Abstract
The introduction of multilocus sequence typing (MLST) in infectious disease research has allowed standardized typing of bacterial clones. Through multiple markers around the genome, it is possible to determine the sequence type (ST) of bacterial isolates to establish the population structure of a species. For the periodontal pathogen, Porphyromonas gingivalis, the MLST scheme has been established at www.pubmlst.org/pgingivalis, and data from the database indicate a high degree of genetic diversity and a weakly clonal population structure comparable with Neisseria menigitidis. The major fimbriae (FimA) have been held responsible for the adhesive properties of P. gingivalis and represent an important virulence factor. The fimA genotyping method (PCR based) indicate that fimA genotype II, IV and Ib are associated with diseased sites in periodontitis and tissue specimens from cardiovascular disease. fimA genotyping of the isolates in the MLST database supports the association of genotypes II and IV with periodontitis. As a result of multiple positive PCR reactions in the fimA genotyping, sequencing of the fimA gene revealed only minor nucleotide variation between isolates of the same and different genotypes, suggesting that the method should be redesigned or re-evaluated. Results from several investigations indicate a higher intraindividual heterogeneity of P. gingivalis than found earlier. Detection of multiple STs from one site in several patients with “refractory” periodontitis, showed allelic variation in two housekeeping genes indicating recombination between different clones within the periodontal pocket.
Keywords: periodontitis, Porphyromonas gingivalis, MLST, population genetics, recombination, fimA genotyping
The development in high throughput sequencing of DNA through the last decades and subsequent full genome sequencing of bacterial species accelerated the possibility for microbiologists to investigate genetic variation of bacterial strains by different typing techniques (1, 2). Three classes of genes can be identified in prokaryotes: informational, core housekeeping, and hypervariable genes. Informational genes are essential for central metabolism and are related to transcription, translational, and regulatory pathways, whereas the core housekeeping genes are ubiquitous and coding for other essential proteins. Both classes evolve at a slow rate and are necessary for survival of the cell. On the contrary, the hypervariable genes encode for surface proteins interacting with the host, and for pathogenic species they may be associated with virulence. This class of genes evolve at an accelerated rate compared to the former due to the necessity for the bacterial cell to adapt to variable host conditions (1). Thus, studies of variation in the two different groups of genes (housekeeping and hypervariable) may reflect different aspects of characterization of pathogenic bacterial strains.
Housekeeping genes
Multilocus enzyme electrophoresis, population genetic models
Before nucleotide sequencing was available, genetic variation could be investigated within a bacterial population by multilocus enzyme electrophoresis (MLEE). The technique was based on variation in electrophoretic mobility of the products of several housekeeping genes, where the mobility variants of the enzymes were directly equated with alleles at the corresponding locus. The variants were used to define an allelic profile of each isolate where the acquired electrophoretic types had the main advantage of being directly related to the variation at the chromosome.
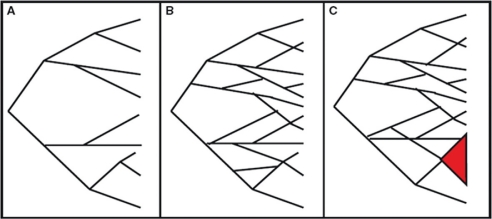
Large-scale MLEE surveys of many pathogens performed in the 1980s suggested that most bacterial populations were highly clonal (Fig. 1) because the majority of isolates seemed to belong to a small number of clusters of closely related genotypes (3, 4). Furthermore, the repeated recovery of identical isolates from diverse geographic origin, many years apart, was also interpreted as a low rate of homologous recombination in natural bacterial populations, also supporting the clonal population structure paradigm (4–6) (Fig. 1). In this model, vertical descent of genetic information from mother to daughter cells occurs, and the changes are only caused by point mutations in the chromosome. Based on the MLEE investigations, the clonal paradigm remained central for the understanding of genetic variation of bacterial populations until the beginning of the 1990s. However, as early as in 1986, results from a MLEE study of isolates of Neisseria meningitidis from subjects with systemic disease and healthy carriers gave clear indications of a substantial rate of recombination within isolates of the species (7).
Fig. 1.
Models of genetic population structures of prokaryotes (5) (figure adapted from Kilian et al. (10)). The population structure in A is fully clonal at all levels where no recombination occurs either between isolates in the same, or in different branches of the tree (strong linkage disequilibrium between alleles). B shows a panmictic population structure with recombination at all levels in the population with random association of alleles. C illustrates an epidemic population structure consisting of a panmictic population with a clonal complex (3) of closely related isolates (marked red).
Horizontal exchange of genetic material in prokaryotes can occur through several mechanisms: Transduction refers to horizontal transfer of DNA among bacteria that have the common property of being infected by a specific bacterial virus; transformation refers to bacterial uptake of DNA from the environment, whereas conjugation involves a cell to cell contact-dependent horizontal gene transfer. When recombinational events occur to a large extent with rearrangement of the bacterial chromosome, in addition to the vertical descent of genetic information, the population structure becomes non-clonal, panmictic or free recombining (Fig. 1). In a non-clonal population structure model, genetic variation is randomly assorted and with no evidence of lineage structure and no congruence of gene trees (5). With the accumulation of MLEE data and later the increasing use of nucleotide sequencing, it became evident that genetic variation at housekeeping loci in many bacterial species accumulated as frequently by homologous recombination as by point mutation (3, 5, 8). It also became evident from these data, in particular from those on N. meningitidis, that closely related clones, designated clonal complexes, could arise from an underlying recombining background population, defining another term, the epidemic population structure (4–6, 9, 10) (Fig. 1). These data and conclusions made the basis for modern population genetics in prokaryotes.
