Abstract
Background
Staphylococcus epidermidis (SE) has emerged as one of the most important causes of nosocomial infections. The SaeRS two-component signal transduction system (TCS) influences virulence and biofilm formation in Staphylococcus aureus. The deletion of saeR in S. epidermidis results in impaired anaerobic growth and decreased nitrate utilization. However, the regulatory function of SaeRS on biofilm formation and autolysis in S. epidermidis remains unclear.
Results
The saeRS genes of SE1457 were deleted by homologous recombination. The saeRS deletion mutant, SE1457ΔsaeRS, exhibited increased biofilm formation that was disturbed more severely (a 4-fold reduction) by DNase I treatment compared to SE1457 and the complementation strain SE1457saec. Compared to SE1457 and SE1457saec, SE1457ΔsaeRS showed increased Triton X-100-induced autolysis (approximately 3-fold) and decreased cell viability in planktonic/biofilm states; further, SE1457ΔsaeRS also released more extracellular DNA (eDNA) in the biofilms. Correlated with the increased autolysis phenotype, the transcription of autolysis-related genes, such as atlE and aae, was increased in SE1457ΔsaeRS. Whereas the expression of accumulation-associated protein was up-regulated by 1.8-fold in 1457ΔsaeRS, the expression of an N-acetylglucosaminyl transferase enzyme (encoded by icaA) critical for polysaccharide intercellular adhesin (PIA) synthesis was not affected by the deletion of saeRS.
Conclusions
Deletion of saeRS in S. epidermidis resulted in an increase in biofilm-forming ability, which was associated with increased eDNA release and up-regulated Aap expression. The increased eDNA release from SE1457ΔsaeRS was associated with increased bacterial autolysis and decreased bacterial cell viability in the planktonic/biofilm states.
Background
The opportunistic pathogen Staphylococcus epidermidis has emerged as an important etiologic agent of nosocomial infections. The ability to form biofilms on the surfaces of medical devices is an important component of S. epidermidis pathogenicity. Biofilm resistance to antibiotics and host defense mechanisms are often regulated by two-component signal transduction systems (TCSs) [1].
Biofilm formation proceeds in two distinct developmental phases: primary attachment of staphylococcal cells to a polystyrene surface followed by bacterial accumulation in multiple layers [2]. The initial adhesion of bacterial cells to a polymer surface is influenced by a variety of factors, including AtlE, Embp, and other staphylococcal surface-associated proteins. During the bacterial accumulation phase in S. epidermidis, biofilm formation is mediated by extracellular polysaccharides and proteins, such as polysaccharide intercellular adhesin (PIA) [3] and accumulation-associated protein (Aap) [4]. In addition to extracellular polysaccharides and proteins, extracellular DNA (eDNA) is a matrix component that is critical for bacterial attachment during the initial stage of biofilm formation [5,6]. Extracellular DNA release from S. epidermidis is related to AtlE-mediated bacterial autolysis [7]. Another autolysin recently identified in S. epidermidis, Aae, also has bacteriolytic activities and adhesive properties [8].
TCSs regulate bacterial adaptation, survival, virulence and biofilm formation [9-12]. TCSs comprise a membrane-associated histidine kinase and a cytoplasmic response regulator. Overall, 16 or 17 TCSs have been identified in the genomes of S. epidermidis ATCC12228 or ATCC35984 [13,14]. In S. epidermidis, the TCS agrC/agrA has been proven to negatively regulate biofilm formation [15,16]. In a previous study of the S. epidermidis saeRS TCS, a saeR deletion mutant exhibited a lower anaerobic growth rate, a significantly reduced rate of nitrate utilization and a slightly higher biofilm-forming ability compared to the parental strain [11]. In S. aureus, the saeRS TCS influences biofilm formation [17] and the expression of virulence-associated factors, such as protein A, α- and β-hemolysins, and coagulase [18]. However, whether saeRS regulates S. epidermidis autolysis and biofilm formation remains unclear.
In the present work, we constructed a SE1457ΔsaeRS mutant with deletion of the genes that encode both the histidine kinase (SaeS) and the response regulator (SaeR) by homologous recombination. The effects of the saeRS deletion on S. epidermidis autolysis, eDNA release, bacterial cell viability, and biofilm formation were investigated.
Methods
Bacterial strains, plasmids, and media
The bacterial strains and plasmids used in this study are listed in Table 1. S. epidermidis cells were grown at 37°C in BM medium (per liter = tryptone 10 g, yeast extract 5 g, NaCl 5 g, K2HPO4 1 g, and glucose 1 g) or tryptic soy broth (TSB) (Oxiod, Basingstoke, Hampshire, England) supplemented with antibiotics when necessary. Antibiotics were used at the following concentrations: erythromycin at 2.5 μg/mL, chloramphenicol at 10 μg/mL, spectinomycin (spc) at 300 μg/mL for S. epidermidis and S. aureus, and ampicillin at 100 μg/mL for E.coli.
Table 1.
Bacterial strains and plasmids used in the present study
| Strain or plasmid | Relevant genotype or characteristic | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | λ- ϕ80dlacΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK- mK-) supE44 thi-1 gyrA relA1 | [49] |
| SE1457 | Biofilm positive strain | [50] |
| S. aureus RN4220 | Restriction-negative, modification-positive isolate | [51] |
| SE1457ΔsaeRS | saeRS deletion mutant of strain 1457, Spcr | This study |
| SE1457saec | 1457ΔsaeRS complemented with saeRS | This study |
| Plasmids | ||
| pET-28a(+) | Expression vector, KanR | Novagen |
| pBT2 pCX19 |
Temperature-sensitive E. coli- Staphylococcus shuttle vector. Apr (E. coli) Cmr (Staphylococcus) Derivate of pCX15 |
[52] [53] |
| pMAD | Escherichia coli/Staphylococcus Shuttle vector | [54] |
| pMAD-saeRS | Vector for allelic gene replacement of saeRS in S. epidermidis | This study |
| pBT2-saeRS | Vector for complementation of saeRS in S. epidermidis 1457ΔsaeRS | This study |
*Abbreviations: Amp, ampicillin; Cm, chloramphenicol; Em, erythromycin.
Determination of the growth curves of S. epidermidis strains
The aerobic growth curves of S. epidermidis strains were determined by measuring the optical density (OD600) as described previously [11]. Briefly, overnight cultures were diluted 1:200 and incubated at 37°C with shaking at 220 rpm. The OD600 of the culture were measured at 60 min intervals for 12 h. At 6, 12, and 24 h time points, colony forming units on TSA plates were further counted with serial dilutions of each sample plated on 6 agar plates. For anaerobic growth conditions, bacteria were cultured in the Eppendorf tubes which were filled up with the TSB medium and sealed with wax.
Detection of biofilm formation
The biofilm-forming ability of S. epidermidis strains was determined by the microtiter-plate test as described by Christensen [19,20]. Briefly, overnight cultures of S. epidermidis were diluted 1:200 and inoculated into wells of polystyrene microtiter plates (200 μL per well) at 37°C for 24 h. At different time points (0, 6, 12, and 24 h), DNase I (Takara Bio, Kyoto, Japan) was added at 28 U/200 μL. After incubation, the wells were gently washed three times with 200 μL PBS and stained with 2% crystal violet for 5 min. Absorbance was determined at 570 nm.
