in JAK1 and JAK2 has been demonstrated in various hematopoietic malignancies (52), and JAK inhibitors are already in different phases of clinical trials for the treatment of these diseases (53). Mutations in the gene encoding gp130, a receptor upstream of JAKs that mediates the action of several cytokines including IL-6, have also been reported in inflammatory hepatocellular tumors (54). In breast cancer, mutations in JAKs have not been described yet, although a recent whole-genome sequencing study of a basal-like breast tumor did identify a JAK2 mutation with unknown functional significance (55). Thus, mutational activation of the JAK2/Stat3 pathway is unlikely to be responsible for its frequent activation in breast cancer. Instead, we hypothesize that CD44+CD24– and even CD44+CD24+ breast cancer cells have high pStat3 due to their expression of genes that increase it, such as IL6, PTGIS, and HAS1, activating an autocrine loop, whereas some CD44–CD24+ and CD44–CD24– breast cancer cells are pStat3+ due to their uptake of IL-6 (or other cytokines) secreted by neighboring CD44+ cells and stromal inflammatory cells and fibroblasts (ref. 56 and Figure 7D). We were not able to derive xenografts from primary breast tumors that contain pStat3+ CD44–CD24+ breast cancer cells; thus, at this time, we cannot determine if therapeutic responses to JAK2 inhibitor treatment correlate with the presence of pStat3 irrespective of the presence of a CD44+ stem cell–like phenotype. Thus, therapeutic inhibition of JAK2/Stat3 signaling might be effective not only in basal-like breast tumors highly enriched in CD44+CD24– breast cancer cells, but also in other tumor subtypes that contain pStat3+ breast cancer cells. Furthermore, we observed a more pronounced effect of the JAK2 inhibitor on tumor cell growth in vivo than in cell culture, potentially due to its ability to interrupt tumor-promoting paracrine epithelial-stromal and stromal-stromal cell interactions important for angiogenesis. Systemic inhibition of the JAK pathway appears to be nontoxic, as several JAK inhibitors are already in clinical trials and have been well tolerated with minimal side effects (53).
In summary, we identified multiple signaling pathways that are specifically required for the viability of CD44+CD24– breast cancer cells and regulation of Stat3 activation in those cells, which are highly represented in basal-like breast tumors. Inhibition of these pathways is a promising strategy for targeting these stem cell–like breast cancer cells in all tumors that contain them. This type of therapy may be effective in conjunction with other treatments designed to eliminate other breast cancer cell types, and such a combined treatment strategy may also help prevent therapeutic resistance and limit side effects of cancer treatment. Because inhibitors against the IL-6/JAK2/Stat3 and CXCL3/CXCR2 pathways are already in clinical trials for other indications (57, 58), our findings may be rapidly translated into breast cancer treatments.
Methods
Cells and tissue specimens.
Tumor samples were collected using protocols approved by the Institutional Review Board at Brigham and Women’s Hospital. Cell lines were obtained from ATCC (except for SUM159PT, which was obtained from S. Ethier, University of Michigan, Ann Arbor, Michigan, USA) and cultured according to their recommendations. Cells were always cultured at 37°C with 5% CO2 in the medium used for the shRNA screen. Cell viability was always measured using CellTiter-Glo (Promega).
shRNA screen and validation.
