Abstract
Genomic copy number alterations (CNA) are common in breast cancer. Identifying characteristic CNAs associated with specific breast cancer subtypes is a critical step in defining potential mechanisms of disease initiation and progression. We used genome-wide array comparative genomic hybridization (aCGH) to identify distinctive CNAs in breast cancer subtypes from 259 young (diagnosed with breast cancer at <55 years) African American (AA) and Caucasian American (CA) women originally enrolled in a larger population-based study. We compared the average frequency of CNAs across the whole genome for each breast tumor subtype and found that estrogen receptor (ER)-negative tumors had a higher average frequency of genome-wide gain (p<0.0001) and loss (p=0.02) compared to ER-positive tumors. Triple negative (TN) tumors had a higher average frequency of genome-wide gain (p<0.0001) and loss (p=0.003) than non-TN tumors. No significant difference in CNA frequency was observed between HER2-positive and negative tumors. We also identified previously unreported recurrent CNAs (frequency >40%) for TN breast tumors at 10q, 11p, 11q, 16q, 20p and 20q. In addition, we report CNAs that differ in frequency between TN breast tumors of AA and CA women. This is of particular relevance because TN breast cancer is associated with higher mortality and young AA women have higher rates of TN breast tumors compared to CA women. These data support the possibility that higher overall frequency of genomic alteration events as well as specific focal CNAs in TN breast tumors might contribute in part to the poor breast cancer prognosis for young AA women.
Keywords: breast cancer, triple negative, genomic alteration, array comparative genomic hybridization, young women
Introduction
Breast cancer is a heterogeneous disease consisting of five major breast tumor subtypes, basal, human epidermal growth factor receptor 2 (HER2)-expressing/estrogen receptor (ER)-negative, luminal A, luminal B and normal-like. These subtypes have been shown to have distinct expression patterns, based on microarray profiling, and clinical outcomes [1–4]. Protein expression of ER, progesterone receptor (PR), and HER2, detected by standard immunohistochemical (IHC) assays, has been used to approximate the gene expression subtypes. Triple negative (TN) breast cancer (ER−, PR−, HER2−) overlaps with basal cancer, ER-positive cancers closely define luminal cancers and the ER-negative, PR-negative and HER+ cancers approximate the HER2-expressing subtype.
The TN breast cancer subtype is not synonymous with the basal phenotype and expression of basal markers such as epidermal growth factor receptor (EGFR) and/or basal cytokeratins highlight the heterogeneity of the TN grouping [5]. However, the TN subset of breast cancers is highly enriched for the basal phenotype [6,7,1,3]. As is the case for basal cancers, TN tumors arise at an earlier age than non-TN cancers, and are almost exclusively high grade tumors [8].
One of the most striking findings related to TN breast cancer is that African American (AA) women exhibit an almost two-fold higher prevalence of this breast cancer subtype than Caucasian American (CA) women. The prevalence of TN breast cancer has been reported as high as 40% in AA women [9,1,10]. Additionally, TN incidence rates have now been reported and are also nearly twice as high among AA compared to CA women [11]. Even though AA have an overall lower incidence of breast cancer than CA women, the higher likelihood of developing TN breast cancer might contribute to the higher mortality from breast cancer experienced by AA women compared with CA women [12–17,10].
In addition to the differentiation of breast cancer subtypes by gene expression profiling, genome-wide array comparative genomic hybridization (CGH) techniques have demonstrated that breast cancer subtypes are associated with characteristic copy number alteration (CNA) profiles [18–21]. These characteristic genomic alterations serve as useful markers for subtype classification and can be applied towards the identification of critical genes that are consistently lost or gained in the specific subtypes to impact tumor biology.
The acquisition of genomic alterations is a critical event in cancer formation, potentially impacting the expression patterns of individual genes or entire biological pathways. For this study of young women we used genome-wide array CGH to identify CNAs that are high frequency and/or different in frequency between breast cancer subtypes defined by protein expression of ER, PR and HER2. We also compared CNA profiles of TN breast cancers in young CA and AA women to identify biological factors that might contribute to the poor breast cancer prognosis for young AA women.
Materials and Methods
Tumor Specimens
Primary breast tumors were from AA or CA women ages 20–54 years previously enrolled in the population-based Atlanta Women’s Interview Study of Health (WISH) cohort, which included 950 women diagnosed with breast cancer between 1990 and1992 [22]. When diagnosed, these women were residents of a 3-county metropolitan region of Atlanta, Georgia. The 259 breast tumors analyzed in the present study are the subset of the women in the WISH study with sufficient tumor tissue for testing. All tumors were reviewed and scored by an expert pathologist (P.L.P.) for tumor grade, ER, PR, and HER2 status [23].
Flow Cytometry Cell Sorting and DNA Extraction and Labeling
Flow cytometry was performed on macro-dissected and dissociated formalin-fixed, paraffin-embedded tumor samples to enrich for tumor cells by removing contaminating stromal and lymphocytic cells as previously described [24].
Genomic DNA was extracted from the flow sorted tumor cells (minimum of 100,000 cells per tumor) as previously described [25]. We quantified tumor genomic DNA by real-time PCR (Applied Biosystems, Foster City, CA) using two chromosome 2 specific probes at 2p25.3 (29,907–30,162) and 2q31.1 (21,407,882–21,408,181) with normal human female genomic DNA (Promega, Madison, WI) as the reference.
Whole-genome amplified and labeled samples were prepared as previously reported [21]. Briefly, ten nanograms each, of tumor and DpnII digested normal female reference DNA (Promega, Madison, WI), were random amplified and labeled with a Cy5- or Cy3-labeled primer, respectively, according to the method of Lieb et al., with modifications [26]. Labeled PCR products purified and combined with blocking agents (50 μg of human Cot-1 DNA and 100 μg of yeast tRNA) and hybridized to the microarray.