To determine whether saeRS affects cell death in biofilms, S. epidermidis cells were cultivated in FluoroDish (FD35-100, WPI, USA) as previously described [7]. Briefly, overnight cultures of S. epidermidis grown in TSB medium were diluted 1:200, inoculated into dishes (2 mL per dish), and then incubated at 37°C for 24 h. The dishes were then carefully washed with PBS and stained with a LIVE/DEAD kit (containing SYTO9 and PI, Invitrogen Molecular Probes, USA) following the manufacturer's instructions. SYTO9 stains viable bacteria green while PI stains dead bacteria red. Biofilms of S. epidermidis 1457 and SE1457ΔsaeRS were observed under a Leica TCS SP5 confocal laser scanning microscope (CLSM) using a 63 ×(zoom ×3) objective lens and the Z-stack composite confocal photomicrographs of viable cells, dead cells, and both cells (viable & dead) were generated by Leica LAS AF softwear (version 1.8.1). The fluorescence quantity of each stack was determained using ImageJ software.
Electron microscopy
For scanning electron microscopy (SEM), biofilms were grown in TSB for 24 h at 37°C with fragments of an introvenous catheter, rinsed with PBS three times, fixed with a 2% (w/v) solution of glutaraldehyde prepared in phosphate-buffered saline, and then observed under a TECNAI- 12 field emission source instrument (Philips, Eindhoven, The Netherlands).
For transmission electron microscopy (TEM), bacteria grown for 24 h were stained by mixing with a 1% (w/v) solution of uranyl acetate on an electron microscope grid covered with a carbon-coated Formvar film. S. epidermidis cells were observed using a Hitachi S-520 electron microscope (Hitachi, Tokyo, Japan).
RNA extraction and microarray analysis
Overnight cultures of S. epidermidis 1457 and 1457 ΔsaeSR were diluted 1:200 into fresh TSB and grown at 37°C to an OD600 of 3.0 (mid-exponential growth). Eight millilitres of bacterial cultures were pelleted, washed with ice-cold saline, and then homogenized using 0.1 mm Ziconia-silica beads in Mini-Beadbeater (Biospec) at a speed of 4800 rpm. The bacterial RNA was isolated using a QIAGEN RNeasy kit according to the standard QIAGEN RNeasy protocol.
The microarray was manufactured by in situ synthesis of 14,527, 60-mer long oligonucleotide probes (Agilent, Palo Alto, CA, USA), selected as previously described [21]. It covers > 95% of all ORFs annotated in strains ATCC12228 (GeneBank accession number NC_004461), ATCC35984 (GeneBank accession number NC_002976), SE1457 (unpublished sequence). Preparations of 10 μg of total S. epidermidis RNA were labeled by Cy-3 dCTP (Perkin-Elmer) using the SuperScript II (Invitrogen, Basel, Switzerland) and purified as previously described [22]. Pool of purified genomic DNA from the reference sequenced strains used for the design of the microarray was labeled with Cy-5 dCTP [21] and used for microarray normalization [23]. Mixtures of Cy5-labeled DNA and Cy3-labeled cDNA were hybridized and scanned as previously described [22] in a dedicated oven. Fluorescence intensities were quantified using Feature Extraction software (Agilent, version 8). Green (Cy3) and red (Cy5) feature extraction processed data were imported in the Partek genomics suite software (Partek Incorporated. St. Louis, USA). Data were normalized to baseline using red channel data as control [23] and mean to estimate baseline. Variance analysis of three biological replicates was processed with a false discovery rate value of 5% (P value cutoff; 0.05) and an arbitrary threshold of 3.0 fold for defining significant differences in expression ratios. The complete raw microarray dataset has been posted on the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), accession number GPL13532 for the platform design and GSE29309 for the original dataset.
Quantitative real-time PCR analysis
DNase-treated RNA was reverse transcribed using M-MLV and a hexamer random primer mix. Appropriate concentration of cDNA sample was then used for real-time PCR using an ABI 7500 real-time PCR detection system, gene-specific primers, and the SYBR Green I mixture (Takara, Dalian, China). Relative expression levels were determined by comparison to the level of gyrB expression in the same cDNA preparations. Gene-specific primers were designed according to GenBank gene sequences (Accession number: CP000029, Table 2). All samples were analyzed in triplicate and normalized against gyrB expression.
Table 2.
Oligonucleotide primers
| Target gene | GenBank accession no. | Primer* | Primer sequence | Location |
|---|---|---|---|---|
| Oligonucleotide primers used for RT real-time PCR | ||||
| gyrB | 57636585 | gyrB-F | CTTATATGAGAATCCATCTGTAGG | 1110-1263 |
| gyrB-R | AGAACAATCTGCCAATTTACC | |||
| lrgA | 57636056 | lrgA-F | TGGACTTGTACTATTATTTATCGC | 165-309 |
| lrgA-R | AAGGATTGGTAAAGAGTTAATGAC | |||
| lytS | 57636054 | lytS-F | CTGTTCAAGATAATGGTCAAGG | 1535-1680 |
| lytS-R | CAGTGCCGATGTTGTTCC | |||
| serp0043 | 57636640 | serp0043-F | CAAGCACAAGCGTCTTCATC | 73-236 |
| serp0043-R | ACTCTTTCACCATTATTTGTTTCAG | |||
| glpQ | 57637130 | glpQ-F | CCGTTACACTGGGTTTAGC | 41-221 |
| glpQ-R | TTACCACTTACTGAGTCTGATTC | |||
| arlR | 57636010 | arlR-F | AGAGAATGATGGAAAGGCAGGT | 90-253 |
| arlR-R | ATGTCTCGCTTTTCGCAGTAAT | |||
| atlE | 57637180 | atlE-F | AACAACCACAGAATCAGTCTAATC | 92-237 |
| atlE-R | TTGAACTTGGGTAGGGTCTTG | |||
| aae | 57637180 | aae-F | AACAAATTGATAAAGCAACG | 1970-2186 |
| aae-R | GTTGTCTTTCCTTTAGTGTC | |||
| aap | 57636451 | aap-F | AATAGAACCTACAACTTCAGAACC | 945-1039 |
| aap-R | TGTTATTGGATGAACTATCAGCAG | |||
| icaA | 57636387 | icaA-F | GGTTGTATCAAGCGAAGTC | 556-754 |
| icaA-R | ACATCCAGCATAGAGCAC | |||
| saeS | 57636974 | saeS-F | GGTATCGTTCCAGAACTTCAATC | 757-881 |
| saeS-R | ATTTGTTGTGCTAACTCATTTGC | |||
| saeR | 57636975 | saeR-F | CTCAAGAACATGACACGATATACG | 245-354 |
| saeR-R | TCTAGCGAGAAGGTTATTAGTACG | |||
| saeQ | 57636990 | saeQ-F | GCAAGTTTCTTTGGAGCCTTC | 268-447 |
| saeQ-R | CTTATCTTCACCTCGGTTATTACG | |||
| saeP | 57636991 | saeP-F | CTAACTCGGAAAGCGATCAC | 71-258 |
| saeP-R | GTCTGGACCTTTAGAAGATTTG | |||
| Oligonucleotide primers used for eDNA quantification | ||||
| gyrA | 57636584 | gyrA-F | CCTTATGAAACTCGGAGATGG | 2382-2489 |
| gyrA-R | TCAGTAGTAGTAGATTGTTGCG | |||
| lysA | 57637514 | lysA-F | TGACAATGGGAGGTACAAGC | 32-107 |
| lysA-R | TGGTCTTCATCGTAAACAATCG | |||
| serp0306 | 57636873 | serp0306-F | ATGCCACATCCACGAAAGA | 203-381 |
| serp0306-R | TGTAACTGACAATGCCCAATC | |||
| leuA | 57638228 | leuA-F | GTGAACGGTATTGGTGAAAGAG | 685-762 |
| leuA-R | GTGGTCCTTCCTTACATATAAAGC | |||
F, forward primer; R, reverse primer
Determination of Triton X-100-induced autolysis
Triton X-100-induced autolysis was performed to determine the potential role of saeRS in autolysis regulation in S. epidermidis, as described elsewhere [24-26]. SE1457ΔsaeRS, SE1457, and SE1457saec cells were diluted in TSB containing 1 M NaCl, grown to mid-exponential phase (OD600 = ~0.6-0.8), washed twice in cold sterile distilled water, resuspended in the same volume of 0.05 M Tris-HCl containing 0.05% Triton X-100 (pH 7.2), and incubated at 30°C. OD600 was measured every 30 min. The Triton X-100-induced autolysis rate was calculated as follows: Ra = OD0-ODt/OD0.