For screening, cells were infected in duplicate with and without puromycin in 8 μg/ml polybrene (H9268; Sigma-Aldrich) 1 day after plating and assessed 6 days later for viability. Specific growth, infection, and selection conditions were used for each cell line (Supplemental Table 9). Quantitative methylation-specific PCR (qMSP) was performed as previously described (9). Inhibitors included JAK inhibitor I (420099; Calbiochem), tranylcypromine (inhibits PTGIS) (59) (P8511; Sigma-Aldrich), 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-1 (inhibits PFKFB3) (47) (provided by J. Chesney, University of Louisville, Louisville, Kentucky, USA), N-(2-Hydroxy-4-nitrophenyl)-N′-(2-bromophenyl)urea (inhibits CXCR2) (60) (559405; Calbiochem), 4-methylumbelliferone (inhibits HAS1) (40) (M1381; Sigma-Aldrich), and 1,4-dimethoxy-2-methylnaphthalene (inhibits NQO1) (61) (provided by D. Criddle, University of Liverpool, Liverpool, United Kingdom). siGENOME SMARTpools and Non-Targeting siRNA #2 were introduced into cells at 0.1 μM using DharmaFECT 1 (Dharmacon). Sequences of the siRNAs in the SMARTpools are listed in Supplemental Table 10. Cells were assayed for caspase activation using the Apo-ONE Homogeneous Caspase-3/7 Assay (Promega) and for cell cycle by PI (propidium iodide) staining. More details on all shRNA screen and validation procedures are included in the Supplemental Data.
IL6 shRNA validation, Stat3 activity, immunofluorescence, immunohistochemistry, immunoblots, and flow cytometry.
Sequences of the IL6 shRNAs and qRT-PCR primers used are listed in Supplemental Table 10. qRT-PCR was performed as previously described (9). IL-6 levels measured by ELISA (EH2IL65; Thermo Scientific) were adjusted for cell number. Recombinant human IL-6 (rhIL-6) (eBioscience 14-8069-62) was added every 24 hours for 3 days. Antibodies for immunoblots, immunohistochemistry, and immunofluorescence were pStat3 (9131; Cell Signaling), Stat3 (sc-482; Santa Cruz Biotechnology Inc.), pStat1 (33-3400; Zymed), pStat5 (9359; Cell Signaling), tubulin (T-5168; Sigma-Aldrich), phosphoserine-727–Stat3 (pserStat3) (62), CD24 (clone SN3b; Thermo Scientific), and CD44 (clone 156-3C11; Thermo Scientific). Antibodies for flow cytometry were PE-CD44 (555479; BD Biosciences) and Alexa Fluor 647–CD24 (311110; BioLegend). For Stat3 transcriptional activity assays, the m67-luc reporter construct (provided by J. Bromberg, Memorial Sloan-Kettering Cancer Center, New York, New York, USA) and pRL-TK (Promega) were introduced into cells with Lipofectamine 2000 (Invitrogen). More details on all IL6 shRNA validation, Stat3 activity, immunofluorescence, immunohistochemistry, immunoblots, and flow cytometry procedures are included in the Supplemental Data.
Xenograft assays.
For xenograft assays, cells were injected with 50% Matrigel (354234; BD Biosciences) in DMEM/F12 or Medium 171 (except for IDC50-X cells, which were injected with 3% FBS and 4 mg/ml collagen gel in Medium 171). Animal experiments were conducted following protocols no. 04330 and no. 04089 approved by the Harvard Medical School Animal Care and Use Committee. Sequences of the STAT3 shRNAs used are listed in Supplemental Table 10. More details on these experiments, including how primary tumor samples were processed prior to xenograft studies, are in the Supplemental Data.
Gene expression profiling and computational and statistical data analyses.
Details of SAGE-Seq (serial analysis of gene expression combined with the Illumina/Solexa 1G platform) implementation and all clustering, clinical outcome, and gene ontology analyses are included in the Supplemental Data. Statistics used in our studies included 2-tailed, 2-sample equal-variance Student’s t tests and generalized linear model score tests adjusted for multiple comparisons. For t test, log-rank test, and χ2 test, P < 0.05 was considered statistically significant. For score tests, P < 0.001 was considered statistically significant. The SAGE-Seq data reported in this paper have been deposited in the SAGE Genie database ( http://cgap.nci.nih.gov/SAGE) and in NCBI GEO (accession number GSE22917).
