Abstract
Intact bovine insulin, with its two chains linked via two disulfide linkages, has been used as a model system to study the incorporation of one or more gold cations as means for facilitating the cleavage of multiple disulfide bonds in a tandem mass spectrometry experiment. Gas-phase ion/ion reactions involving Au(I)Cl2− or Au(III)Cl4− were used to incorporate either one or two gold cations into multiply-protonated insulin cations, followed by ion trap collision-induced dissociation (CID) of the products. The incorporation of a single gold cation followed by CID showed little evidence for disulfide bond cleavage. Rather, the CID spectra were similar to those acquired for the same charge state with only excess protons present. However, the incorporation of two gold cations, regardless of oxidation state, resulted in efficient cleavage of the disulfide bonds connecting the two chains of insulin. Furthermore, ion trap CID of the insulin complexes containing two gold cations showed more sequence information compared to the complexes containing only one gold cation or no gold cations. The partitioning of the gold cations between the two chains upon CID proved to be largely asymmetric, as both gold cations tended to stay together. There appeared to be a slight preference for both gold cations to partition into the B-chain. However, the relatively low contribution from single chain ions with only one gold ion suggests a degree of cooperativity in the overall mechanism for separation of the two chains.
Keywords: gas-phase insulin ions, gold (I) and gold (III) cationization, collision induced dissociation, ion-ion chemistry
INTRODUCTION
Disulfide linkages are post translational modifications involved in stabilizing the native structures of many proteins and peptides. The identification and localization of disulfide bonds is therefore an important objective in primary structure characterization of biomolecules [1]. The presence of disulfide linkages in gaseous peptide and protein ions can have a major influence on the dissociation chemistry of such bio-ions. Various dissociation methods have been employed in the study of multiply- or singly-protonated or deprotonated biologically relevant molecules with disulfide bonds, including collision-induced dissociation (CID) [2,3], electron-induced dissociation (EID) [4] post source decay associated with matrix assisted laser desorption ionization (MALDI) [5], infrared multi-photon dissociation (IRMPD) [6], electron capture dissociation (ECD) [7,8], electron transfer dissociation (ETD) [9,10,11], electron detachment dissociation (EDD) [6], and UV-photodissociation [12]. However, no single dissociation method provides all of the structural information that might be sought. A conventional CID experiment of multiply protonated, disulfide-containing peptide or protein ions most often generates backbone cleavages with little or no apparent evidence for cleavage of the disulfide bond. Additionally, the disulfide linkage has been noted to stabilize the region that falls within the loop(s) formed by the disulfide bond(s), limiting the primary structural information forthcoming from these regions of the bio-ion [13,14]. However, when the proton mobility in an ion is limited, the disulfide bond cleavage is the main dissociation channel in the CID of cations [15] and ordinarily dominant in the CID of multiply deprotonated anions [2].
Various alternative techniques have been developed to map disulfide bonds within biomolecules. In addition to traditional, condensed-phase reduction, an electrolytic reduction of disulfide linkages without chemical reagents has been described [16]. Recently, Xia and coworkers have demonstrated a novel technique for cleaving disulfide linkages in polypeptides using low temperature helium plasma in air [17]. Furthermore, the incorporation of metal ions with known affinity for sulfur has proved to be effective in disulfide bond dissociation. Transition metal ions, such as Fe+ and Cu+ have been used to cleave bonds in organic molecules in the gas phase [18], while Fe− and Co− have specifically been used to cleave disulfide bonds [19]. A gas phase ion/ion reaction between Fe+ and multiply charged insulin anions has been reported where selective disulfide bond cleavage was achieved. However, it is unclear whether the disulfide bond dissociation was due to Fe+-selective chemistry or due to the excitation of the insulin anions from the exothermicity of the ion/ion reaction [20]. Cleavage of disulfide bonds in insulin anions under CID conditions has been demonstrated [2]. More recently, disulfide bond cleavage in proteins and peptides complexed with different transition metal ions has been examined employing ECD [21] and CID [22,23]. Polypeptide cationization by various alkaline earth metals in conjunction with low energy CID has also been demonstrated to be effective in disulfide bond cleavage [24,25].