Zymogram
The murein hydrolase activities of SE1457, SE1457ΔsaeRS, SE1457saec, and SE1457ΔatlE were detected by zymographic analysis as described elsewhere [26,27]. Extracts from lysostaphin- and SDS-treated S. epidermidis (Ex-Lys and Ex-SDS, respectively) and the concentrated supernatants of the bacterial culture (Ex-Sup) were used to analyze the murein hydrolase activities of each strain. Ex-Lys were obtained by treating S. epidermidis cells with 30 μg/mL of lysostaphin for 2 h at 37°C and subsequently centrifuged at 8,000 g for 30 min. Ex-SDS were obtained by treating S. epidermidis cells in 100 μL of 100 mM phosphate buffer containing 4% SDS at 37°C for 30 min and centrifuged (10,000 g) for 10 min. Ex-Sup were acquired by concentrating supernatants of overnight S. epidermidis cultures to 10% initial volume using a centrifugal filter device (Millipore, Billerica, MA).
S. epidermidis cell extracts were separated on a SDS-PAGE gel (10% acrylamide, pH 8.8) containing 0.2% (wt/vol) lyophilized Micrococcus luteus (M. luteus) or S. epidermidis cells. After electrophoresis, the gels were washed four times with distilled water for 30 min at room temperature, incubated in 25 mM Tris-HCl containing 1% Triton X-100 (pH 8.0) at 37°C for 6 h, and then stained with methylene blue.
Quantification of eDNA
Extracellular DNA isolation from biofilms was performed as described by Rice et al. [7,19,28]. Briefly, SE1457, SE1457ΔsaeRS, and SE1457saec biofilms (grown for 24 h) were chilled at 4°C for 1 h and treated with 1.0 μL of 0.5 M EDTA. Supernatants were discarded, and the unwashed biofilms were resuspended in 50 mM TES buffer (Tris-HCl (pH 8.0), 10 mM ETDA, 500 mM NaCl). Extracellular DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1), precipitated with 100% ethanol, and dissolved in 20 μL of TE buffer.
Extracellular DNA was quantified by qPCR using gyrA (gyrase A), serp0306 (ferrichrome transport ATP-binding protein A), lysA (diaminopimelate decarboxylase A), and leuA (2-isopropylmalate synthase) primers as listed in Table 2. Each sample was diluted to 1:10, and PCRs were performed with SYBR Premix Ex TaqTM (TaKaRa, Japan) and primers (2 μM), according to the manufacturer's recommendations. The average OD600 of each unwashed biofilm was determined for calculating potential differences in biomass. The amount of eDNA per relative biomass of each biofilm was then calculated as follows: total eDNA (ng)/ relative OD600.
Initial bacterial attachment assays
Initial cell attachment was detected as described by Heilmann et al. [29]. Briefly, mid-exponential phase cells were diluted to OD600 = 0.1 in PBS and then incubated in wells (1 mL per well) of cell-culture polystyrene chambers (Nunc, Denmark) with DNase I (140 U/mL) for 2 h at 37°C. Numbers of attached cells were counted under a microscope. Three independent experiments were carried out.
Detection of Aap expression
Concentrations of lysostaphin-treated whole bacterial proteins from SE1457ΔsaeRS, SE1457, and SE1457saec were determined by the Bradford method. For the detection of Aap in all samples by Western blot assay, proteins were separated on a 7% SDS-PAGE gel and then transferred to polyvinylidene fluoride (PVDF) membranes (Whatman, D-37586 Dassel, Germany) by electroblotting with a Mini-Transfer system (Bio-Rad, Mississauga, Canada) at 200 mA for 2 h (4°C). Monoclonal antibodies against the Aap B-repeat region (prepared by Abmart, Shanghai, China) were diluted 1:6000, and horseradish peroxidase-conjugated goat anti-mouse IgG antibodies (Sino-American Biotech) were diluted 1:2000. The gray scale of the bands corresponding to Aap was quantified using the Quantity-one software (Bio-Rad, USA).
Semi-quantitative detection of PIA
PIA was detected as described elsewhere [30-32]. Briefly, S. epidermidis strains were grown in 6-well plates (Nunc, DK-4000 Roskitde, Denmark) under static conditions at 37°C for 24 h. The cells were scraped off and resuspended in 0.5 M EDTA (pH 8.0). The supernatant was treated with proteinase K (final concentration 4 mg/mL; Roche, MERCK, Darmstadt, Germany) for 3 h (37°C). Serial dilutions of the PIA extract were then transferred to a nitrocellulose membrane (Millipore, Billerica, MA) using a 96-well dot blot vacuum manifold (Gibco). The air-dried membrane was blocked with 3% (wt/vol) bovine serum albumin and subsequently incubated with 3.2 μg/mL wheat germ agglutinin coupled to horseradish peroxidase (WGA-HRP conjugate; Lectinotest Laboratory, Lviv, Ukraine) for 1 h. Horseradish peroxidase (HRP) activity was visualized via chromogenic detection. The gray scale of the spots corresponding to PIA was quantified using the Quantity-one software.
Statistical analysis
Experimental data were analyzed with the SPSS software and compared using the Student's t-test. Differences with a P value of < 0.05 were considered statistically significant.
Results
Effect of saeRS deletion on S. epidermidis biofilm formation
In order to explore the influence of saeR and saeS on S. epidermidis biofilm formation, an S. epidermidis 1457ΔsaeRS mutant (SE1457ΔsaeRS) and a complemented strain (SE1457saec) were constructed using the shuttle plasmids pMAD and pBT2, respectively. The biofilm-forming ability of SE1457ΔsaeRS on polystyrene plates was higher compared to the parental strain. Although it did not reach the level of the wild-type strain, complementation of saeRS resulted in decreased biofilm formation (Student's t-test, P < 0.05) (Figure 1). The growth curves of SE1457ΔsaeRS and the parental strain were similar in either aerobic or anaerobic growth conditions (Additional file 1: Fig. S1).
Figure 1.
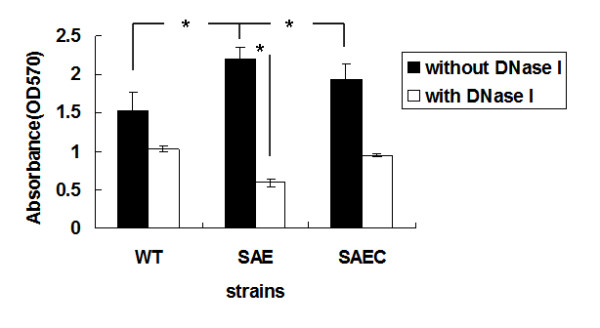
Effect of DNaseI on SE1457ΔsaeRS, SE1457, and SE1457saec biofilm formation. SE1457ΔsaeRS, SE1457, and SE1457saec biofilms were washed and then stained with crystal violet. Their retained biomass was quantified by measuring the absorbance of each well at 570 nm. Biofilms were formed in the absence (black bars) or presence of DNase I (28 U/200 μL/well) (white bars). Mean values and standard deviations from three independent experiments are shown. (*), P < 0.05. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec.