Multilocus sequence typing
The introduction of the multilocus sequence typing (MLST) method in infectious disease research has allowed the identification of bacterial clones in a standardized manner, by making data comparable between laboratories, using high throughput sequencing of fragments of housekeeping genes as determinants. The method derived from MLEE, with use of multiple markers, scattered physically around the genome, requires knowledge of approximately 450 bp fragments of at least seven ubiquitous, core housekeeping genes (1, 11). Gene fragments are amplified by PCR and sequenced for determination of the nucleotide sequence. Thus, the MLST detects allelic variation at multiple housekeeping loci accumulating slowly in the bacterial population. The discriminatory power is achieved by the analysis of the DNA sequence of fragments detecting the degree of genetic diversity within the population of interest. The combination of the different alleles for a selected strain will assign the strain to a certain sequence type (ST) number according to earlier recordings of other STs.
Soon after the introduction of the MLST for N. meningitidis (11), other schemes were developed for several prokaryotic (6) and eukaryotic pathogens such as Candida albicans (12) and Candida glabrata (13). Since then MLST schemes have also been created for numerous bacteria, among them Streptococcus pneumoniae, Campylobacter jejuni, Streptococcus pyogenes, Salmonella spp., Staphylococcus aureus, Haemophilus influenzae and Bacillus cereus (14–20).
MLST data generated throughout the last decade have contributed to both epidemiologic surveillance and fundamental studies of pathogen biology (2, 8, 21). Thus, MLST is a well-established and good method to detect genetic diversity of bacterial pathogens and well suited to study large strain collections of broad origin for long-term molecular epidemiology. Furthermore, the method has provided numerous insights into epidemiology and population genetics of bacterial populations (2). An overview of the MLST schemes for pathogenic organisms can be accessed at several MLST websites existing on the Internet (http://pubmlst.org/databases.shtml), and MLST databases are hosted at several institutions around the world (the University of Oxford, UK (http://pubmlst.org) (22); Imperial College, London, UK (http://www.mlst.net/databases/) (9); the Environmental Resarch Institute, Cork, Ireland, (http://mlst.ucc.ie), at the Pasteur Institute, Paris, France (http://www.pasteur.fr/recherche/genopole/PF8/mlst), at the University of Oslo, Oslo, Norway (http://mlstoslo.uio.no), and at The University of Hong Kong).
The largest MLST database, for Neisseria spp. (http://pubmlst.org/neisseria), contains at date 8,972 distinct allelic profiles or STs, differentiated among 18,584 isolates.
However, recent advances in technology have further improved DNA sequence capability and increased the discriminatory power of sequence-based data beyond the MLST method, in bacterial populations where levels of nucleotide diversity are too low. These improvements have resulted in the detection of single nucleotide polymorphisms, small insertions, deletions, and other detailed genetic variation (2, 23).
The periodontal pathogen Porphyromonas gingivalis
Porphyromonas gingivalis is a small black-pigmented anaerobic rod-shaped bacteria, belonging to the oral microbiota. Most commonly, it can be detected in subgingival biofilms in coaggregation with other species, among them Fusobacterium spp. and Streptococcus gordonii (24, 25). It is one of the main periodontal pathogens involved in the initiation and progression of periodontal diseases and can be detected in patients with various forms of periodontitis (26–30). Furthermore, it belongs to the ‘red complex’ bacteria together with Treponema denticola and Tannerella forsythia recognized as a microbial indicator of progression of chronic periodontitis (31, 32).
For a long time, it was assumed to be related only to diseased periodontal sites because it was not possible to culture from healthy gingival sulci. However, improved techniques have made detection possible also in healthy sites (33–35) as well as in other parts of the oral cavity (lateral part and dorsum of the tongue), located to niches with growth conditions favoring its special requirements (36).
The species harbors a number of virulence factors, e.g. fimbriae, lipopolysaccharide, collagenase, and cysteine proteinases (gingipains) (27, 28, 37–41) and is capable of invading host tissue cells that may protect it from the host's immune system. Furthermore, new investigations have also shown in vitro that the organism can survive intracellularly in a nutritionally rich environment (28, 42, 43) and can be transmitted between different types of host cells such as epithelial, endothelial, and smooth muscle cells (43, 44).
Because of the large pathogenic potential of P. gingivalis, it is referred to as a molecular vampire (41). In addition to being a major periodontal pathogen, it is suggested to be a mediator in the development of cardiovascular disease by spreading from the periodontal pocket to the blood stream (45–49).