Gold cationization ion/ion reactions have previously been utilized for selective and efficient cleavage of disulfide linkages in polypeptide ions arising from the digestion of disulfide linked peptides [26]. Cleavage of an intramolecular disulfide linkage in a small, intact peptide has also been studied in association with gold (I) cations [27]. This ion/ion reaction approach avoids complications arising from adding the salt directly to the peptide solution, which can introduce variability in the number of metal adducts, variability in charge state, and possible matrix effects. However, gold cationization in solution via the addition of salts is an alternative means for generating the gaseous ions of interest. In this report, we further extend the study of selective disulfide bond cleavage via gold cationization to multiply protonated intact bovine insulin ions, which contain multiple disulfide linkages. We further demonstrate the incorporation of multiple gold cations in the structure of insulin in order to cleave multiple disulfide bonds in a single dissociation period. Subsequent collisional activation of the protein-gold complexes shows improved sequence coverage for the protein, relative to that accessible via CID of multiply-protonated species.
EXPERIMENTAL SECTION
Materials
Acetonitrile, glacial acetic acid, hydrochloric acid, and methanol were purchased from Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Bovine insulin was purchased from Sigma Aldrich (St. Louis, MO). The protein solution was prepared at a concentration of approximately 50 µM in 49.5/49.5/1 (v/v/v) solvent mixture of water/ methanol/ acetic acid for positive ESI. Gold (III) chloride trihydrate (chloroauric (III) acid trihydrate) (Mallinckrodt Baker, Inc., Phillipsburg, NJ) was dissolved in acetonitrile with hydrochloric acid (1% by volume) at a concentration of approximately 20 mM for negative ESI. Negative ESI of the gold chloride solution generates AuCl2− and AuCl4− ions, where the gold valency is (I) and (III), respectively.
Mass Spectrometry
All experiments were performed using a prototype version of a QTRAP mass spectrometer [28] (AB Sciex, Concord, ON, Canada) equipped with a home-built dual nano-electrospray ionization source [29]. The experiments were controlled using the research software Daetalyst 3.14 provided by AB Sciex. The procedure for the cation-switching ion/ion reactions has been described previously [26,27]. Briefly, the positively charged biomolecule and negatively charged reagent ions were sequentially injected and independently mass selected in Q1. Following isolation, the oppositely charged reactants were sent to Q2, the collision cell, and were allowed to react for various times (50–100 ms) in mutual storage mode [30,31]. Multiple gold cations incorporation was achieved by increasing the mutual storage reaction time (100–200 ms). The formed biomolecule-gold chloride