Array CGH
The array consists of 4320 human bacterial artificial chromosomes (BAC) with a median spacing of 413 kb when pericentric heterochromatic regions and the short arms of acrocentric chromosomes are excluded [21]. BAC clone locations are based on NCBI Build 36.1 of the human genome.
The Cy3- and Cy5- labeled genomic DNA and blocking agents were hybridized to the BAC array as described previously [21]. Arrays were scanned with a GenePix 4000A scanner (Axon Instruments Inc., Union City, CA); fluorescence data were processed with GenePix 3.0 image analysis software (Axon Instruments, Inc.). For each spot, log2 ratio=log2(Cy5/Cy3) and average log2 intensity=[log2(Cy5) + log2(Cy3)]/2 were calculated, where Cy5 and Cy3 refer to the median foreground fluorescent signals of the tumor and reference DNA, respectively. The log2 ratios on each array were normalized and corrected for intensity-based location adjustment with a block-level lowess algorithm [27].
Statistical Analysis
Normalized aCGH data were processed using wavelet along the genome [28]. The processed aCGH values were then categorized into copy number loss, no change, and gain events using the cut-off log2ratio −0.34 and 0.38, for loss and gain respectively, where the cut-off values were chosen based on X-chromosome titration experiments, previously reported [21].
Sampling weights were incorporated into the analysis based on the larger cohort of 950 cases for analysis of the 259 cases that were analyzed by aCGH. (See Supplemental Data 1 for additional details of statistical analysis; Supplemental Data Table 1). We calculated the weighted average overall genome-wide frequencies of copy number gain or loss by race (AA/CA) and the following tumor subtypes: ER status (positive/negative), triple negative (yes/no), and HER2 status (positive/negative), where the genome-wide copy number gain (or loss) for a tumor was defined as number of clones showing gains (or losses) divided by the total number of clones. To adjust for possible confounding effects of age and stage, weighted multivariable logistic regression was performed to examine whether each comparison group differs in gains and losses at each of the 4320 clones, respectively. Given some clones may have no or few events of gains or losses, the p-values based on asymptotic distributions of the test statistics would be biased. To correct for this bias, the bootstrap method was used to obtain exact p-values. A total of 1000 bootstrap samples were used for each comparison.
Hierarchical clustering was performed using clones that show statistical significance in any of the comparisons to identify whether subtypes of tumors would cluster based on the profiles of copy number alterations. For the heatmap clustering, we used the eucledian distance as the dissimilarity function and complete linkage.
All the analyses were done using statistical software R version 2.6.0. The wavelet smoothing required package of ‘waveslim’; the weighted logistic regression required package of ‘survey’ (http://www.r-project.org/). Throughout the paper, a p-value < 0.05 is considered statistically significant.
Results
Breast Tumor Characteristics of Young African American and Caucasian American Women
The individual and tumor characteristics of the 259 women (AA n=53 and CA n=206) such as age, vital status, ER, PR, HER2 expression status, grade, and stage are shown for all tumors and separately by race (Table 1). The samples in the present study, as observed in previous reports of this population-based Atlanta WISH cohort, show a significant racial difference in the distribution of tumors based on ER, PR, and HER2 expression status, as well as vital status, stage and grade for young AA compared to CA women [10,23,29] and, as described in the methods and Supplemental Data, a weighted analysis was performed to more accurately reflect the make-up of the original cohort.
Table 1.
Tumor Characteristics of Cases Analyzed with aCGH
| All aCGH n=259 | African American n=53 | Caucasian American n=206 | p-value* | |
|---|---|---|---|---|
| n | n (%) | n (%) | ||
| Estrogen Receptor | ||||
| negative | 105 | 36 (68) | 69 (34) | <.0001 |
| positive | 154 | 17 (32) | 137 (66) | |
| Progesterone Receptor | ||||
| negative | 99 | 32 (60) | 67 (33) | 0.0004 |
| positive | 160 | 21 (40) | 139 (67) | |
| HER2 | ||||
| negative | 224 | 44 (83) | 180 (87) | 0.5467 |
| positive | 35 | 9 (17) | 26 (13) | |
| Triple Negative | ||||
| Yes | 65 | 22 (42) | 43 (21) | 0.004 |
| No | 194 | 31 (59) | 163 (79) | |
| Tumor grade | ||||
| Low | 50 | 1 (2) | 49 (24) | <.0001 |
| Intermediate | 96 | 15 (28) | 81 (39) | |
| High | 113 | 37 (70) | 76 (37) | |
| AJCC Stage | ||||
| I | 89 | 7 (13) | 82 (40) | 0.0009 |
| IIA | 79 | 17 (32) | 62 (30) | |
| IIB | 54 | 16 (30) | 38 (18) | |
| III/IV | 37 | 13 (25) | 24 (12) | |
p-values were determined with a chi-square test
Genome-wide Copy Number Alterations in Subtypes of Breast Cancer and Racial Groups
We compared the average genome-wide frequency of copy number gain and loss for individual tumors of IHC-defined breast cancer subtypes (Table 2). In the comparison between ER-positive and ER-negative tumors, ER-negative tumors had a significantly higher frequency of both copy number gain (gain: 6.9% versus 4.9% p<0.0001) and loss (8.5% versus 7.3% p=0.02) events. TN breast tumors similarly had a significantly higher overall frequency of both genome-wide copy number gain (7.3% versus 5.2%; p<0.0001) and loss (8.9% versus 7.4%; p=0.003) than non-TN tumors, respectively. There was no statistically significant difference in the overall genome-wide frequency of copy number gain (5.6% versus 5.8%; p=0.68) and loss (7.3% versus 7.9%; p=0.37) when comparing HER2-positive to HER-2-negative tumors, respectively. When we compared overall frequency of genomic alterations in breast tumors of AA to CA women, we observed higher frequencies of both gain (6.9% versus 5.2%; p=0.0002) and loss (9.0% versus 7.3%; p=0.003) in AA than CA women, respectively. This difference is consistent with the higher percentages of ER negative and TN breast tumors in AA compared to CA women (Table 1), suggesting that breast tumor subtypes differ with respect to frequency of genome-wide alteration events.