Scanning electron microscopy (SEM) of biofilms on catheters showed that SE1457ΔsaeRS biofilms contained more extracellular matrix compared to SE1457 and SE1457saec biofilms (Figure 2A). In planktonic cultures, intercellular adhesion of the SE1457ΔsaeRS and the wild-type strain was observed using transmission electron microscopy (TEM). While thread-like material between SE1457ΔsaeRS cells was observed, such material was rarely found between parental strain cells (Figure 2B).
Figure 2.
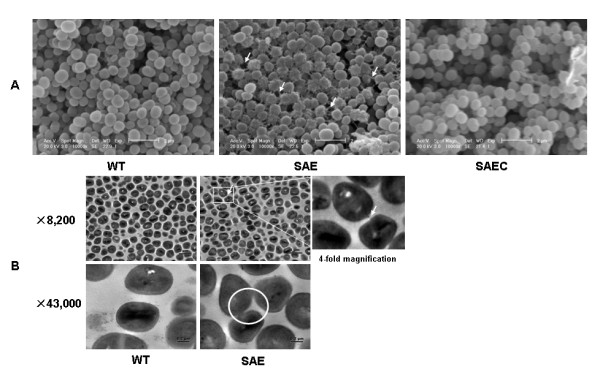
SEM and TEM observations of SE1457ΔsaeRS and wild-type strain. (A) Biofilms of SE1457ΔsaeRS, SE1457, and SE1457saec after 24 h of growth on hydroxyapatite disks were observed by SEM. Arrows show the extracellular polymeric substances (EPSs) (10,000× magnification). (B) Planktonic cells of SE1457ΔsaeRS and SE1457 cultured for 24 h were observed by TEM. Cell-cell accumulations in SE1457ΔsaeRS are circled; arrow indicates the thread-like material linking neighboring cells. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec.
Effect of saeRS deletion on the autolysis of S. epidermidis
To examine the effect of saeRS deletion on autolysis, Triton X-100-induced autolysis of SE1457ΔsaeRS, SE1457, and SE1457saec was analyzed. Bacterial cells were harvested at the mid-exponential phase grown in TSB medium containing 1 M NaCl. Following the addition of 0.05% Triton X-100, SE1457ΔsaeRS cultures exhibited a much higher autolysis rate (approximately 3-fold) compared to the wild-type strain; decreased autoloysis was partially restored in the complementation strain SE1457saec (Figure 3).
Figure 3.
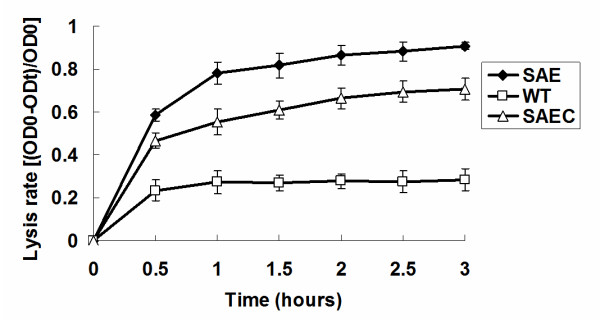
Effect of saeRS deletion on Triton X-100-induced autolysis. SE1457ΔsaeRS, SE1457, and SE1457saec cells were diluted in TSB medium containing 1 M NaCl, grown to mid-exponential phase (OD600 = ~0.6-0.8), and resuspended in the same volume of 0.05 M Tris-HCl solution containing 0.05% Triton X-100 (pH 7.2). OD600 readings were measured every 30 min. The autolysis rate induced by Triton X-100 was calculated as follows: lysis rate = OD0 - ODt/OD0. The experiments were carried out in triplicate independently. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec.
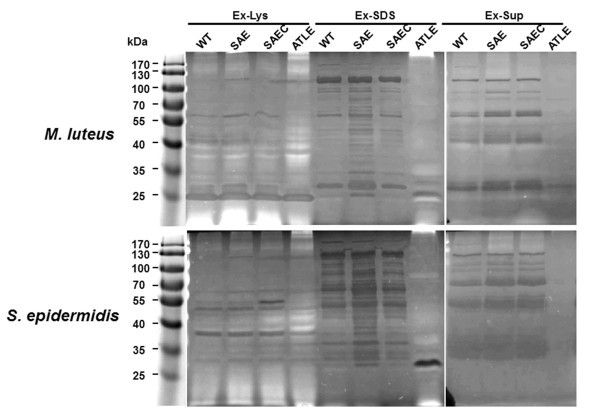
The effect of saeRS deletion on murein hydrolase activity was determined by zymographic analysis using lyophilized Micrococcus luteus (M. luteus) or S. epidermidis cells as substrates [26,33]. Briefly, extracts from lysostaphin- and SDS-treated S. epidermidis (Ex-Lys and Ex-SDS, respectively) cells and concentrated supernatants of the bacterial culture (Ex-Sup) were used to assess the murein hydrolase activities of each strain. As a control, extracts from the S. epidermidis atlE deletion mutant SE1457ΔatlE were used and resulted in only one lytic band (~30 kDa). In contrast, extracts from SE1457, SE1457ΔsaeRS and SE1457saec displayed multiple bacteriolytic bands. The zymogram profiles of Ex-SDS from SE1457ΔsaeRS extracts showed more lytic bands (from 25 to 90 kDa) compared to the zymogram profiles of SE1457 and SE1457saec extracts, indicating that autolysins may contribute to the increased autolysis of the mutant strain. The Ex-Lys and Ex-Sup zymogram profiles of SE1457ΔsaeRS were similar to the profiles observed for SE1457 and SE1457saec (Figure 4).
Figure 4.
Zymographic analysis of autolytic enzyme extracts. Bacteriolytic enzyme profiles were analyzed on SDS gels (10% separation gel) containing lyophilized M. luteus cells (0.2%) or S. epidermidis cells (0.2%) as substrates. After electrophoresis, the gels were washed for 30 min in distilled water, incubated for 6 h at 37°C in a buffer containing Triton X-100, and then stained with methylene blue. The S. epidermidis atlE mutant was used as a negative control. Bands with lytic activity were observed as clear zones in the opaque gel. The clear zones appeared as dark bands after photography against a dark background. The molecular mass standard is shown on the left of the gels. Ex-Lys, cell-wall extracts of lysostaphin-treated S. epidermidis; Ex-SDS, cell-wall extracts of SDS-treated S. epidermidis; Ex-Sup, concentrated S. epidermidis culture supernatants; WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec; ATLE, SE1457ΔatlE.
Effect of saeRS deletion on S. epidermidis viability in planktonic and biofilm states
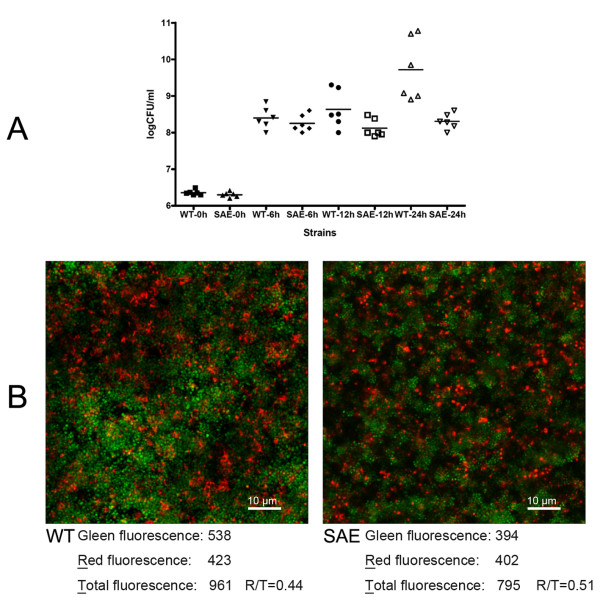
To investigate whether the increased autolysis that resulted from saeRS deletion affected S. epidermidis cell viability, colony-forming unit (CFU) counts of the SE1457 and SE1457ΔsaeRS strains in the planktonic state were determined. Cultures were inoculated with approximately 104 CFU/mL of each strain and incubated under normal conditions. At 6 h, SE1457ΔsaeRS and SE1457 had log CFU/mL counts of 8.2 of and 8.4, respectively. CFU counts were also similar at 12 h post-inoculation, with log CFU/mL counts of 8.1 and 8.6 for SE1457ΔsaeRS and SE1457 respectively. However, after 24 h, SE1457ΔsaeRS cultures had a lower CFU count (8.3 log CFU/mL) compared to the wild-type strain (9.7 log CFU/mL) (P = 0.002) (Figure 5A).