product complexes were subsequently transferred from Q2 to Q3. In the transfer step, the insulin-gold chloride complexes were accelerated to collisionally dissociate residual adduct ions via loss of two or more molecules of HCl. The resulting gold containing complexes were isolated in Q3 prior to collisional activation, and the product ions generated by ion trap CID were analyzed using mass selective axial ejection (MSAE) [32]. An ion/ion reaction experiment involving multiply-protonated insulin and AuCl4− anions was conducted using a quadrupole/time-of-flight tandem mass spectrometer to provide better mass measurement accuracy than that available with the ion trap instrument. The experiment was conducted to verify the assignments of the B-chain ions. This instrument, its modifications, and the procedure for conducting ion/ion reactions have recently been described [33].
RESULTS AND DISCUSSION
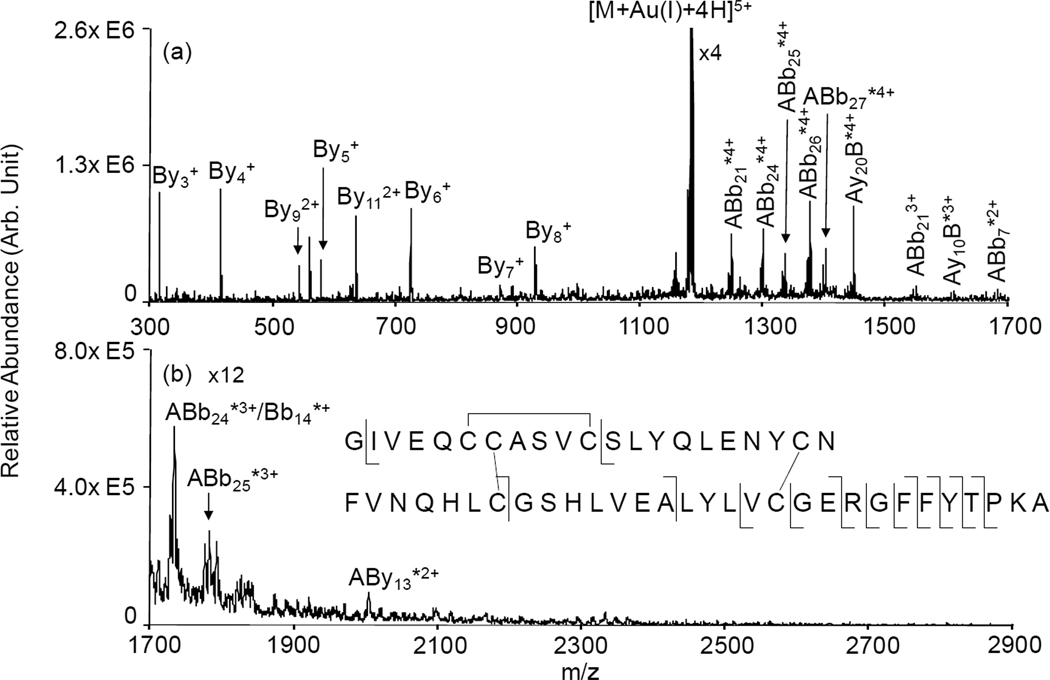
Intact bovine insulin is well-suited as a model protein to demonstrate the cleavage of two disulfide bonds in a single activation period via double gold cationization. The structure of bovine insulin consists of two chains linked together via two disulfide bonds, and a third disulfide linkage is contained within the A-chain. The ion/ion reaction between [Insulin+6H]6+ and AuCl2− resulted in the formation of [Insulin+Au(I)+4H]5+ and [Insulin+2Au(I)+2H]4+ complexes. The normal operating mass range of the instrument (100 Da-1700 Da) was extended via the use of a decreased MSAE ejection frequency in order to access an m/z region that encompasses any singly charged A-chain ions that might be formed. When only one gold(I) ion is incorporated into the ion, the ion trap CID spectrum in Figure 1 shows mostly y-type ions from the B-chain of the protein region that is outside of the disulfide stabilized portion. Additionally, some larger fragment ions with a gold adduct are also observed containing the entire A-chain and fairly large portions of the B-chain. However, aside from the appearance of the gold cation in the larger fragments, the product ion spectrum of the [Insulin+Au(I)+4H]5+ ion differs very little from the product ion spectrum derived from the multiply protonated protein, (see Figure S1 in Supplementary Information). Evidence in the product ion spectrum for cleavage of any disulfide bonds is minor, as the signals that are consistent with cleavages from within the inter-chain loop (i.e., the products assigned as ABb7*2+, Bb14*+, and ABy13*2+) are modest.
Figure 1.
Product ion spectrum derived from ion trap CID of [Insulin+Au(I)+4H]5+ : (a) mass-to-charge range from 300–1700; (b) mass-to-charge range from 1700–2900 . * denotes ions with a gold adduct; Ion nomenclature: for example, By4 is a y4 ion from B-chain; ABb21 is an ion that contains entire A-chain and b21 ion from B-chain.
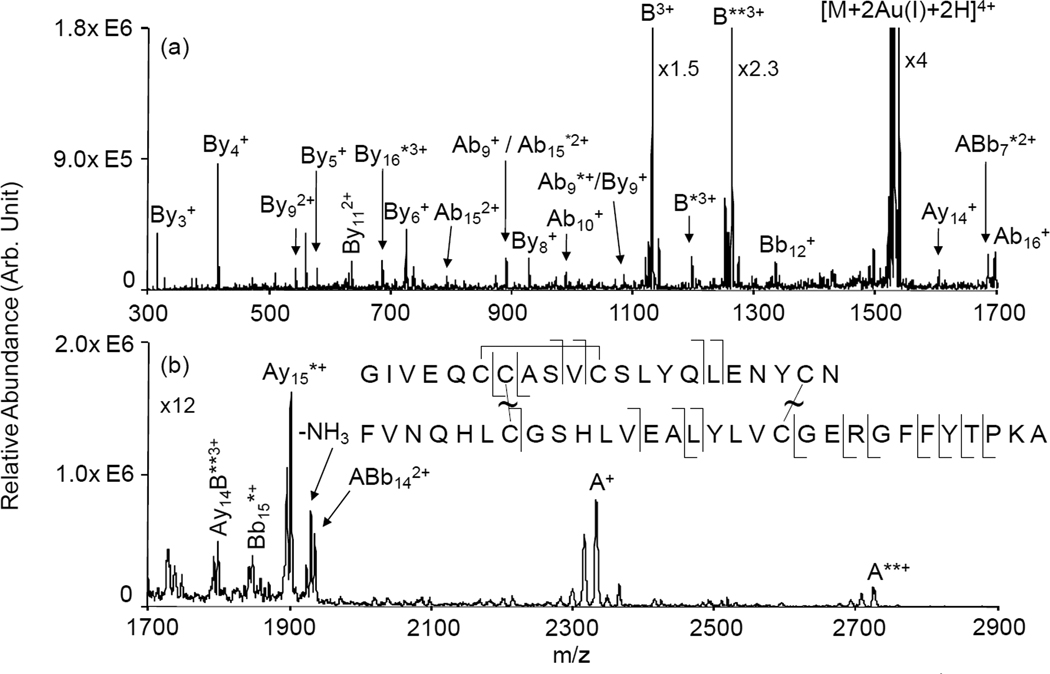