Table 2.
Comparison of the Weighted Average Frequency* of Genome-wide Copy Number Alterations
| ER-negative (n=105) | ER-positive (n=154) | p-value** | |
|---|---|---|---|
| Gain | 0.069 | 0.049 | <0.0001 |
| Loss | 0.085 | 0.073 | 0.02 |
| HER2-positive (n=35) | HER2-negative (n=224) | p-value | |
|---|---|---|---|
| Gain | 0.056 | 0.058 | 0.68 |
| Loss | 0.073 | 0.079 | 0.37 |
| Triple Negative (n=65) | Non-Triple Negative (n=194) | p-value | |
|---|---|---|---|
| Gain | 0.073 | 0.052 | <0.0001 |
| Loss | 0.089 | 0.074 | 0.003 |
| African American (n=53) | Caucasian American (n=206) | p-value | |
|---|---|---|---|
| Gain | 0.069 | 0.052 | 0.0002 |
| Loss | 0.090 | 0.073 | 0.003 |
Genome-wide gain or loss events were calculated for each tumor then averaged (weighted) across tumors in each comparison group.
p-values were generated with a weighted t-test
Frequent Copy Number Alterations in Breast Cancer Subtypes and Racial Groups
To identify the specific genomic regions that were more frequently altered (copy number gain or loss) in the breast cancer subtypes, we classified CNAs as a “high-frequency” event if the gain or loss events for the specific probe on the array occurred in over 40% of the tumors in the specified subtype. We mapped these probes indicating high-frequency CNAs for the specific breast cancer subtypes to chromosome arms (Table 3) and cytogenetic bands (Supplemental Data Table 2) to profile the distribution of high frequency events. We identified high-frequency CNAs that were shared by the subtypes, as well as high-frequency CNAs that differed by subtype. The high frequency events common to all subtypes included gain events on chromosomes 1q and 8q, and high-frequency loss events on chromosomes 8p, 10q, 11q, 12q, and 16q. Subtype specific high-frequency CNAs included copy number loss events on 4p and 11p, which were observed in ER-negative, but not ER-positive tumors. Interestingly, HER2-positive tumors had the widest distribution of high-frequency loss events across the genome, i.e., the most chromosome arms with high-frequency loss events. We also observed high-frequency loss on 13q and 20q in TN breast cancers that were not observed in other non-TN tumors. Taken together, these results support the presence of high-frequency subtype-specific CNA events.
Table 3.
High Frequency Copy Number Alterations in Subtypes of Breast Cancer*
| Chromosomal arm | ER-positive (n=154) | ER-negative (n=105) | HER2-positive (n=35) | Triple Negative (n=65) |
|---|---|---|---|---|
| 1p | Gain | Gain | Gain/Loss | |
| 1q | Gain | Gain | Gain/Loss | Gain |
|
| ||||
| 2p | Loss | |||
| 2q | Loss | |||
|
| ||||
| 3p | Loss | Loss | ||
| 3q | Loss | |||
|
| ||||
| 4p | Loss | Loss | Loss | |
| 4q | Loss | |||
|
| ||||
| 5p | ||||
| 5q | Loss | Loss | ||
|
| ||||
| 6p | Loss | |||
| 6q | Loss | |||
|
| ||||
| 7p | Loss | |||
| 7q | Loss | |||
|
| ||||
| 8p | Loss | Loss | Loss | Loss |
| 8q | Gain | Gain | Gain/Loss | Gain |
|
| ||||
| 9p | Loss | |||
| 9q | Loss | |||
|
| ||||
| 10p | Loss | |||
| 10q | Loss | Loss | Loss | Loss |
|
| ||||
| 11p | Loss | Loss | Loss | |
| 11q | Loss | Loss | Loss | Loss |
|
| ||||
| 12p | Loss | |||
| 12q | Loss | Loss | Loss | Loss |
|
| ||||
| 13q | Loss | |||
|
| ||||
| 14q | Loss | Loss | ||
|
| ||||
| 15q | Loss | Loss | ||
|
| ||||
| 16p | Loss | |||
| 16q | Loss | Loss | Loss | Loss |
|
| ||||
| 17p | Loss | |||
| 17q | Gain/Loss | |||
|
| ||||
| 18p | ||||
| 18q | Loss | |||
|
| ||||
| 19p | Loss | |||
| 19q | Loss | |||
|
| ||||
| 20p | Loss | Loss | ||
| 20q | Loss | |||
|
| ||||
| 21p | ||||
| 21q | ||||
|
| ||||
| 22p | ||||
| 22q | Loss | Loss | ||
|
| ||||
| Xp | ||||
| Xq | Loss | |||
High-frequency CNA (weighted fraction >40% of tumors) mapped by chromosomal arm.