Supplementary Material
Acknowledgments
We thank the members of the Broad RNAi Platform, the DFCI RNAi Screening Facility, and the Wistar Institute who provided technical assistance with this work, and we thank A. Partridge and J. Bahen for their help with collecting tissue samples. We also thank M. Brown, J. Brugge, and D. Pellman and members of the Polyak lab for their critical reading of the manuscript and advice during the course of this project. We thank C. Pissot-Soldermann, F. Baffert, and T. Radimerski (Novartis Institutes for BioMedical Research, Basel, Switzerland) for providing BSK805 compound and advice on its use. This work was supported by Novartis Pharmaceuticals Inc. (to K. Polyak), the National Cancer Institute (P50 CA89383-07 to K. Polyak and A.L. Richardson; NS50830 and MH079826 to D.A. Frank; and R33 CA12862 to W.C. Hahn), the American Cancer Society (RSG-05-154-01-MGO to K. Polyak), US Army Congressionally Directed Medical Research Program (W81XWH-07-1-04-8 to W.C. Hahn; BC087579 to A. Marusyk), the Breast Cancer Research Foundation (to K. Polyak, A.L. Richardson, and D.A. Frank), the Snyder Medical Foundation (to W.C. Hahn), the Susan G. Komen Foundation (to D.A. Frank, M. Shipitsin, and R. Maruyama), the Terri Brodeur Foundation (to S.A. Choudhury), and the Cellex Foundation (to V. Almendro). We thank the late Candice Maxwell Bahen for her donation.
Footnotes
Conflict of interest: Kornelia Polyak and William C. Hahn receive research support from Novartis Oncology and are consultants to Novartis Oncology. Kornelia Polyak serves on the Scientific Advisory Board of Metamark Genetics Inc. and Theracrine Inc. and holds AVEO Pharmaceuticals Inc. stocks.
Citation for this article: J Clin Invest. 2011;121(7):2723–2735. doi:10.1172/JCI44745.
References
- 1.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 2.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28(25):4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120(1):41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 9.Bloushtain-Qimron N, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105(37):14076–14081. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K. Heterogeneity for stem cell-related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res. 2010;16(3):876–887. doi: 10.1158/1078-0432.CCR-09-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honeth G, et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider BP, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14(24):8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 14.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120(2):636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3(9):715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 16.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baffert F, et al. Potent and selective inhibition of polycythemia by the quinoxaline JAK2 inhibitor NVP-BSK805. Mol Cancer Ther. 2010;9(7):1945–1955. doi: 10.1158/1535-7163.MCT-10-0053. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19(2):283–294. doi: 10.1016/j.ccr.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths DS, et al. LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat Cell Biol. 2011;13(1):13–21. doi: 10.1038/ncb2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Shaw PE. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J Biol Chem. 2002;277(20):17397–17405. doi: 10.1074/jbc.M109962200. [DOI] [PubMed] [Google Scholar]
- 21.Berishaj M, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9(3):R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker SR, et al. Reciprocal effects of STAT5 and STAT3 in breast cancer. Mol Cancer Res. 2009;7(6):966–976. doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 23.Hedvat M, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16(6):487–497. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolsky Y, Kirillov E, Zuev R, Rakhmatulin E, Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated “knowledge-based” platform. Methods Mol Biol. 2009;563:177–196. doi: 10.1007/978-1-60761-175-2_10. [DOI] [PubMed] [Google Scholar]
- 25.Devarajan E, Huang S. STAT3 as a central regulator of tumor metastases. Curr Mol Med. 2009;9(5):626–633. doi: 10.2174/156652409788488720. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 27.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 28.Chang HY, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15(2):79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117(12):3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda T, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18(15):4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knupfer H, Preiss R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res Treat. 2007;102(2):129–135. doi: 10.1007/s10549-006-9328-3. [DOI] [PubMed] [Google Scholar]