When two Au(I) cations are incorporated in the insulin structure, the fragmentation behavior of the [Insulin+2Au(I)+2H]4+ complex changes markedly. The ion trap CID spectrum of [Insulin+2Au(I)+2H]4+ in Figure 2 shows the separation of the two chains as the most dominant fragmentation channel. This suggests that the two Au(I) cations have cleaved the two disulfide linkages holding the two chains together. The most abundant fragment ion is triply-charged B-chain containing the two Au(I) cations. The next most abundant fragment is triply-charged B-chain with no gold adducts. The B-chain with one Au(I) adduct is also observed but at much lower abundance, which suggests that there must be a degree of cooperativity in the overall mechanism for cleavage of both disulfide linkages. If the cleavages of each disulfide bond were independent of each other, the partitioning of one gold ion into each fragment would be the most abundant channel from a statistical point of view. The mass measurements associated with the ions derived from the two chains are consistent with the mechanism described by Lioe et al. [22] to account for the cleavage of a disulfide bond with a metal coordinated to it. However, the detailed mechanism, be it concerted or step-wise, which leads to both gold ions remaining with the same chain is unclear. The complementary singly-charged A-chain ions are also observed, although at lower abundances, presumably due to instrumental discrimination effects such as poorer detector responses for the singly-charged ions. In addition to a series of b- and y-type ions from the B-chain, several fragment ions from the A-chain also appear in the spectrum. Fragment ions such as Ab9 (with no Au(I) and with one Au(I)) and Ay14 suggest that the disulfide linkage within the A-chain is cleaved in addition to one of the bonds holding the two chains together. The rest of the fragment ions that originate from the A-chain contain the intra-molecular disulfide linkage. Multiply protonated insulin ions were also reacted with AuCl4− reagent ions. After beam type CID of the protein-gold chloride complexes, [Insulin+Au(III)+2H]5+ or [Insulin+2Au(III)-2H]4+ complexes are formed. Ion trap CID of these complexes leads to results that are generally similar to those generated from CID of the Au(I) containing complexes. Somewhat more cleavage of the two chains with one gold ion in each chain is noted for the Au(III) complex but the asymmetric partitioning of the gold ions between the chains is dominant (See Figure S2 and Figure S3 in Supplementary Information).
Figure 2.
Product ion spectrum derived from ion trap CID of [Insulin+2Au(I)+2H]4+ : (a) mass-to-charge range from 300–1700; (b) mass-to-charge range from 1700–2900. * denotes ions with a gold adduct; ** denotes ions with two gold adducts. Ion nomenclature: for example, By4 is a y4 ion from B-chain; ABb21 is an ion that contains entire A-chain and b21 ion from B-chain.