The major fimbriae (FimA)
P. gingivalis’ ability to adhere to salivary components, host cells, solid surfaces, and bacterial cells is facilitated by its fimbriae (42, 48–51). These are curly and filamentous appendages arranged peritrichously on the cell surface (50) and are essential for the early establishment of infection by its adhesive properties. The fimbriae have been classified into major and minor types (50–54) and have been extensively studied throughout the last decades. It is constructed from a fimbrillin monomer subunit protein of approximately 43 kDa as the main building block (50), coded from the fimA gene. FimA has been held directly responsible for many of the adhesive properties of the organism because of specifically binding to and activation of various host cells (human epithelial, endothelial, spleen, and peripheral blood monocytes), resulting in the release of cytokines and several adhesion molecules (28, 30, 42). The fimA gene occurs as a single copy in the chromosome (29, 51, 53, 55), and it has been classified into six types based on variation in nucleotide sequence (genotype I, Ib, II, III, IV, and V) (49, 55–58).
Several clinical studies have indicated that the variation of the fimA gene is related to the virulence of different strains of the species. In chronic periodontitis, P. gingivalis isolates with fimA genotypes II, IV, and Ib are significantly more prevalent than isolates with other genotypes (56–61). Studies concerning the pathogenic potential of distinct genotypes also indicate that genotype II is more prevalent in patients suffering from aggressive periodontitis (62). In contrast, isolates with fimA genotype I are most prevalent among P. gingivalis positive healthy adults, followed by genotype III and V (28).
Furthermore, it has been demonstrated that recombinant FimA protein corresponding to fimA genotype II has greater ability to adhere to and invade human epithelial cells than the FimA protein corresponding to other genotypes (63). The pathogenic difference between different genotypes has also been verified in animal models supporting the hypothesis of variable virulence because fimA II, Ib, and IV types are causing stronger infectious symptoms and inflammatory changes than strains harboring genotypes I and III (51, 64–66). Furthermore, mutants where the type I gene was substituted with type II showed an enhanced bacterial adhesion/invasion of epithelial cells and vice versa (67). However, a study of adhesion and invasion abilities of different fimA genotypes of P. gingivalis in KB cells (originally derived from epidermal carcinoma of the mouth) did not indicate any significant difference between different genotypes, suggesting that other characteristics besides the variation of the fimA gene also could be responsible for the observations (10, 51, 68). In addition, Inaba et al. (69) reported a heterogenic virulence among fimA genotype II strains, indicating that variation in pathogenic potentials and invasive efficiency could be related to gingipain activities and correlated to extracellularly secreted gingipains.
Frequently detected fimA type II and IV clones from cardiovascular specimens of P. gingivalis-positive patients also indicate a possible involvement of these in the initiation and progression of cardiovascular disease (70). Recent results (49) also indicate that fimA II, IV, and Ib genotypes may be involved in the formation of aortic aneurysms, and that the major characteristics of aortic tissue with P. gingivalis infection were smooth muscle cell proliferation in association with intense, local inflammation.
Results from studies of periradicular infections also indicate an association between P. gingivalis-specific fimA genotypic clones (types II, IV, and Ib) in primary endodontic infections as well as chronic, apical periodontitis (71, 72).
MLST – P. gingivalis
The genetic diversity among strains of bacteria is a reflection of nucleotide sequence differences in their chromosomal genes. Before 2003, several molecular typing methods (MLEE, random amplified polymorphic DNA fingerprinting, and ribotyping) were used to characterize P. gingivalis strains (73–76). The results of these studies were contradictory when it came to population structure of the species. However, in 2003, MLST was first introduced for the species (77). To test the conclusions from this study which indicated a panmictic population structure of the organism, we investigated 38 isolates from various geographical origins (78). In additional studies (79), the overall results of the MLST analysis represented a total of 84 isolates from human periodontitis with an even broader geographic origin. Due to the limited variation of one selected gene (nah) observed both by Koehler et al. (77) and by Enersen et al. (78), the MLST results presented in Enersen et al. (79) were based on a seven loci scheme without the nah gene. The proposed MLST modification in this paper with exclusion of the nah gene, is an example of how important the selection of the right genes is for keeping the discriminatory power of the method. The modified MLST scheme forms the basis for the MLST website, http://pubmlst.org/pgingivalis, set up in collaboration with Dr. K. Jolly, University of Oxford and professor D. A. Caugant, National Institute of Public Health, Oslo (79).
Population structure
The evaluation of a new molecular technique on isolates that have been studied earlier with other techniques is important for assessing the discriminatory power of the new method and for testing the strength of the conclusions made earlier. In this comparison, the conclusion from Enersen et al. (79) is in accordance with earlier studies (73, 74), indicating that genetic exchange through recombination may not be as extensive in P. gingivalis as suggested by other investigators (76, 77).
Several methods can be used in the analysis of data in population genetic studies. The MLST analysis software S.T.A.R.T. 2 (Sequence Type Analysis and Recombinational Tests) is a single package available at the http://www.pubmlst.org (80). The standard MLST statistics, number of polymorphic sites, average G+C contents, allele frequencies, phylogenetic trees and the calculation of pairwise ratios of non-synonymous to synonymous substitutions (dN/dS) can be calculated with the software, as well as the index of association (78–80).
The index of association, IA, has been used to statistically estimate the degree of association between alleles at different loci using one representative of each ST in a bacterial population (5). Originally, from MLEE data the IA value for Salmonella species was calculated to be 1, a fully clonal species, while Neisseria gonorrhoeae was estimated to be a panmictic species with IA close to 0 (5). Index of association can also be used in MLST calculations (5, 22) (http://pubmlst.org/software/analysis/start/manual/index_of_association.shtml).