Differential Copy Number Alterations in Breast Cancer Subtypes
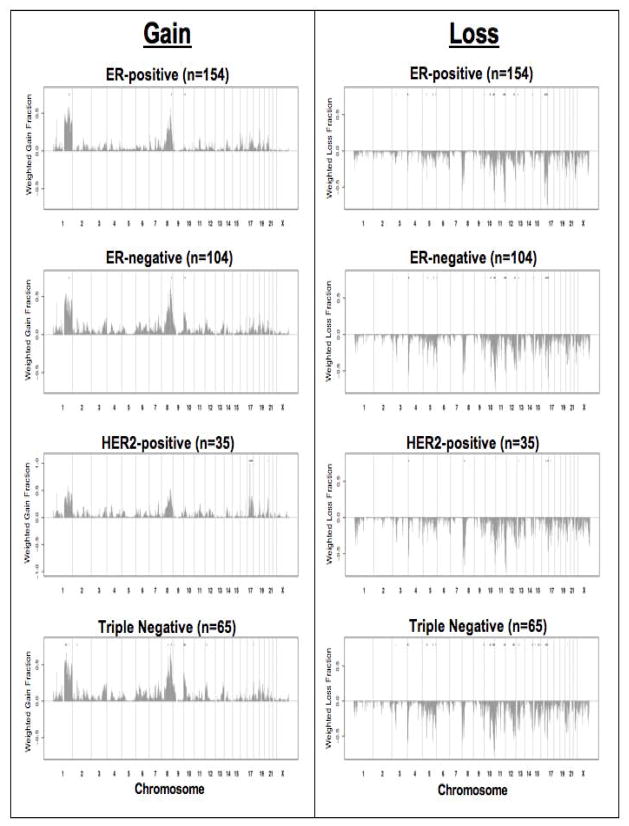
In addition to the CNA frequency differences at the genome-wide and chromosomal arm level, we identified subtype specific CNA events (gain and loss) at individual BAC clones. Differential CNAs (CNAs that differ in frequency between groups (p<0.05)) by individual BAC clones were identified based on comparisons of age- and stage-adjusted tumors in each comparison and restricted to clones with the difference of CNA frequency 0.20 (Table 4 and Supplemental Data Table 3). In the comparison between ER-positive (n= 154) and ER-negative (n=105) breast cancers, we found 90 (79 loss and 11 gain) differential CNAs. ER-positive tumors were characterized by differential gain events on 1q, and loss events on 11q and 16q, whereas, ER-negative tumors were characterized by differential gain events on 8q and 10p, and loss events on 3p, 4p, 5q, 9q, 10q, 12q, 13q, and 14q (Figure 1). In HER2-positive (n=35) vs. HER2-negative (n=224; including both ER-positive and ER-negative) tumors, a total of 49 (25 loss and 24 gain) CNAs were identified to be differential. HER2-positive tumors were characterized by differential gain on 17q and 20q, and loss on 4p, 8p, and 13q. HER2-negative tumors exhibited differential loss only on 16q (Figure 1). When we compared TN (n=65) to all non-TN tumors (n=194), we found a total of 203 (145 loss and 58 gain) differential CNAs. Triple-negative tumors were characterized by differential gain events on 1q, 2p, 8q, 10p, 12p, and 18p, and loss events on 3p, 4p, 5q, 9q, 10q, 12q, 13q, 14q, 15q, and 20q. Non-TN tumors had differential gain on 1q, and loss on 11q and 16q (Figure 1).
Table 4.
Differential Copy Number Alterations in Breast Cancer Subtypes and Racial Groups
| Cytogenetic Band* | ER-Negative vs ER-Positive | HER2 Positive vs HER2 Negative | Triple Negative (TN) vs Non-Triple Negative | African American (AA) vs Caucasian American (CA) | African American (AA) vs Caucasian American (CA) in Triple Negative Only** |
|---|---|---|---|---|---|
| 1p36 | AA | ||||
| 1p36 | CA | ||||
| 1p21 | CA | ||||
| 1q23 | TN | AA | |||
| 1q24 | TN | AA | |||
| 1q25 | TN | AA | |||
| 1q31 | AA | ||||
| 1q32 | ER-pos | non-TN | AA | ||
|
| |||||
| 2p25 | AA | ||||
| 2p16 | TN | ||||
| 2p15 | TN | ||||
|
| |||||
| 3p21 | ER-neg | TN | AA | ||
|
| |||||
| 4p16 | ER-neg | HER2 pos | TN | ||
| 4p15 | TN | CA | |||
| 4q13 | AA | ||||
| 4q31 | AA | ||||
|
| |||||
| 5p15 | CA | ||||
| 5p14 | AA | ||||
| 5p13 | CA | ||||
| 5q11 | ER-neg | TN | |||
| 5q13 | TN | ||||
| 5q31 | ER-neg | TN | AA | ||
| 5q35 | ER-neg | TN | AA | ||
|
| |||||
| 6p21 | CA | ||||
| 6q11 | AA | ||||
| 6q12 | AA | ||||
|
| |||||
| 7q31 | CA | ||||
| 7q36 | CA | ||||
|
| |||||
| 8p21 | HER2 pos | ||||
| 8q21 | TN | AA | |||
| 8q22 | AA | ||||
| 8q23 | AA | CA | |||
| 8q24 | ER-neg | TN | AA | ||
|
| |||||
| 9p21 | TN | ||||
| 9q32 | CA | ||||
| 9q33 | CA | ||||
| 9q34 | ER-neg | TN | AA | AA | |
|
| |||||
| 10p15 | ER-neg | TN | |||
| 10p14 | TN | ||||
| 10p13 | ER-neg | TN | |||
| 10p12 | ER-neg | TN | |||
| 10q22 | ER-neg | TN | AA | CA | |
| 10q23 | CA | ||||
| 10q25 | ER-neg | TN | AA | CA/AA | |
| 10q26 | ER-neg | TN | AA | AA | |
|
| |||||
| 11p15 | AA | ||||
| 11p11 | AA | ||||
| 11q13 | CA | ||||
| 11q13 | AA | ||||
| 11q23 | ER-pos | non-TN | |||
| 11q24 | ER-pos | non-TN | |||
| 11q25 | ER-pos | non-TN | CA | ||
|
| |||||
| 12p13 | TN | AA | |||
| 12p12 | TN | AA | |||
| 12q12 | CA | ||||
| 12q14 | CA | ||||
| 12q15 | CA | ||||
| 12q21 | CA | ||||
| 12q23 | ER-neg | TN | CA | ||
| 12q24 | ER-neg | TN | |||