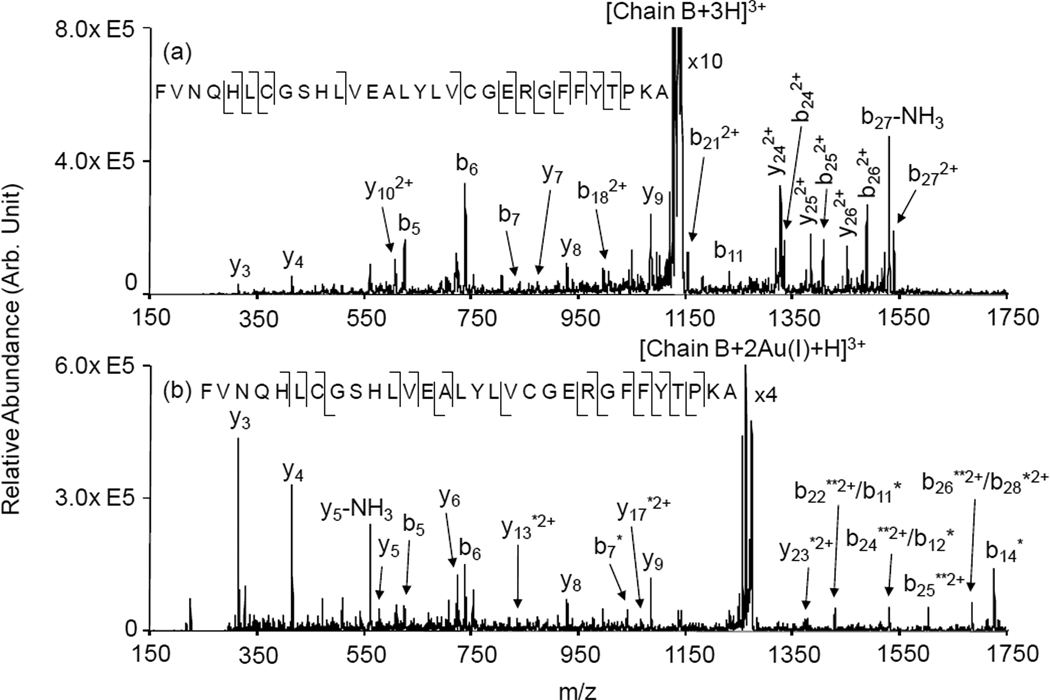
The incorporation of two gold cations into insulin, either in the form of Au(I) or Au(III), is clearly effective in separating the two chains of insulin upon CID and leads to the generation of relatively abundant single-chain products with either two or no gold ions. Figure 3 compares the ion trap CID of the B-chain fragments with either no (Figure 3(a)) or two (Figure 3(b)) Au(I) cations. Based on the ‘electrophilic’ mechanism proposed by Lioe et al. [22], the triply charged B-chain without gold ions is expected to contain a five-membered ring at each cysteine location with the sulfur of the cysteine bound to the C-terminal amide nitrogen. The B-chain, therefore, differs in mass from the triply-protonated reduced form of insulin B-chain (free cysteines) by 4 Da due to the formation of the cycles, which lack the thiol hydrogen and the hydrogen at the amide nitrogen. Nevertheless, the spectrum of Figure 3(a) is very similar, in terms of observed fragments and abundances, to that of the published ion trap CID spectrum of triply protonated reduced insulin B-chain [13]. The triply-charged B-chain ions with two Au(I) cations is expected to contain two strong gold thiolate bonds that localize the gold cations at the cysteine side-chains. The product ion spectrum of Figure 3(b) is consistent with this expectation.
Figure 3.
Product ion spectra derived from MS3 of: (a) [B-Chain+3H]3+ and (b) [B-Chain+2Au(I)+H]3+. * denotes ions with one Au(I) adduct; ** denotes ions with two Au(I) ions.
CONCLUSIONS
The incorporation of a single gold cation, either as Au(I) or Au(III), into multiply protonated insulin shows little evidence upon CID for separation of the precursor ion into chains A and B. A single gold cation is ineffective in cleaving the two disulfide linkages that bind the two chains. In fact, the fragmentation behaviors of the 5+ ions with and without one gold are sufficiently similar that it is not clear how much single disulfide cleavage occurs due to the presence of a single gold cation. The addition of a second gold cation, however, upon CID leads to extensive chain separation with the –S-S- bonds cleaved preferentially. While all three combinations of gold ion partitioning were noted, a strong preference for the two gold atoms to partition together was observed. The statistically favored partitioning of one gold ion into each chain occurred only to a relatively minor extent, particularly with Au(I). The strong preference for asymmetric gold partitioning is suggestive of a cooperative mechanism for the cleavage of the two disulfide bonds. The insulin results described here suggest that the incorporation of multiple gold cations into proteins with multiple disulfide linkages might be useful in the structural characterization of disulfide linked proteins via tandem mass spectrometry.