For the P. gingivalis isolates in the MLST database, IA is calculated to be 0.3704 (81), indicating a significant linkage disequilibrium pointing to a weakly clonal population structure. The standardized index of association ( ) (82) (Haubold and Hudson, 2000) which is a result of further development of the simple IA calculation, takes into account the number of genes investigated in the MLST and gives n even better estimate of linkage equilibrium than the simple IA value (0.062). Further support for a weakly clonal population structure is verified when comparing the corresponding value estimated for the human pathogen N. meningitidis (0.14) with the value estimated for N. gonorrhoeae (0.005), characterized as a panmictic species undergoing extensive recombination (5).
According to population genetic theory (83), the recovery of strains with identical or similar genotypes sampled in different geographic areas as well as no evidence of intragenic recombination (no mosaic genes) support a significant degree of clonality of the species. It is reasonable to assume that the explanation for the results in our MLST investigations (79) could be the inclusion of a larger strain population with a much broader geographic origin, comparable to the studies by Loos et al. and Menard and Mouton (73, 74), than in the study by Koehler et al. (77).
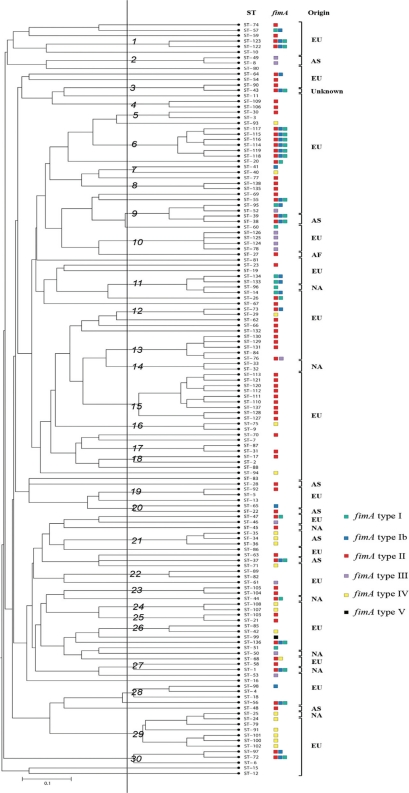
At the moment, there are 138 STs in the MLST database for P. gingivalis including the strains collected from patients with ‘refractory’ periodontitis (79, 81, 84). The UPGMA dendrogram presented in Fig. 2 shows the genetic relationships among all the STs (one representative of each) in the database, and 30 clusters were identified. Though the overall number of isolates of P. gingivalis analyzed by MLST still is small compared to databases of other bacterial pathogens, the dendrogram verifies clusters of identical or closely related STs from diverse geographic origins, which may be sampled years apart. This is an observation that further supports the substantial clonality and a low rate of homologous recombination for the species.
Fig. 2.
Genetic relationships among 138 isolates of P. gingivalis in the MLST database (www.pubmlst.org/pgingivalis). A distance matrix was calculated by the unweighted pair group method with arithmetic averages (UPGMA) on the basis of the differences in the allelic profiles. Clusters at a genetic distance of 0.1 were designated 1 to 30 at a cutoff level of about 50%. Columns to the right of the tree designate ST number, fimA genotyping results (see below), and continents of origin (EU, Europe; NA, North America; AF, Africa; AS, Asia).Multiple colored squares mark isolates with multiple positive fimA genotyping reactions. fimA genotyping was not performed for the German isolates (see later).
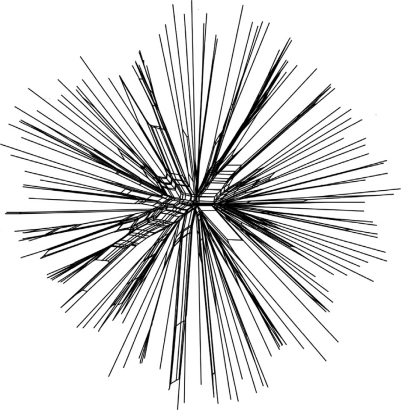
Split decomposition analysis (85) is a tool for graphical illustration of whether recombination plays a role in the evolutionary history of the separate genes investigated in the MLST. When utilizing the allelic profiles of the isolates (seven gene fragments) (79) the obtained splitstree graph visualizes the degree of recombination between the isolates in the respective bacterial populations. The algorithm utilized in the analysis create a tree-like structure when the descent is clonal and bush-like structure when recombination plays a major role in the evolutionary history of the genes investigated. The test can be performed for all isolates with each of the gene fragments individually or for the concatenated allele sequences. Based on the allelic profiles of 138 STs in the MLST database (Fig. 3), this analysis also strongly indicates the clonality of P. gingivalis and is in accordance with the results from the other presented methods. However, it is important to emphasize that our results altogether only reflect a small population of P. gingivalis strains.
Fig. 3.
Split decomposition analysis graph (85) constructed by SplitsTree (v. 4.6) (www.splitstree.org) from the allelic profiles of the 138 STs of P. gingivalis in the MLST database. Recombination is illustrated by the net- or bush-like structures to the center. The star- or tree-like structures indicate clonality.