- 33.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15(2):91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6(2):130–140. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- 38.Holmes MD, Chen WY, Li L, Hertzmark E, Spiegelman D, Hankinson SE. Aspirin intake and survival after breast cancer. J Clin Oncol. 2010;28(9):1467–1472. doi: 10.1200/JCO.2009.22.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wai PY, Kuo PC. Osteopontin: regulation in tumor metastasis. Cancer Metastasis Rev. 2008;27(1):103–118. doi: 10.1007/s10555-007-9104-9. [DOI] [PubMed] [Google Scholar]
- 40.Götte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66(21):10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 41.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 42.Adamia S, Maxwell CA, Pilarski LM. Hyaluronan and hyaluronan synthases: potential therapeutic targets in cancer. Curr Drug Targets Cardiovasc Haematol Disord. 2005;5(1):3–14. doi: 10.2174/1568006053005056. [DOI] [PubMed] [Google Scholar]
- 43.Ginestier C, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010;120(2):485–497. doi: 10.1172/JCI39397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bieche I, et al. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr Relat Cancer. 2007;14(4):1039–1052. doi: 10.1677/erc.1.01301. [DOI] [PubMed] [Google Scholar]
- 45.Bektas N, et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008;10(4):R58. doi: 10.1186/bcr2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu M, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13(5):394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clem B, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol Cancer Ther. 2008;7(1):110–120. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 48.Pen A, Moreno MJ, Durocher Y, Deb-Rinker P, Stanimirovic DB. Glioblastoma-secreted factors induce IGFBP7 and angiogenesis by modulating Smad-2-dependent TGF-beta signaling. Oncogene. 2008;27(54):6834–6844. doi: 10.1038/onc.2008.287. [DOI] [PubMed] [Google Scholar]
- 49.Lo RK, Wise H, Wong YH. Prostacyclin receptor induces STAT1 and STAT3 phosphorylations in human erythroleukemia cells: a mechanism requiring PTX-insensitive G proteins, ERK and JNK. Cell Signal. 2006;18(3):307–317. doi: 10.1016/j.cellsig.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 50.Bourguignon LY, Peyrollier K, Xia W, Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem. 2008;283(25):17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10(12):1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vainchenker W, Dusa A, Constantinescu SN. JAKs in pathology: role of Janus kinases in hematopoietic malignancies and immunodeficiencies. Semin Cell Dev Biol. 2008;19(4):385–393. doi: 10.1016/j.semcdb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Pardanani A. JAK2 inhibitor therapy in myeloproliferative disorders: rationale, preclinical studies and ongoing clinical trials. Leukemia. 2008;22(1):23–30. doi: 10.1038/sj.leu.2404948. [DOI] [PubMed] [Google Scholar]
- 54.Rebouissou S, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457(7226):200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Studebaker AW, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68(21):9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- 57.Nakahara H, Nishimoto N. Anti-interleukin-6 receptor antibody therapy in rheumatic diseases. Endocr Metab Immune Disord Drug Targets. 2006;6(4):373–381. doi: 10.2174/187153006779025694. [DOI] [PubMed] [Google Scholar]
- 58.Chapman RW, Phillips JE, Hipkin RW, Curran AK, Lundell D, Fine JS. CXCR2 antagonists for the treatment of pulmonary disease. Pharmacol Ther. 2009;121(1):55–68. doi: 10.1016/j.pharmthera.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Wada M, et al. Purification and characterization of recombinant human prostacyclin synthase. J Biochem. 2004;135(4):455–463. doi: 10.1093/jb/mvh059. [DOI] [PubMed] [Google Scholar]
- 60.Bakshi P, Margenthaler E, Laporte V, Crawford F, Mullan M. Novel role of CXCR2 in regulation of gamma-secretase activity. ACS Chem Biol. 2008;3(12):777–789. doi: 10.1021/cb800167a. [DOI] [PubMed] [Google Scholar]
- 61.Criddle DN, et al. Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. J Biol Chem. 2006;281(52):40485–40492. doi: 10.1074/jbc.M607704200. [DOI] [PubMed] [Google Scholar]
- 62.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100(12):3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.