Supplementary Material
Acknowledgement
This research was supported by the National Institutes of Health under Grant GM 45372. Ian Webb and Will McGee are acknowledged for assistance in acquiring the quadrupole/time-of-flight results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wu J, Watson JT. Methods Mol. Biol. 2002;194:1. doi: 10.1385/1-59259-181-7:001. [DOI] [PubMed] [Google Scholar]
- 2.Chrisman PA, McLuckey SA. J. Proteome Res. 2002;1:549. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- 3.Zhang MX, Kaltashov IA. Anal. Chem. 2006;78:4820. doi: 10.1021/ac060132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lioe H, O’Hair RAJ. Anal. Bioanal. Chem. 2007;389:1429. doi: 10.1007/s00216-007-1535-1. [DOI] [PubMed] [Google Scholar]
- 5.Jones MD, Patterson SD, Lu HS. Anal. Chem. 1998;70:136. doi: 10.1021/ac9707693. [DOI] [PubMed] [Google Scholar]
- 6.Kalli A, Hakansson K. Int. J. Mass Spectrom. 2007;263:71. [Google Scholar]
- 7.Kocher T, Engstrom A, Zubarev RA. Anal. Chem. 2005;77:172. doi: 10.1021/ac0489115. [DOI] [PubMed] [Google Scholar]
- 8.Zubarev RA, Kruger NA, Fredriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. J. Am. Chem. Soc. 1999;121:2857. [Google Scholar]
- 9.Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. J. Am. Soc. Mass Spectrom. 2005;16:1020. doi: 10.1016/j.jasms.2005.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Gunawardena HP, Huang T-Y, McLuckey SA. Int. J. Mass Spectrom. 2008;276:160. [Google Scholar]
- 11.Gunawardena HP, Gorenstein L, Erickson DE, Xia Y, McLuckey SA. Int. J. Mass Spectrom. 2007;265:130. [Google Scholar]
- 12.Fung YME, Kjeldsen F, Silivra OA, Chan TWD, Zubarev RA. Angew. Chem. Int. Edn. 2005;44:6399. doi: 10.1002/anie.200501533. [DOI] [PubMed] [Google Scholar]
- 13.Stephenson JL, Cargile BJ, McLuckey SA. Rapid Commun. Mass Spectrom. 1999;13:2040. doi: 10.1002/(SICI)1097-0231(19991030)13:20<2040::AID-RCM754>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Hogan JM, McLuckey SA. J. Mass Spectrom. 2003;38:245. doi: 10.1002/jms.458. [DOI] [PubMed] [Google Scholar]
- 15.Wells JM, Stephenson JL, McLuckey SA. Int. J. Mass Spectrom. 2000;203:A1. [Google Scholar]
- 16.Zhang Y, Dewald HD, Chen H. J. Proteome Res. 2011;10:1293. doi: 10.1021/pr101053q. [DOI] [PubMed] [Google Scholar]
- 17.Xia Y, Cooks RG. Anal. Chem. 2010;82:2856. doi: 10.1021/ac9028328. [DOI] [PubMed] [Google Scholar]
- 18.Eller K, Schwarz H. Chem. Rev. 1991;91:1121. [Google Scholar]
- 19.Sallans L, Lane KR, Freiser BS. J. Am. Chem. Soc. 1989;111:865. [Google Scholar]
- 20.Payne AH, Glish GL. Int. J. Mass Spectrom. 2001;204:47. doi: 10.1016/S1044-0305(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 21.Kleinnijenhuis AJ, Mihalca R, Heeren RMA, Heck AJR. Int. J. Mass Spectrom. 2006;253:217. [Google Scholar]
- 22.Lioe H, Duan M, O’Hair RAJ. Rapid Commun. Mass Spectrom. 2007;21:2727. doi: 10.1002/rcm.3122. [DOI] [PubMed] [Google Scholar]
- 23.Mihalca R, van der Burgt YEM, Heck AJR, Heeren RMA. J. Mass Spectrom. 2007;42:450. doi: 10.1002/jms.1175. [DOI] [PubMed] [Google Scholar]
- 24.Kim HI, Beauchamp JL. J. Am. Chem. Soc. 2008;130:1245. doi: 10.1021/ja075698w. [DOI] [PubMed] [Google Scholar]
- 25.Kim HI, Beauchamp JL. J. Am. Soc. Mass Spectrom. 2009;20:157. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Gunawardena HP, O’Hair RAJ, McLuckey SA. J. Proteome Res. 2006;5:2087. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- 27.Mentinova M, McLuckey SA. Rapid Commun. Mass Spectrom. 2009;23:2647. doi: 10.1002/rcm.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hager JW. Rapid Commun. Mass Spectrom. 2002;16:512. [Google Scholar]
- 29.Xia Y, Liang X, McLuckey SA. J. Am. Soc. Mass Spectrom. 2005;16:1750. doi: 10.1016/j.jasms.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Liang X, Xia Y, McLuckey SA. Anal. Chem. 2006;78:3208. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. J. Am. Soc. Mass Spectrom. 2005;16:71. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Londry FA, Hager JW. J. Am. Soc. Mass Spectrom. 2003;14:1130. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 33.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Anal. Chem. 2006;78:4146. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.