Clonal heterogeneity within individuals, recombination
The general acceptance that periodontal patients harbor a single clonal type of P. gingivalis (86–88) changed when Loos et al. (86) discovered by DNA fingerprinting that one out of nine patients was infected with two clonal types. Later, similar findings (intraindividual detection of two or three clones) were interpreted as transient coinfections (89, 90) of additional clones. The studies by Enersen et al. (78, 79), through MLST analysis, revealed an even larger clonal heterogeneity within different periodontitis patients. In three Indonesians, two, three and four distinct STs were detected, respectively, from infected sites, and in a Finnish patient two distinct STs were found. However, the sampled sites in these studies were unknown.
The fact that P. gingivalis can be detected in diseased periodontal sites and in healthy gingival sulci (33, 64, 86, 87) suggests that it is a species of variable pathogenic potential, and single sites have been hypothesized to be colonized by more virulent clones (49, 51, 87).
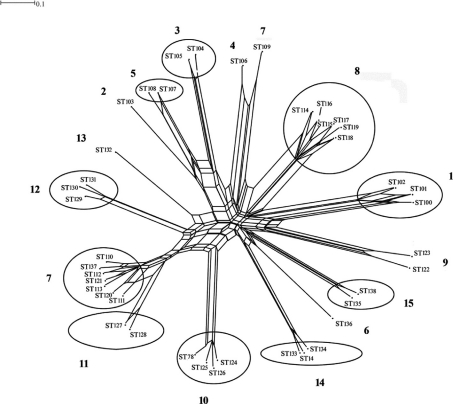
The diversity of P. gingivalis in single sites was investigated by MLST in patients with ‘refractory’ periodontitis (84) (Fig. 4). The diagnosis represents cases of treatment-resistant disease (91, 92) where a variable number of sites may harbor clones of higher virulence. Furthermore, the knowledge of the complexity of the subgingival microbiota in periodontal disease could (87, 88) allow for the possibility of DNA exchange between different strains in the same periodontal pocket. With increasing number of cells over time in one infected site, such events could represent fitness selection of surviving clones with regard to local growth conditions as well as the hosts immune response, which may contribute to further progression of the disease (88).
Fig. 4.
The degree of recombination between 93 isolates of P. gingivalis sampled from 15 single sites of ‘refractory’ periodontitis patients is illustrated by a SplitsTree graph with superimposed results of the eBurst analysis to the graph. Patient/site numbers are marked in bold. Se&v;eral STs detected in each periodontal site are located in groups (circles/ellipses) except patients 2, 4, 6, 9 and 13 where only one ST was found per site. ST 109/patient 7; see text (84).
The clonal diversity in single sites (84) demonstrated colonization by multiple STs. In each of 15 ‘refractory’ periodontal pockets, it was detected from 8–1 STs (Fig. 4), and these results raise the question of a much larger clonal diversity within a patient than earlier reported. The study (84), in regard to virulence and diagnosis, also shows the presence of two STs from two different subjects that were previously recorded in the MLST isolate database and represented strains sampled from cases of aggressive periodontitis (92) in Germany (ST14 and ST78) (www.pubmlst.org/pgingivalis). These may well correspond to more virulent clones, as is the case with other human pathogens (87).
Genetic variation accumulates in many bacterial species at housekeeping loci as frequently by homologous recombination (replacement of corresponding small chromosomal segments from related strains) as by point mutations (3, 88, 93, 94). Based on many MLST data sets from several species, the eBURST program (www.mlst.net) was developed to explore the diversification of closely related bacterial strains on a short time scale (3, 9, 95).
The analysis by eBURST of the allelic profiles in single periodontal pockets where more than three clones were sampled (84) showed 10 clonal complexes defined by single locus variants (9, 95) (Fig. 4). With the exception of one subject (7) (see later) (Fig. 4), the variation in each periodontal site was only related to pepO and recA genes. The alleles of these genes at each site showed multiple identical polymorphic sites in non-adjacent nucleotides with only one variable nucleotide between the alleles (unpublished alignments) (84). Because recombination generally occurs and involves larger segments of chromosomal DNA than the fragments analyzed by MLST, it is most likely that the same polymorphic sites in different alleles of the genes actually reflected recombination rather than point mutations occurring within the investigated site. Furthermore, it is unlikely that multiple point mutations would arise in the same polymorphic sites in the different alleles. These findings are in accordance with other studies concluding that recombination seems to occur to a larger extent among bacterial species than previously expected (93–95). By calculation of the average values of dN/dS which were <1 for both genes (average dN/dS recA=0.000, dN/dS pepO=0.2924; unpublished data), the results of the variation indicated synonymous substitutions with no change in gene products.
An especially noteworthy observation was detected in one periodontal site. A very distinct clone (ST 109) was observed in a clonal complex together with variants of seven closely related others. (Fig. 4). The observation could be explained as an example of a recombinational event in the allele of the affected recA gene of ST 109 giving rise to the other variants. This is supported by Tribble et al. (88) who demonstrated the ability of different P. gingivalis strains to transfer chromosomal DNA to each other by conjugation, representing an example of the underlying mechanism for allele swapping and the genetic variation of P. gingivalis (88). Furthermore, Frandsen et al. (76) showed signs of frequent recombination by identifying a random distribution of two virulence-associated mobile genetic elements among their investigated strains.
fimA gene; genotyping and sequencing
Because the results of the studies summarized here (79, 81) provided further evidence of a high genetic diversity and a clear indication of some degree of clonality of P. gingivalis, an eventual association of virulence genes with specific clones could be a valuable supplement to our knowledge of the characteristics of the species. Thus, all strains were investigated by fimA genotyping.