Figure 5.
Viability of S. epidermidis 1457 in biofilms and the planktonic state. (A) CFU counts of SE1457ΔsaeRS and SE1457. After 0, 6, 12, and 24 h of incubation, CFUs for SE1457 and SE1457ΔsaeRS cultures were calculated using serial dilutions of each sample plated on 6 agar plates. (B) CLSM images of S. epidermidis biofilms. SE1457 and SE1457ΔsaeRS were incubated in glass-bottomed cell culture dishes. After incubation at 37°C for 24 h, SE1457ΔsaeRS and SE1457 cells in biofilms were stained with LIVE/DEAD reagents that indicate viable cells by green fluorescence (SYTO9) and dead cells by red fluorescence (PI). Results depict a stack of images taken at approximately 0.3 μm depth increments and represent one of the three experiments. Fluorescence intensities were quantified using ImageJ software. WT, SE1457; SAE, SE1457ΔsaeRS.
The viability of SE1457ΔsaeRS and the wild-type strain in 24 h biofilm was determined by confocal laser scanning microscopy (CLSM) with LIVE/DEAD staining [34]. More dead cells were observed in the SE1457ΔsaeRS biofilm compared to the wild-type strain (Figure 5B).
Effect of saeRS deletion on eDNA release from S. epidermidis
Extracellular DNA is an important component of the S. epidermidis biofilm matrix [7,35], and its relative concentration in 24 h biofilms formed by SE1457, SE1457ΔsaeRS and SE1457saec was measured utilizing qPCR for gyrA, lysA, serp0306, and leuA [19,28]. Extracellular DNA concentrations were increased in the SE1457ΔsaeRS biofilms compared to the complementation strain and the wild-type strain (Figure 6).
Figure 6.
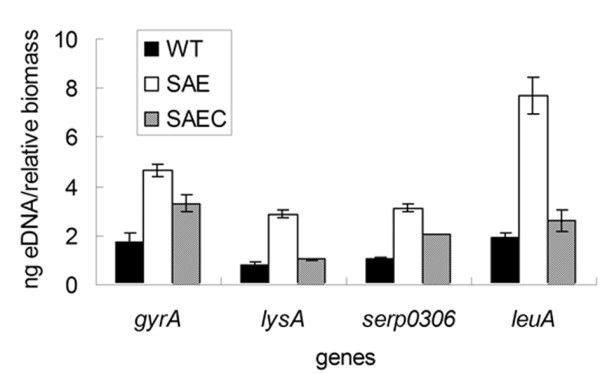
Quantification of eDNA in SE1457ΔsaeRS, SE1457, and SE1457saec biofilms. eDNA was extracted from the unwashed 24 h biofilms of SE1457ΔsaeRS (white bars), SE1457 (black bars), and SE1457saec (gray bars). The eDNA in each biofilm was quantified by qPCR using primers specific for gyrA, serp0306, lysA, and leuA [19,28]. The quantity of eDNA was calculated as follows: total eDNA (ng)/relative OD600. Results represent the mean ± SD of three independent experiments. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec.
When DNase I (28 U/200 μL/well) was added prior to biofilm formation, the biomass of the SE1457ΔsaeRS biofilms was decreased by 4-fold (P < 0.05); in contrast, the biomasses of SE1457 and SE1457saec biofilms were decreased by 1.5-fold (Figure 1).
Effect of eDNA release on SE1457ΔsaeRS primary attachment of SE1457ΔsaeRS
Extracellular DNA is a critical component for bacterial adhesion during the initial stage of biofilm development [5,6]. S. epidermidis cells attached to the polystyrene surface were counted under a microscope at 400× magnification. While 6.8 × 102, 1.2 × 103, and 4.2 × 102 cells per field were adhered for SE1457, SE1457ΔsaeRS, and SE1457saec strains, respectively, few attached SE1457ΔatlE cells were observed. When DNase I (140 U/mL) was added at the time of the attachment assay, SE1457ΔsaeRS cell attachment was significantly reduced by 85%. In contrast, following DNase I addition SE1457 and SE1457saec attachment was reduced by 31% and 48%, respectively (Figure 7).
Figure 7.
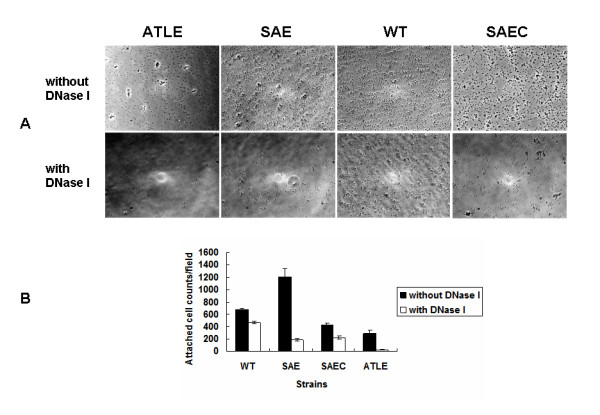
S. epidermidis attachment to polystyrene surfaces in the presence or absence of DNase I. (A) Attached SE1457ΔatlE, SE1457ΔsaeRS, SE1457 and SE1457saec cells were observed by microscopy. Briefly, cell suspensions from the mid-exponential phase were diluted to OD600 = 0.1 in PBS and then incubated in wells (1 mL per well) of cell-culture polystyrene chambers (Nunc, Roskilde, Denmark) with DNase I (140 U/mL) for 2 h at 37°C. S. epidermidis cells attached to the polystyrene surface were counted under microscope (400× magnification). (B) The number of attached bacteria per field was then counted. Results represent the mean ± SD of three independent experiments. *, P < 0.05; WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457 saec; ATLE, SE1457ΔatlE.
Effect of saeRS deletion on PIA production and Aap expression of S. epidermidis
PIA in the extracellular matrix of biofilms was detected using a dot blot assay with the WGA-HRP conjugate. PIA production levels were not significantly different in the SE1457ΔsaeRS strain compared to the SE1457 and SE1457saec strains (Additional file 2: Fig. S2). When assessed by comparative proteomic analysis, expression of accumulation-associated protein (Aap), an important factor for intercellular adhesion, was up-regulated in SE1457ΔsaeRS compared to the wild-type strain (Additional File 3: Fig. S3). Aap in lysostaphin-treated whole bacterial lysates of SE1457ΔsaeRS, SE1457 and SE1457saec strains was detected by Western blot using an anti-Aap monoclonal antibody. The SE1457ΔsaeRS strain expressed more Aap (1.85-fold up-regulation) compared to the wild-type and the complementation strains (Additional file 4: Fig. S4).