|
| |||||
| 13q12 | ER-neg | HER2 pos | TN | ||
| 13q14 | TN | Ca | |||
| 13q32 | AA | ||||
| 13q33 | AA | ||||
| 13q34 | AA | ||||
|
| |||||
| 14q32 | ER-neg | TN | AA | AA | |
|
| |||||
| 15q12 | TN | AA | |||
| 15q21 | CA | ||||
| 15q22 | TN | AA | |||
| 15q24 | TN | AA | AA | ||
| 15q25 | TN | ||||
| 15q26 | TN | AA | |||
|
| |||||
| 16p13 | AA | ||||
| 16p13 | CA | ||||
| 16q12 | ER-pos | HER2 neg | AA | ||
| 16q12 | AA | ||||
| 16q13 | ER-pos | HER2 neg | |||
| 16q21 | ER-pos | non-TN | |||
| 16q22 | ER-pos | HER2 neg | non-TN | ||
| 16q23 | ER-pos | HER2 neg | non-TN | ||
| 16q24 | ER-pos | HER2 neg | non-TN | AA | |
|
| |||||
| 17q11 | HER2 pos | AA | |||
| 17q12 | HER2 pos | ||||
| 17q21 | HER2 pos | ||||
| 17q22 | HER2 pos | ||||
| 17q23 | HER2 pos | ||||
| 17q24 | HER2 pos | ||||
| 17q25 | HER2 pos | ||||
|
| |||||
| 18p11 | TN | CA | |||
| 18q23 | CA | ||||
|
| |||||
| 19p13 | CA | ||||
|
| |||||
| 20p11 | AA | ||||
| 20q12 | TN | ||||
| 20q13 | HER2 pos | ||||
|
| |||||
| 22q11 | AA | ||||
| 22q12 | AA | ||||
|
| |||||
| Xp22 | CA | ||||
| Xq13 | AA | ||||
Cytobands containing differential BAC clones with p-values <0.05 & weighted fraction difference ≥ 0.20 (with the exception of the race comparison in TN only)
Cytobands containing differential BAC clones with weighted fraction difference ≥ 0.20
 Copy Number Loss
Copy Number Loss
 Copy Number Gain
Copy Number Gain
Figure 1.
Genome-wide copy number alteration (gain and loss) frequencies by subtype. The whole-genome plots show the frequency (y-axis) of gain (gray bars above the x-axis) and loss (gray bars below the x-axis) events for breast cancer subtypes. The black circle at the top of each plot represents the genomic location of a BAC clone demonstrating a differential copy number alteration for the specific breast cancer subtype.
When we compared CNA frequencies by race in breast tumors from AA (n=53) and CA (n=206) women, we found a total of 22 (20 loss and 2 gain) differential CNAs. Breast tumors from AA women, regardless of subtype, were characterized by differential gain at 8q, and loss at 5q, 9q, 10q, 14q, 15q. In this comparison, there were no differential gain or loss events characteristic for CA women. When we compared CNA events in TN only breast tumors of AA (n=22) and CA women (n=43), we found 216 CNAs (130 loss and 86 gain) with a frequency difference >20% by race (see Materials and Methods; Table 4). Overall, these data support the observation that there are differences in frequency and location of CNA events for TN tumors in AA and CA women.
Hierarchical Clustering of Breast Cancer Subtypes Based on Differential Copy Number Alterations
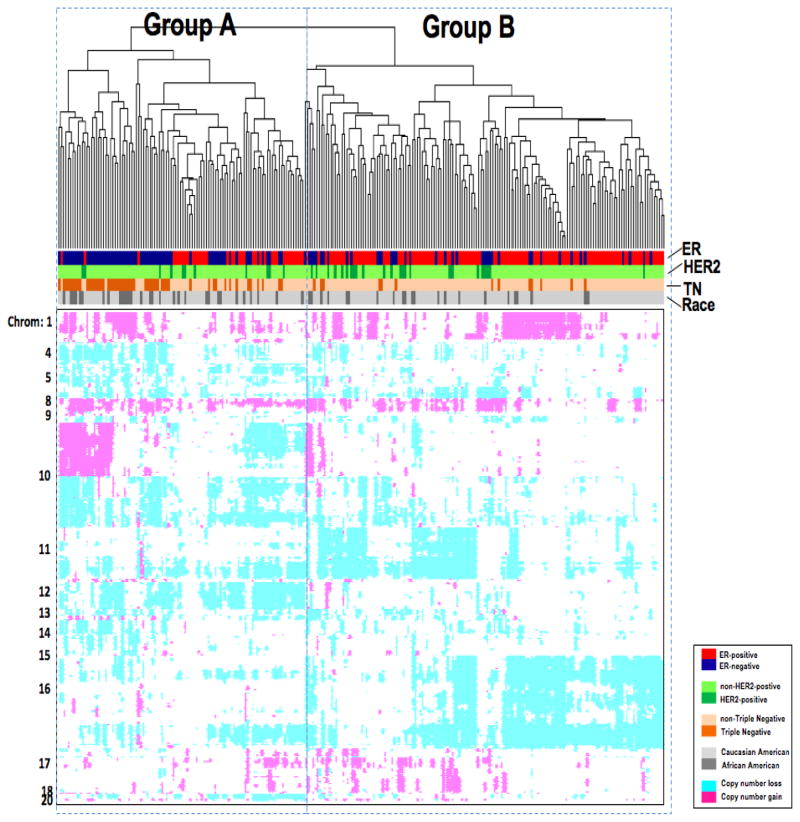
Hierarchical clustering was performed with the 320 statistically significant differential BAC clones that were identified in the subgroup comparisons. We were interested in determining if breast cancer subtypes would cluster based on patterns of their characteristic CNAs. The dendrogram shows that the 259 tumors clustered into two major groups, one enriched for ER-negative tumors (Figure 2. Group A) and the other enriched for ER-positive tumors (Figure 2. Group B), regardless of HER2 status. The Group A cluster contained the majority of TN breast tumors, 52 of the 65 TN tumors (80%) TN tumors were characterized by gains in 1q, 8q, and 10p, and loss in 4p, 5q, 14q, and 15q. Gains at 10p15-10p12 and loss at 14q32 were the most predominant differentiating regions of CNA associated with the TN tumors. Group B on the dendrogram contained the majority of ER-positive tumors (115/154, 75%) and HER2-positive tumors (24/53, 69%).