Eighty two P. gingivalis isolates were investigated by the fimA genotyping method, a method used in a number of studies with direct PCR technique on clinical material for detection of prevalence of fimA genotypes from sites with various periodontal conditions (56–62). The results from the cultured isolates confirmed the results from the clinical investigations that fimA II and IV genotypes seem to be predominant in chronic periodontitis-affected sites (56, 60, 79).
The association of these genotypes to cardiovascular disease has also been described earlier.
A relatively high prevalence of isolates positive in PCR for multiple fimA genotypes was observed in the two bacterial populations (25.6 and 30.1%, respectively) (79), similar to the reports from the clinical material in other investigations (56–61). Because the fimA gene occurs as a single copy in the chromosome (53), detection of multiple fimA genotypes in the clinical studies has been explained by (1) the presence of several different genotypes colonizing the same site (56, 61, 2) the possible existence of new unidentified genotypes (57, 58, 61), or (3) the existence of fimA non-typable strains (presence of new genotypes) (56–60). All the investigated isolates in our studies were typable. Because the investigation from cultured isolates resulted in similar findings as in clinical PCR studies, we anticipated that the reason for the multiple positive PCR reactions could be a previously not identified variation within the fimA gene.
In an attempt to explain these results (fimA genotyping), a set of universal primers were designed for the purpose of sequencing the whole gene in a selected number of isolates. These isolates were chosen based on the results from the genotyping method, focusing mainly on those with multiple positive PCR reactions to the specific primers for fimA genotype I, Ib, and II because of the high prevalence of these combinations. The reference strains for genotype fimA III and V were also selected for sequencing.
The analysis verified a conserved fimA gene with only minor variations among the selected isolates, mainly point mutations. Only one sign of recombination could be detected for an isolate, indicating that a fragment from fimA type III was inserted into the fimA type II gene of another isolate. However, our multiple sequence alignments of the total fimA gene sequences resembled the alignments published by Fujiwara et al. (55), also showing minor variations in single nucleotides of the respective isolates.
The fimA genotyping from the clinical study of single ‘refractory’ periodontitis sites also revealed some sites harboring isolates reacting to multiple specific PCR primer sets. It is likely that the observations of multiple reactions for a number of investigated strains in both studies can be explained partly by the primer design of the fimA genotyping method and partly by the small variation of the fimA gene between isolates. These results put a question mark to the fimA genotyping method, suggesting either redesign or reevaluation. Furthermore, results from alignment of the different fimA sequences revealed generally a conserved gene with only small variations in certain segments (79, 81).
fimA genotyping vs. MLST
The relationship between the distribution of fimA genotypes and MLST data of P. gingivalis has not been investigated earlier. The tendency of clustering of the fimA genotypes (79, 81) with isolates of the same or closely related STs harboring the same fimA genotype strengthens our conclusion of a weakly clonal population structure of P. gingivalis, but with no strong relationship between fimA genotypes and the STs identified (Fig. 2).
Acknowledgements
I thank Professor Ingar Olsen and Professor Dominique A. Caugant for excellent support and advice to my PhD projects.
Conflict of interest and funding
There is no conflict of interest in this study for the author.
References
- 1.Cooper JE, Feil EJ. Multilocus sequence typing – what is resolved? Trends Microbiol. 2004;12:373–7. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Baker S, Hanage W, Holt K. Navigating the future of bacterial molecular epidemiology. Curr Opin Microbiol. 2010;13:640–5. doi: 10.1016/j.mib.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu Rev Microbiol. 2001;55:561–90. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- 4.Feil EC, Enright MC. Analyses of clonality and the evolution of bacterial pathogens. Curr Opin Microbiol. 2004;7:308–13. doi: 10.1016/j.mib.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Maynard Smith J, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–8. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maynard Smith J, Feil E, Smith NH. Population structure and evolutionary dynamics of pathogenic acteria. Bioessays. 2000;22:1115–22. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 7.Caugant DA, Bøvre K, Gaustad P, Bryn K, Holten E, Høiby EA. Multilocus genotypes determined by enzyme electrophoresis of Neisseria meningitidis isolated from patients with systemic disease and from healthy carriers. J Gen Microbiol. 1986;132:641–52. doi: 10.1099/00221287-132-3-641. [DOI] [PubMed] [Google Scholar]
- 8.Maiden MCJ. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–88. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 9.Spratt BG, Hanage WP, Li B, Aanensen D, Feil E. Displaying the relatedness among isolates of bacterial species – the eBURST approach. FEMS Microbiol Lett. 2004;214:129–34. doi: 10.1016/j.femsle.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Kilian M, Frandsen EVG, Haubek D, Poulsen K. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000–2006;42:158–79. doi: 10.1111/j.1600-0757.2006.00159.x. [DOI] [PubMed] [Google Scholar]