Analysis of the autolysis-related gene transcription in SE1457ΔsaeRS
To investigate whether the transcription of autolysis-related genes was regulated by saeRS, DNA microarray and RT-qPCR of total RNAs from the SE1457ΔsaeRS and the wild-type strains were performed. Expression of numerous autolysis-related genes including lytS (two-component sensor histidine kinase LytS), lrgA (holin-like protein), serp0043 (1,4-beta-N-acetylmuramidase), glpQ (glycerophosphoryl diester phosphodiesterase), arlR (DNA-binding response regulator), atlE (autolysin), and aae (autolysin/adhesin) was found to be up-regulated in SE1457ΔsaeRS strain (Table 3). Transcription of the genes in the saeRS operon (saeP, saeQ, saeR, and saeS) in the SE1457ΔsaeRS strain was not detected.
Table 3.
Genes expression regulated by saeRS in S. epidermidis
| Genbank accession no. | Genes/ORF | Description | Expression ratio mutant/WT | P-valueb | Functions | References | |
|---|---|---|---|---|---|---|---|
| Microarraya | RT-qPCR | ||||||
| Autolysis-related genes | |||||||
| AAW52842 | lytS | two-component sensor histidine kinase LytS | 3.87 | 2.33 ± 0.35 | 0.0097 | Negatively modulating the expression of murein hydrolases and positively regulates the expression of the lrgAB operon in S. aureus | [27,43,44] |
| AAW52844 | lrgA | holin-like protein LrgA | 2.28 | 2.75 ± 0.05 | < 0.0001 | Encoding a murein hydrolase exporter similar to bacteriophage holin proteins; may be required for the activity or transport of this cell wall-associated murein hydrolase in S. aureus | [44] |
| AAW53428 | serp0043 | 1,4-beta-N-acetylmuramidase | 4.86 | 2.25 ± 0.20 | 0.0016 | Having lysozyme activity in peptidoglycan catabolic process in S. aureus | [14] |
| AAW53918 | glpQ | glycerophosphoryl diester phosphodiesterase GlpQ, putative | 2.98 | 1.80 ± 0.20 | 0.0080 | Having glycerophosphodiester phosphodiesterase activity in lipid and glycerol metabolic process in S. aureus | [55] |
| AAW54343 | arlR | DNA-binding response regulator | 8.30 | 3.20 ± 0.45 | 0.0015 | Regulating extracellular proteolytic activity; may be involved in the modulation of expression of genes associated with growth and cell division; positively regulating a two-component system lytRS in S. aureus | [18,25,26,56-58] |
| AAW53968 | atlE | S. epidermidis autolysin | UDc | 1.45 ± 0.10 | 0.0053 | Having amidase activity to cleave the amide bond between N-acetyl muramic acid and L-alanine; mediating lysis of a subpopulation of the bacteria and extracellular DNA release in S. epidermidis | [7,29,46] |
| AJ250905 | aae | S. epidermidis autolysin/adhesin | UD | 2.32 ± 0.38 | 0.0088 | Having bacteriolytic activity and binding to fibrinogen, fibronectin and vitronectin in S. epidermidis | [8] |
| Biofilm-forming related genes | |||||||
| AAW53175 | icaA | a gene of ica operon | UD | 1.22 ± 0.13 | 0.20 | Encoding N-acetyglucosaminyltransferase for synthesis of polysaccharide intercellular adhesin (PIA) which is important for biofilm formation of S. epidermidis | [2,31,59] |
| AAW53239 | aap | accumulation-associated protein | UD | 1.62 ± 0.06 | 0.0008 | Contributing to intercellular adhesion and biofilm formation of S. epidermidis | [4,60,61] |
| sae operon | |||||||
| AAW53762 | saeS | sensor histidine kinase SaeS | 0.26 | UD | Encoding a histidine kinase; involving in the tight temporal control of virulence factor expression in S. aureus | [18,47,62] | |
| AAW53763 | saeR | DNA-binding response regulator SaeR | 0.14 | UD | The response regulator SaeR binding to a direct repeat sequence in S. aureus; involving in anaerobic growth and nitrate utilization in S. epidermidis | [11,48] | |
| AAW53764 | saeQ | conserved hypothetical protein | UD | UD | Encoding a membrane protein, function unknown in S. epidermidis | [62] | |
| AAW53765 | saeP | lipoprotein, putative | UD | UD | Encoding a lipoprotein, function unknown in S. epidermidis | [62] | |
a The complete raw microarray dataset has been posted on the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), accession number GPL13532 for the platform design and GSE29309 for the original dataset.
b P-values of RT-qPCR results were caculated using Student's t-test.
c UD: under detection level in microarray analysis or by RT-qPCR.
Discussion
As Staphylococci biofilm formation is influenced by external factors such as glucose, NaCl, temperature, aerobiosis-anaerobiosis, static-dynamic conditions, and pH [36-39], it suggests that there are mechanisms that can sense environmental signals and regulate bacterial biofilm formation. In S. epidermidis, the agrC/A TCS has been proven to negatively regulate biofilm formation [15,16], while the lytS/R TCS has been shown to positively regulate bacterial autolysis [40]. In S. aureus, the saeRS TCS influences biofilm formation [17] and the expression of virulence-associated factors [18], whereas in S. epidermidis, a mutant with saeR deletion showed a slightly higher biofilm-forming ability compared to the parental strain [11].
In the present study, SE1457ΔsaeRS, a saeR and saeS deletion mutant from S. epidermidis 1457, was constructed by homologous recombination. Although saeRS in S. epidermidis ATCC 35984 and S. aureus Newman are similar both at nucleotide sequence level (75% for saeR and 67% for saeS) and at the amino acid level (84% for SaeR and 70% for SaeS), both biofilm formation and autolysis were up-regulated in SE1457ΔsaeRS, suggesting that saeRS in S. epidermidis plays a different role from that in S. aureus. Additionally, when examined by SEM, increased quantities of extracellular polymeric substances (EPSs) were observed in the SE1457ΔsaeRS biofilm compared to the SE1457 and SE1457saec biofilms (Figure 2A).
Aap expression and PIA synthesis are important for biofilm formation. Therefore, we examined the contribution of Aap and PIA to SE1457ΔsaeRS biofilm formation. In S. epidermidis, Aap plays an important role in biofilm formation, and biofilm-positive strains that express aap show higher biofilm forming abilities than strains that lack the Aap protein [41]. In SE1457ΔsaeRS, Aap up-regulation was detected using 2-DE and confirmed by Western blot, suggesting that Aap is a factor associated with the enhanced biofilm formation capacity of SE1457ΔsaeRS. PIA plays a major role in intercellular adhesion in S. epidermidis biofilms [42]. However, no obvious differences in either PIA production or transcription of icaA, the gene that encodes an N-acetylglucosaminyl transferase enzyme critical for PIA synthesis, were observed between SE1457ΔsaeRS and SE1457 (Table 3). These results are consistent with the findings reported for a saeR deletion mutant by Handke et al. [11].
The enhanced S. epidermidis biofilm formation may be correlated with the increased amounts of eDNA released in the biofilm matrix [19,25,28]. Quantitative PCR revealed that eDNA release from S. epidermidis 1457ΔsaeRS was up-regulated (Figure 6). Furthermore, the biomass of SE1457ΔsaeRS biofilms was markedly decreased compared to SE1457 and SE1457saec biofilms when DNase I was added prior to biofilm formation.