Figure 2.
Hierarchical clustering based on differential copy number alterations. Individual tumors (columns) clustered into two major groups (A and B). Tumor characteristics such as estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) and racial background are indicated in the legend. Chromosome locations of copy number alteration events (gain in pink and loss in turquoise) are indicated to the left of the heatmap.
Discussion
In the current study, we used a genome-wide aCGH approach to profile CNAs from breast cancers for 259 young women from a previously reported population-based case-control study in Atlanta, GA [22]. We identified characteristic CNAs associated with breast cancer subtypes (ER+/−, HER2+/−, TN) and found statistically significant differences in the average overall frequency of genome-wide CNAs in subtype comparisons, as well as frequency differences in CNAs occurring at specific genomic sites. We also observed differences in the frequency (>20%) and genomic locations of CNA events for TN tumors in AA and CA women.
Our results demonstrate that TN tumors had marked genomic instability with the highest average frequency of genome-wide CNAs compared to the other breast cancer subtypes. Chin et al. and Bergamaschi et al. also reported similar findings, with TN/basal tumors having the highest frequency of both copy number gains and losses compared to other subtypes [18,19]. Fridlyand et al. observed a subset of ER- tumors associated with poor outcomes and extensive genomic instability, classifying this molecular subtype as the “complex” subtype. These “complex” tumors were found to have a high degree of similarity for CNA profiles when compared to BRCA1 hereditary tumors [20]. We also observe our TN tumor samples to have CNAs in genomic regions that are characteristically altered in BRCA1 hereditary tumors specifically at 5q, 10p, 12p, 12q, and 20q [30,31].
Copy number gain at 10p has been reported to be a distinguishing CNA for TN/basal tumors compared to other breast cancer subtypes [32,18,33,19]. We observed a copy number gain in the region of 10p spanning 10p15-10p12 in our set of TN tumors. There are numerous genes spanning this region, with several confirmed to have increased protein expression associated with TN/basal tumors. Up-regulation of gene expression for several genes in this region (10p13), specifically, C10orf7, UPF2, HSPA14, RPP38 and CAMK1D has been confirmed to correlate with copy number gain [32,34]. The region of 10p13 also contains the gene for vimentin (VIM) that has been associated with increased expression with TN/basal tumors and plays a role in the epithelial-mesenchymal transition [35]. Although we do not present corresponding gene expression data for our samples, we see a significantly higher frequency of copy number gain at 10p13, corresponding to the genomic region containing the gene for VIM in TN tumors.
Amplification at 8q24 is common in breast cancer and has been previously observed in TN/basal and BRCA1 breast tumors and associated with poor outcomes [36,37]. We observed a significant difference in the frequency of gain events in the genomic region (8q24) containing the C-MYC gene in ER-negative, primarily TN tumors, with >50% of the TN tumors compared to non-TN tumors (30%) having copy number gain in this region. We also compared the frequency of CNAs in TN tumors of AA and CA at 8q24, and observed a negligible CNA frequency difference between AA women (54%) and CA women (52%) for gains in this region, indicating that copy number gain in this region, containing the C-MYC, is not a distinguishing feature between tumors of AA and CA women.
There was twice the frequency of copy number gain in 13q31-13q34 for TN tumors for AA (20%) versus CA (9%) women. Amplification in the region of 13q31-13q34 has been previously reported to be associated with TN/basal tumors (20%) and BRCA1-associated breast tumors (8.1%) in a study reported by Melchor et al. [38]. They identified two “driver” genes in 13q34 that facilitate tumor progression, cullin4A (CUL4A) and transcription factor Dp-1 (TFDP1). Both were demonstrated to have increased protein expression in tumors with amplification at 13q34. Both CUL4A and TFDP1 overexpression in breast cancers have been associated with shorter overall and disease-free survival [39,40]. The study conducted by Melchor et al. included a total of 188 familial and 277 sporadic breast cancer samples, most of which came from cancer centers of predominantly Latino/Hispanic patients in Spain and Ecuador. Both AA and Hispanic women with TN/basal tumors have poorer outcomes compared to CA women [9,41]. Additional studies are needed to evaluate events associated with amplification of 13q31-13q34 in relation to race and ethnicity and clinical outcome for AA, Latino/Hispanic, and Caucasian women.
ER-negative, specifically TN tumors, had a statistically significant differential frequency of copy number loss at 14q32.2 (p=0.001) when compared to the ER-positive and non-TN tumors, respectively and rarely occurred in HER2-positive tumors (5%) (Table 4). In addition, this CNA occurred more than twice as often in TN tumors of AA women compared to TN tumors of CA women (59% vs. 21%, respectively). The 14q32.2 region contains the gene for the microRNA, miR-342. MiR-342 has a critical role in proliferation, differentiation, development, and metabolism (reviewed in [42]) and functions as a pro-apoptotic tumor suppressor in colon tumors [43]. For breast cancer, a recent study demonstrated that miR-342 expression was highest in ER-positive and HER2-positive breast tumors and lowest in TN/basal tumors. This expression pattern is consistent with the CNA profiles they we found at 14q32.2 for the breast tumor subtypes, suggesting that copy number loss at 14q32.2 in TN tumors may lead to the downregulation of miR-342 expression, particularly in tumors of AA women.
Although the current literature has been inconsistent with respect to biological differences between tumors of AA and CA women, two recent reports support the hypothesis that biological differences exist and find that in women with breast tumors of similar ER status, AA women have poorer survival than CA women, even after adjustment for socioeconomic factors [44,45]. In addition, a separate study showed that there are biological differences impacting angiogenesis, chemotaxis, and immunobiology pathways in breast tumors of AA and CA women based on the comparison of gene expression profiles of tumor and stromal tissue from breast tumors of these two racial populations [46]. Our preliminary findings of differences in CNA frequencies in TN tumors from AA and CA women support the observations that there may be biological differences in the TN tumors. It is still unknown how these differences contribute to prognosis for AA and CA women.