- 11.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U S A. 1998;95:3140–5. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bougnoux ME, Aanensen DM, Morand S, Théraud M, Spratt BG, d'Enfert C. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol. 2004;3:243–52. doi: 10.1016/j.meegid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol. 2003;41:5709–17. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–60. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 15.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright MC, Spratt BG, Kalia A, Cross JH, Bessen DE. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect Immun. 2001;69:2416–27. doi: 10.1128/IAI.69.4.2416-2427.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotetishvili M, Stine OC, Kreger A, Morris JG, Jr, Sulakvelidze A. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J Clin Microbiol. 2002;40:1626–35. doi: 10.1128/JCM.40.5.1626-1635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, et al. Characterisation of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–36. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolstø A-B. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl Environ Microbiol. 2004;70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urwin R, Maiden MCJ. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11:479–87. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Jolley K, Chan MS, Maiden MC. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Kolenbrander PE, Palmer RJ, JR, Rickard AH, Jakubovics N, Chalmers NI, Diaz P. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 26.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 27.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol. 1999;20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 28.Amano A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003;74:90–6. doi: 10.1902/jop.2003.74.1.90. [DOI] [PubMed] [Google Scholar]
- 29.Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J Bacteriol. 2003;185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holt S, Ebersole J. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the ‘red complex’, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000–2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 31.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 32.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol. 2000–2005;38:135–87. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 33.Dzink JL, Socransky SS, Haffajee AD. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J Clin Periodontol. 1988;15:316–23. doi: 10.1111/j.1600-051x.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 34.Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, et al. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–53. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 35.Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol. 2000–2006;40:50–76. doi: 10.1111/j.1600-0757.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 36.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. 2003;30:644–54. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 37.Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 38.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–5. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 39.Lamont RJ, Jenkinson H. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–63. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–8. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Prot Pept Sci. 2003;4:409–26. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 42.Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15:341–9. doi: 10.1034/j.1399-302x.2000.150601.x. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Michel R, Cohen J, Decarlo A, Kozarov E. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol. 2008;8:26. doi: 10.1186/1471-2180-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hajishengallis G. Porphyromonas gingivalis – host interactions: open war or intelligent guerilla tactics? Microbes and Infection. 2009;11:637–45. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scannapieco FA, Bush RB, Paju S. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann Periodontol. 2003;8:38–53. doi: 10.1902/annals.2003.8.1.38. [DOI] [PubMed] [Google Scholar]
- 46.Kozarov EV, Dorn BR, Shelburne CE, Dunn WA, Jr, Progulske-Fox A. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler Thromb Vasc Biol. 2005;25:17–8. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- 47.Kozarov E, Sweier D, Shelburne C, Progulske-Fox A, Lopatin D. Detection of bacterial DNA in atheromatous plaques by quantitative PCR. Microbes Infect. 2006;8:687–93. doi: 10.1016/j.micinf.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Wada K, Kamisaki Y. Roles of oral bacteria in cardiovascular diseases. From molecular mechanisns to clinical cases: involvement of Porphyromonas gingivalis in the deveopment of human aortic aneurysm. J Pharmacol Sci. 2010;113:115–9. doi: 10.1254/jphs.09r22fm. [DOI] [PubMed] [Google Scholar]
- 49.Nakano K, Wada K, Nomura R, Nemoto H, Inaba H, Kojima A, et al. Characterization of aortic aneurysms in cardiovascular disease patients harboring Porphyromonas gingivalis. Oral Diseases. 2011;17:370–8. doi: 10.1111/j.1601-0825.2010.01759.x. [DOI] [PubMed] [Google Scholar]
- 50.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–38. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 51.Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39:136–42. doi: 10.1111/j.1600-0765.2004.00719.x. [DOI] [PubMed] [Google Scholar]
- 52.Hamada N, Sojar HT, Cho M, Genco RJ. Isolation and characterization of minor fimbria from Porphyromonas gingivalis. Infect Immun. 1996;59:4788–94. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickinson DP, Kubiniec MA, Yoshimura F, Genco RJ. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–65. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JY, Sojar HT, Bedi GS, Genco RJ. Porphyromonas (Bacteroides) gingivalis fimbrillin: size, amino-terminal sequence, and antigenic heterogeneity. Infect Immun. 1991;59:383–9. doi: 10.1128/iai.59.1.383-389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujiwara T, Morishima S, Takahashi I, Hamada S. Molecular cloning and sequencing of the fimbrillin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem Biophys Res Commun. 1993;197:241–7. doi: 10.1006/bbrc.1993.2467. [DOI] [PubMed] [Google Scholar]