Extracellular DNA is known to be released following bacterial autolysis [19]. SE1457ΔsaeRS showed a higher level of Triton X-100-induced autolysis compared to the wild-type strain in TSB medium containing 1 M NaCl. In accordance with the enhanced autolysis of SE1457ΔsaeRS, extracts from SDS-treated SE1457ΔsaeRS cells exhibited more bacteriolytic bands compared to extracts from the wild-type strain. These results indicate that saeRS influenced the activity of autolysins that bind non-covalently to the S. epidermidis cell wall. In S. aureus, autolysis is a complicated process regulated by the lytSR TCS [43] and global regulators such as mgrA and sarA [44,45]. Autolysis is influenced by a variety of different factors such as NaCl, pH, temperature, and growth phase, suggesting the existence of a mechanism that can sense environmental conditions [36-39]. However, Zhu et al. have demonstrated that the lytSR TCS in S. epidermidis is not involved in Triton X-100-induced autolysis and does not alter the zymogram profile [40], indicating that a different mechanism for autolysis regulation exists in S. epidermidis. The findings in the present study suggest that the saeRS TCS may regulate S. epidermidis autolysis.
The increased autolysis rate observed in SE1457ΔsaeRS may also be associated with the up-regulated expression of autolysins. In S. epidermidis, AtlE and Aae are important autolysins [8,46]. AtlE is expressed as a 138 kDa precursor protein that is proteolytically processed to release the GL (51 kDa) and AM domains (62 kDa) [13,14,23]. Aae, a 35 kDa protein, contains three repetitive sequences in its N-terminal portion. These repeats comprise features of a putative peptidoglycan binding domain (LysM domain) found in several enzymes that are involved in cell-wall metabolism. Aae from S. epidermidis O-47 exhibited bacteriolytic activity in zymographic analysis using S. carnosus or S. epidermidis cells as a substrate. In the present study, atlE and aae transcription was up-regulated in SE1457ΔsaeRS (Table 3), which may account for the increase in bacteriolytic bands in the zymogram assay. In addition, expression of numerous autolysis-related genes in SE1457ΔsaeRS, such as lytS, lrgA, arlR, serp0043 and glpQ, were also up-regulated, suggesting that S. epidermidis autolysis mediated by saeRS may be influenced by other factors that remain to be defined.
Transcriptional profile analysis of the saeRS mutant and the wild-type strain found 135 differentially expressed genes in the present study, whereas in the Handke's study, only 65 genes in the saeR mutant were differentially expressed compared to the wild-type strain. The deletion of saeRS in S. epidermidis affects genes with a variety of functions, including bacterial autolysis (lrgA, arlR, lytS), biofilm formation (ebhA), leucine biosynthesis (leuD), protein hydrolysis (clpP), stress resistance (asp23), and cell viability (yycH). Three genes with increased expression, pflB (formate acetyltransferase), pflA (formate acetyltransferase-activating enzyme) and lrgA (holin protein) in SE1457ΔsaeRS, overlapped with the saeR deletion mutant. The discrepancies of the microarray data between the saeR mutant and the saeRS mutant may result from crosstalk between saeS and the response regulators of other TCSs. When the transcriptional profiles of the saeRS deletion mutant was compared to the S. aureus strains N315, COL, and Newman, only three differentially expressed genes, geh (glycerol ester hydrolase), efb (fibrinogen-binding protein) and lrgA (holin-like protein LrgA), were found to overlap [18,47]. Taken together, these results suggest a different role for saeRS in S. epidermidis from that in S. aureus.
Through the use of regulatory sequence analysis tools (http://rsat.ulb.ac.be/rsat), we further analyzed the upstream regions of the genes that were differentially expressed in SE1457ΔsaeRS compared to the wild-type strain for the GTTAAN6GTTAA SaeR-binding motif in S. aureus reported by Sun et al. [48]. Only Eight genes involved in metabolic process [SERP2414, SERP2360, SERP2192 (cysH), SERP1745 (deoC), SERP0721 (pheS), SERP0371, SERP0365 (saeR), and SERP0164] that contained the direct repeat sequence with no more than one mismatch were found (Table 4), suggesting that the potential role of saeRS in autolysis regulation in S. epidermidis may be different from its role in S. aureus.
Table 4.
Genes containing the direct repeat sequence with no more than one mismatch
| Gene IDa | Name | Startb | Sequencec | Endb | Product |
|---|---|---|---|---|---|
| SERP0164 | -1 | GTTAAATTTAATTTAA | -16 | ATP:guanido phosphotransferase family protein | |
| SERP0365 | saeR | -488 | GTTAAATCATATTTAA | -503 | DNA-binding response regulator SaeR |
| SERP0371 | -575 | GTTAATCTTCATTTAA | -590 | exsD protein | |
| SERP0721 | pheS | -648 | GATAACATGATGTTAA | -663 | phenylalanyl-tRNA synthetase, alpha subunit |
| SERP1745 | deoC | -1091 | GTAAAAATAAAGTTAA | -1106 | deoxyribose-phosphate aldolase |
| SERP2192 | cysH | -172 | GATAATCAAAAGTTAA | -187 | phosophoadenylyl-sulfate reductase |
| SERP2360 | -114 | GTTAAACCACCGTCAA | -129 | 3-hydroxyacyl-CoA dehydrogenase family protein | |
| SERP2414 | -270 | GTTAACAGATAGTAAA | -285 | lipoprotein, putative |
a These genes are identified in microarray analysis.
b The start point and end point are the distance from the translation start codon.
c Conserved repeat sequences are underlined.
Conclusions
The deletion of saeRS in S. epidermidis resulted in the alteration of bacterial autolysis, increased eDNA release, and decreased bacterial cell viability in the planktonic/biofilm states. Further, Aap expression and the transcription of autolysin genes such as atlE and aae were up-regulated. Overall, these alterations were associated with the increased biofilm-forming ability of the saeRS deletion mutant. The present study suggests that in S. epidermidis, the saeRS TCS plays an important role in regulating bacterial autolysis, which is related to biofilm formation.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
QL performed the molecular genetic studies, participated in the sequence alignment, and drafted the manuscript. TZ helped to construct the saeRS deletion mutant. JH performed the autolysis and zymogram analysis. HB participated in the 2-DE study. JY performed the RT-qPCR analysis. FY participated in the CLSM analysis. JL participated in the RNA extractions. YW participated in the design of the study, performed the statistical analysis and edited the manuscript. AF, PF, and JS performed and analyzed microarray experiments. DQ participated in the study design and coordination and helped to draft and edit the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Fig. S1. Growth curves of SE1457ΔsaeRS and the parental strain in aerobic (A) or anaerobic (B) growth conditions. Overnight cultures were diluted 1:200 and incubated at 37°C with shaking at 220 rpm. The OD600 of the cultures was measured at 60 min intervals for 12 h. For anaerobic growth conditions, bacteria were cultured in the Eppendorf tubes that were filled up with the TSB medium and sealed with wax. WT, SE1457; SAE, SE1457ΔsaeRS.
Fig. S2. PIA detection in S. epidermidis biofilms. S. epidermidis strains were grown in 6-well plates under static conditions at 37°C for 24 h. Next, the cells were removed by scraping and collected by centrifugation before being resuspended in 0.5 M EDTA (pH 8.0). After proteinase K treatment (20 mg/mL) for 3 h at 37°C, serial dilutions of the PIA extracts were spotted onto PVDF membranes. Spots corresponding to PIA were quantified using the Quantity-one software. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec; 35984, S. epidermidis ATCC35984.
Fig. S3. SE1457ΔsaeRS and wild-type strain 2-DE profiles. SE1457ΔsaeRS and SE1457 were grown in TSB medium at 37°C until the post-exponential growth phase; the bacteria were then separated by centrifugation. Bacteria cell pellets were dissolved in lysis buffer and sonicated on ice. The 2-DE gels were performed using 24 cm immobilized dry strips (IPG, nonlinear, pH 4-7, GE Healthcare) and analyzed by ImageMaster 2D platinum 6.0 software (Amersham Biosciences). Protein spots were identified using a 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, California, USA).