One potential study limitation was selection bias in the array-tested samples. Therefore, we conducted a weighted analysis to address the issue of selection bias, but cannot be certain that this weighting completely addressed that issue. In addition, although there were limitations in the use of the moderate resolution BAC array for the identification of CNAs, we successfully demonstrated that we could confirm previously identified CNAs associated with specific breast cancer subtypes and identify additional novel CNAs not previously reported, particularly for the TN/basal subtype.
In this report we found characteristic genomic alterations associated with subtypes of breast cancer. The breast cancer samples included in this study were a part of a larger cohort of young women, and included the largest aCGH study on both breast tumors from young women and on number of TN tumors analyzed by aCGH. Further replication studies will need to be performed to confirm these findings. These results can be applied to future studies to increase our understanding of the biology of the different breast cancer subtypes, particularly TN tumors, and differences by race, ultimately leading towards the identification of improved targeted therapeutic strategies and breast cancer survival.
Supplementary Material
References
- 1.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295 (21):2492–2502. doi: 10.1001/jama.295.21.2492. 295/21/2492 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98 (19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. 0932692100 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu K, Lee CH, Tan PH, Tan P. Conservation of breast cancer molecular subtypes and transcriptional patterns of tumor progression across distinct ethnic populations. Clin Cancer Res. 2004;10(16):5508–5517. doi: 10.1158/1078-0432.CCR-04-0085. 10/16/5508 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, Perou CM, Nielsen TO. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14 (5):1368–1376. doi: 10.1158/1078-0432.CCR-07-1658. 14/5/1368 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cdna expression analyses. Int J Cancer. 2005;116 (3):340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 7.Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203 (2):661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- 8.Diaz LK, Cryns VL, Symmans WF, Sneige N. Triple negative breast carcinoma and the basal phenotype: From expression profiling to clinical practice. Adv Anat Pathol. 2007;14(6):419–430. doi: 10.1097/PAP.0b013e3181594733. 00125480-200711000-00004 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (er)-negative, progesterone receptor (pr)-negative, and her2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the california cancer registry. Cancer. 2007;109 (9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 10.Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, Flagg EW, O’Regan RM, Gabram SG, Eley JW. Race and triple negative threats to breast cancer survival: A population-based study in atlanta, ga. Breast Cancer Res Treat. 2009;113 (2):357–370. doi: 10.1007/s10549-008-9926-3. [DOI] [PubMed] [Google Scholar]
- 11.Lund MJ, Butler EN, Bumpers HL, Okoli J, Rizzo M, Hatchett N, Green VL, Brawley OW, Oprea-Ilies GM, Gabram SG. High prevalence of triple-negative tumors in an urban cancer center. Cancer. 2008;113 (3):608–615. doi: 10.1002/cncr.23569. [DOI] [PubMed] [Google Scholar]
- 12.Amend K, Hicks D, Ambrosone CB. Breast cancer in african-american women: Differences in tumor biology from european-american women. Cancer Res. 2006;66 (17):8327–8330. doi: 10.1158/0008-5472.CAN-06-1927. 66/17/8327 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Chlebowski RT, Chen Z, Anderson GL, Rohan T, Aragaki A, Lane D, Dolan NC, Paskett ED, McTiernan A, Hubbell FA, Adams-Campbell LL, Prentice R. Ethnicity and breast cancer: Factors influencing differences in incidence and outcome.[see comment] Journal of the National Cancer Institute. 2005;97 (6):439–448. doi: 10.1093/jnci/dji064. [DOI] [PubMed] [Google Scholar]
- 14.Dayal HH, Power RN, Chiu C. Race and socio-economic status in survival from breast cancer. J Chronic Dis. 1982;35 (8):675–683. doi: 10.1016/0021-9681(82)90020-0. [DOI] [PubMed] [Google Scholar]
- 15.Eley JW, Hill HA, Chen VW, Austin DF, Wesley MN, Muss HB, Greenberg RS, Coates RJ, Correa P, Redmond CK, et al. Racial differences in survival from breast cancer. Results of the national cancer institute black/white cancer survival study. JAMA. 1994;272 (12):947–954. doi: 10.1001/jama.272.12.947. [DOI] [PubMed] [Google Scholar]
- 16.Newman LA, Griffith KA, Jatoi I, Simon MS, Crowe JP, Colditz GA. Meta-analysis of survival in african american and white american patients with breast cancer: Ethnicity compared with socioeconomic status. J Clin Oncol. 2006;24 (9):1342–1349. doi: 10.1200/JCO.2005.03.3472. 24/9/1342 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Shavers VL, Harlan LC, Stevens JL. Racial/ethnic variation in clinical presentation, treatment, and survival among breast cancer patients under age 35. Cancer. 2003;97 (1):134–147. doi: 10.1002/cncr.11051. [DOI] [PubMed] [Google Scholar]
- 18.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes & Cancer. 2006;45 (11):1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 19.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, Chen F, Feiler H, Tokuyasu T, Kingsley C, Dairkee S, Meng Z, Chew K, Pinkel D, Jain A, Ljung BM, Esserman L, Albertson DG, Waldman FM, Gray JW. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies.[see comment] Cancer Cell. 2006;10 (6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, McLennan J, Ziegler J, Chin K, Devries S, Feiler H, Gray JW, Waldman F, Pinkel D, Albertson DG. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, Malone KE, Daling JR, Delrow JJ, Trask BJ, Hsu L, Porter PL. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Research. 2004;64 (23):8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 22.Brinton LA, Potischman NA, Swanson CA, Schoenberg JB, Coates RJ, Gammon MD, Malone KE, Stanford JL, Daling JR. Breastfeeding and breast cancer risk. Cancer Causes Control. 1995;6 (3):199–208. doi: 10.1007/BF00051791. [DOI] [PubMed] [Google Scholar]