- 56.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37:1426–30. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakagawa I, Amano A, Kimura RK, Nakamura T, Kawabata S, Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J Clin Microbiol. 2000;38:1909–14. doi: 10.1128/jcm.38.5.1909-1914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, et al. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37:425–32. doi: 10.1034/j.1600-0765.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- 59.Amano A, Kubinowa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79:1664–8. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 60.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Fleming TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111:390–4. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 61.Missailidis CG, Emeda JE, Ota-Tsuzuki C, Anzai D, Mayer MPA. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19:224–9. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 62.Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J Periodontal Res. 2005;40:147–52. doi: 10.1111/j.1600-0765.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 63.Nakagawa I, Amano A, Kubinowa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect Immun. 2002;70:277–85. doi: 10.1128/IAI.70.1.277-285.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neiders ME, Chen PB, Suido H, Reynolds HS, Zambon JJ, Shlossman M, et al. Heterogeneity of virulence among strains of Bacteroides gingivalis. J Periodontal Res. 1989;24:192–8. doi: 10.1111/j.1600-0765.1989.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 65.Genco CA, Van Dyke T, Amar S. Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 1998;6:444–9. doi: 10.1016/s0966-842x(98)01363-8. [DOI] [PubMed] [Google Scholar]
- 66.Nakano K, Kubinowa M, Nakagawa I, Yamamura T, Nomura R, Okahashi N, et al. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol Immunol. 2004;19:205–9. doi: 10.1111/j.0902-0055.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 67.Kato T, Kawai S, Nakano K, Inaba H, Kubinowa M, Nakagawa I, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9:753–65. doi: 10.1111/j.1462-5822.2006.00825.x. [DOI] [PubMed] [Google Scholar]
- 68.Umeda JE, Missailidis C, Longo PL, Anzai D, Wikstrom M, Mayer MPA. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol. 2006;21:415–9. doi: 10.1111/j.1399-302X.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- 69.Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kubinowa M, et al. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol. 2008;23:29–35. doi: 10.1111/j.1399-302X.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 70.Nakano K, Inaba H, Nomura R, Nemoto H, Takeuchi H, Toda K, et al. Distribution of Porphyromonas gingivalis fimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol. 2008;23:170–72. doi: 10.1111/j.1399-302X.2007.00406.x. [DOI] [PubMed] [Google Scholar]
- 71.Rôças IN, Siqueira JF., JR Distribution of Porphyromonas gingivalis fimA genotypes in primary endodontic infections. Oral Surg Oral Med Oral Path Oral Radiol Endod. 2010;10:474–8. doi: 10.1016/j.tripleo.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Zhou X, Zheng Q, Wang Y, Tang L, Huang D. Distribution of Porphyromonas gingivalis fimA genotypes in chronic apical periodontitis associated with symptoms. J Endodontics. 2010;36:1790–5. doi: 10.1016/j.joen.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Loos BG, Dyer DW, Whittam TS, Selander RK. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–12. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–31. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ali RW, Martin L, Haffajee AD, Socransky SS. Detection of identical ribotypes of Porphyromonas gingivalis in patients residing in the United States, Sudan, Romania and Norway. Oral Microbiol Immunol. 1997;12:106–11. doi: 10.1111/j.1399-302x.1997.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 76.Frandsen EV, Poulsen K, Curtis MA, Kilian M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect Immun. 2001;69:4479–85. doi: 10.1128/IAI.69.7.4479-4485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koehler A, Karch H, Beikler T, Flemmig TF, Suerbaum S, Schmidt H. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology. 2003;149:2407–15. doi: 10.1099/mic.0.26267-0. [DOI] [PubMed] [Google Scholar]
- 78.Enersen M, Olsen I, Caugant DA. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J Clin Microbiol. 2006;44:35–41. doi: 10.1128/JCM.44.1.35-41.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46:31–42. doi: 10.1128/JCM.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
- 81.Enersen M. Thesis. Oslo, Norway: University of Oslo; 2008. Population structure of the periodontal pathogen Porhyromonas gingivalis: diversity in housekeeping genes and the major fimbrae gene. [Google Scholar]
- 82.Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics. 2000;16:847–8. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 83.Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 84.Enersen M, Olsen I, Caugant DA. Genetic diversity of Porphyromonas gingivalis isolates from single ‘refractory’ periodontitis sites. Appl. Environ. Microbiol. 2008;74:5817–21. doi: 10.1128/AEM.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 86.Loos BG, Van Winkelhoff AJ, Dunford RG, Genco RJ, De Graaff J, Dickinson DP, et al. A statistical approach to the ecology of Porphyromonas gingivalis. J Dent Res. 1992;71:353–8. doi: 10.1177/00220345920710020201. [DOI] [PubMed] [Google Scholar]
- 87.Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–31. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 88.Tribble GA, Lamont GJ, Progulske-Fox A, Lamont RJ. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J Bacteriol. 2007;189:6382–8. doi: 10.1128/JB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang YJ, Yasui S, Yoshimura F, Ishikawa I. Multiple restriction fragment length polymorphism genotypes of Porphyromonas gingivalis in single periodontal pockets. Oral Microbiol Immunol. 1995;10:125–8. doi: 10.1111/j.1399-302x.1995.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 90.Leys EJ, Smith JH, Lyons SR, Griffen AL. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J Clin Microbiol. 1999;37:3906–11. doi: 10.1128/jcm.37.12.3906-3911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haffajee AD, Uzel NG, Arguello EI, Torresyap G, Guerrero DM, Socransky SS. Clinical and microbiological changes associated with the use of combined antimicrobial therapies to treat ‘refractory’ peridontitis. J Clin Periodontol. 2004;31:869–71. doi: 10.1111/j.1600-051X.2004.00573.x. [DOI] [PubMed] [Google Scholar]
- 92.Armitage GC. The development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 93.Hanage WP, Fraser C, Spratt BG. The impact of homologous recombination on the generation of diversity in bacteria. J Theor Biol. 2006;239:210–9. doi: 10.1016/j.jtbi.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 94.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–80. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]