- 23.Porter PL, Lund MJ, Lin MG, Yuan X, Liff JM, Flagg EW, Coates RJ, Eley JW. Racial differences in the expression of cell cycle-regulatory proteins in breast carcinoma. Cancer. 2004;100 (12):2533–2542. doi: 10.1002/cncr.20279. [DOI] [PubMed] [Google Scholar]
- 24.Glogovac JK, Porter PL, Banker DE, Rabinovitch PS. Cytokeratin labeling of breast cancer cells extracted from paraffin-embedded tissue for bivariate flow cytometric analysis. Cytometry. 1996;24 (3):260–267. doi: 10.1002/(SICI)1097-0320(19960701)24:3<260::AID-CYTO9>3.0.CO;2-L [pii] 10.1002/(SICI)1097-0320(19960701)24:3<260::AID-CYTO9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Loo LW, Ton C, Wang YW, Grove DI, Bouzek H, Vartanian N, Lin MG, Yuan X, Lawton TL, Daling JR, Malone KE, Li CI, Hsu L, Porter PL. Differential patterns of allelic loss in estrogen receptor-positive infiltrating lobular and ductal breast cancer. Genes Chromosomes Cancer. 2008;47 (12):1049–1066. doi: 10.1002/gcc.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28(4):327–334. doi: 10.1038/ng569. ng569 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Yang MC, Ruan QG, Yang JJ, Eckenrode S, Wu S, McIndoe RA, She JX. A statistical method for flagging weak spots improves normalization and ratio estimates in microarrays. Physiol Genomics. 2001;7(1):45–53. doi: 10.1152/physiolgenomics.00020.2001. 7/1/45 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Hsu L, Self SG, Grove D, Randolph T, Wang K, Delrow JJ, Loo L, Porter P. Denoising array-based comparative genomic hybridization data using wavelets. Biostatistics. 2005;6 (2):211–226. doi: 10.1093/biostatistics/kxi004. 6/2/211 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Trivers KF, Lund MJ, Porter PL, Liff JM, Flagg EW, Coates RJ, Eley JW. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 2009;20 (7):1071–1082. doi: 10.1007/s10552-009-9331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joosse SA, van Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, Ligtenberg MJ, Wessels LF, Axwijk P, Verhoef S, Hogervorst FB, Nederlof PM. Prediction of brca1-association in hereditary non-brca1/2 breast carcinomas with array-cgh. Breast Cancer Res Treat. 2009;116 (3):479–489. doi: 10.1007/s10549-008-0117-z. [DOI] [PubMed] [Google Scholar]
- 31.Wessels LF, van Welsem T, Hart AA, van’t Veer LJ, Reinders MJ, Nederlof PM. Molecular classification of breast carcinomas by comparative genomic hybridization: A specific somatic genetic profile for brca1 tumors. Cancer Res. 2002;62 (23):7110–7117. [PubMed] [Google Scholar]
- 32.Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, Schoenmakers E, Viens P, Jacquemier J, Birnbaum D, Bertucci F, Chaffanet M. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67 (24):11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. 67/24/11565 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, Kang JJ, Bae JY, Jeon YK, Park IA, Nicolau M, Jeffrey SS, Noh DY. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47 (6):490–499. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 34.Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, Langerod A, Han W, Noh DY, Huntsman DG, Jeffrey SS, Borresen-Dale AL, Pollack JR. Camk1d amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol. 2008;2 (4):327–339. doi: 10.1016/j.molonc.2008.09.004. S1574-7891(08)00124-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME, et al. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150 (3):534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- 36.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: A meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83(12):1688–1695. doi: 10.1054/bjoc.2000.1522. S0007092000915222 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, Esterni B, Geneix J, Finetti P, Zemmour C, Viens P, Charafe-Jauffret E, Jacquemier J, Birnbaum D, Chaffanet M. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. 1471-2407-6-245 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, Palacios J, Nathanson KL, Garcia MJ, Benitez J. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals tfdp1 and cul4a as likely candidate target genes. Breast Cancer Res. 2009;11 (6):R86. doi: 10.1186/bcr2456. bcr2456 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abba MC, Fabris VT, Hu Y, Kittrell FS, Cai WW, Donehower LA, Sahin A, Medina D, Aldaz CM. Identification of novel amplification gene targets in mouse and human breast cancer at a syntenic cluster mapping to mouse ch8a1 and human ch13q34. Cancer Res. 2007;67 (9):4104–4112. doi: 10.1158/0008-5472.CAN-06-4672. 67/9/4104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27 (2):949–952. [PubMed] [Google Scholar]
- 41.Parise CA, Bauer KR, Brown MM, Caggiano V. Breast cancer subtypes as defined by the estrogen receptor (er), progesterone receptor (pr), and the human epidermal growth factor receptor 2 (her2) among women with invasive breast cancer in california, 1999–2004. Breast J. 2009;15 (6):593–602. doi: 10.1111/j.1524-4741.2009.00822.x. TBJ822 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. S0092867404000455 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135 (4):1079–1099. doi: 10.1053/j.gastro.2008.07.076. S0016-5085(08)01451-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albain KS, Unger JM, Crowley JJ, Coltman CA, Jr, Hershman DL. Racial disparities in cancer survival among randomized clinical trials patients of the southwest oncology group. J Natl Cancer Inst. 2009;101 (14):984–992. doi: 10.1093/jnci/djp175. djp175 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menashe I, Anderson WF, Jatoi I, Rosenberg PS. Underlying causes of the black-white racial disparity in breast cancer mortality: A population-based analysis. J Natl Cancer Inst. 2009;101 (14):993–1000. doi: 10.1093/jnci/djp176. djp176 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, Tsai YC, Williams EH, Lee DH, Stephens RM, Weissman AM, Ambs S. Differences in the tumor microenvironment between african-american and european-american breast cancer patients. PLoS One. 2009;4 (2):e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.