Fig. S4. Detection of Aap expression. Aap in lysostaphin-treated bacterial cells of SE1457ΔsaeRS, SE1457, and SE1457saec was detected by Western blot using an anti-Aap monoclonal antibody (made in our laboratory). Proteins were separated on 7% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes by electroblotting. Bands corresponding to Aap were quantified using the Quantity-one software. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457sae.
Contributor Information
Qiang Lou, Email: qiang_lou@126.com.
Tao Zhu, Email: 071101036@fudan.edu.cn.
Jian Hu, Email: 081101043@fudan.edu.cn.
Haijing Ben, Email: benhaijing@foxmail.com.
Jinsong Yang, Email: yjsong123@126.com.
Fangyou Yu, Email: wzjxyfy@163.com.
Jingran Liu, Email: 082101071@fudan.edu.cn.
Yang Wu, Email: yangwu@fudan.edu.cn.
Adrien Fischer, Email: adrien.fischer@genomic.ch.
Patrice Francois, Email: patrice.francois@genomic.ch.
Jacques Schrenzel, Email: Jacques.Schrenzel@genomic.ch.
Di Qu, Email: dqu@fudan.edu.cn.
Acknowledgements
We thank Prof. Friedrich Götz (University of Tübingen) for his academic advice regarding zymogram analysis, PIA detection, and microarray analysis. We appreciate the suggestions and support of Prof. Søren Molin (Technical University of Denmark) regarding biofilm CLSM observation. We also thank Prof. Michel Débarbouillé (Institut Pasteur) for providing the pMAD plasmid for the construction of the SE1457ΔsaeRS strain.
This work was supported by the National High Technology Research and Development Program (863 Program) (2006AA02A253), the Scientific Technology Development Foundation of Shanghai (10410700600, 09DZ1908602, 08JC1401600), the National Natural Science Foundation of China (30800036, J0730860), National Science and Technology Major Project (2009ZX09303-005, 2008ZX10003-016, 2009ZX10004-502), the Program of Ministry of Science and Technology of China (2010DFA32100), and the IBS Open Research Grant (IBS09064).
References
- von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infectious Diseases. 2002;2(11):677–685. doi: 10.1016/S1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- Rohde H, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JKM, Heilmann C, Herrmann M, Mack D. The repetitive domain B of the accumulation associated protein Aap mediates intercellular adhesion and biofilm formation in Staphylococcus epidermidis. Int J Med Microbiol. 2004;294:128–128. [Google Scholar]
- Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol. 2010;76(10):3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Qin ZQ, Ou YZ, Yang LA, Zhu YL, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiol-Sgm. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149(Pt 10):2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189(22):8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pinar R, Ramos JL, Rodriguez-Herva JJ, Espinosa-Urgel M. A two-component regulatory system integrates redox state and population density sensing in Pseudomonas putida. J Bacteriol. 2008;190(23):7666–7674. doi: 10.1128/JB.00868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke LD, Rogers KL, Olson ME, Somerville GA, Jerrells TJ, Rupp AE, Dunman PA, Fey PD. Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infect Immun. 2008;76(1):141–152. doi: 10.1128/IAI.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Jiang DR. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiol-Sgm. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49(6):1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. Fems Microbiol Lett. 2007;270(2):179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect Immun. 2008;76(4):1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. 2006;188(22):7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4(6):e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of Coagulase-Negative Staphylococci to Plastic Tissue-Culture Plates - a Quantitative Model for the Adherence of Staphylococci to Medical Devices. Journal of Clinical Microbiology. 1985;22(6):996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnier Y, Gettler B, Francois P, Bento M, Renzoni A, Vaudaux P, Schlegel W, Schrenzel J. A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus. BMC Genomics. 2005;6:95. doi: 10.1186/1471-2164-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Francois P, Charbonnier Y, Deshusses JM, Koessler T, Huyghe A, Bento M, Stahl-Zeng J, Fischer A, Masselot A, Vaezzadeh A, Gallé F, Renzoni A, Vaudaux P, Lew D, Zimmermann-Ivol CG, Binz PA, Sanchez JC, Hochstrasser DF, Schrenzel J. Exploring glycopeptide-resistance in Staphylococcus aureus: a combined proteomics and transcriptomics approach for the identification of resistance-related markers. BMC Genomics. 2006;7:296. doi: 10.1186/1471-2164-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat AM, Howard ST, Hale Wt, Lyons R, Garner H, Johnston SA. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 2002;30(20):e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignac A, Sieradzki K, Tomasz A. Perturbation of cell wall synthesis suppresses autolysis in Staphylococcus aureus: evidence for coregulation of cell wall synthetic and hydrolytic enzymes. J Bacteriol. 2007;189(21):7573–7580. doi: 10.1128/JB.01048-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182(14):3955–3964. doi: 10.1128/JB.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Yu C, Bayles K, Lasa I, Ji Y. Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus. J Bacteriol. 2007;189(7):2734–2742. doi: 10.1128/JB.01806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzoni A, Barras C, Francois P, Charbonnier Y, Huggler E, Garzoni C, Kelley WL, Majcherczyk P, Schrenzel J, Lew DP, Vaudaux P. Transcriptomic and functional analysis of an autolysis-deficient, teicoplanin-resistant derivative of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Ch. 2006;50(9):3048–3061. doi: 10.1128/AAC.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- Al Laham N, Rohde H, Sander G, Fischer A, Hussain M, Heilmann C, Mack D, Proctor R, Peters G, Becker K, von Eiff C. Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with the small-colony-variant phenotype. J Bacteriol. 2007;189(12):4494–4501. doi: 10.1128/JB.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger U, Ulrich M, Steinhuber A, Doring G, Mack D, Landmann R, Goerke C, Wolz C. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect Immun. 2005;73(3):1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso M, Garcia E, Lopez R. Biofilm formation by Streptococcus pneumoniae: Role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188(22):7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Gotz F, Otto M. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun. 2000;68(3):1048–1053. doi: 10.1128/IAI.68.3.1048-1053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagensen JA, Klausen M, Ernst RK, Miller SI, Folkesson A, Tolker-Nielsen T, Molin S. Differentiation and distribution of colistin- and sodium dodecyl sulfate-tolerant cells in Pseudomonas aeruginosa biofilms. J Bacteriol. 2007;189(1):28–37. doi: 10.1128/JB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci USA. 2010;107(32):14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin RW, Chatterjee AN, Young FE. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J Bacteriol. 1972;111(1):272–283. doi: 10.1128/jb.111.1.272-283.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabu K, Kaneda S. Salt-induced cell lysis of Staphylococcus aureus. Curr Microbiol. 1995;30(5):299–303. doi: 10.1007/BF00295505. [DOI] [PubMed] [Google Scholar]
- Wells JE, Russell JB. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl Environ Microbiol. 1996;62(4):1342–1346. doi: 10.1128/aem.62.4.1342-1346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin PJ, Mani N, Jayaswal RK. Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie Van Leeuwenhoek. 1994;65(1):71–78. doi: 10.1007/BF00878281. [DOI] [PubMed] [Google Scholar]
- Zhu T, Lou Q, Wu Y, Hu J, Yu F, Qu D. Impact of the Staphylococcus epidermidis LytSR two-component regulatory system on murein hydrolase activity, pyruvate utilization and global transcriptional profile. Bmc Microbiol. 2010;10:287. doi: 10.1186/1471-2180-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher-Perdreau F, Heilmann C, Peters G, Gotz F, Pulverer G. Comparative analysis of a biofilm-forming Staphylococcus epidermidis strain and its adhesion-positive, accumulation-negative mutant M7. Fems Microbiol Lett. 1994;117(1):71–78. doi: 10.1111/j.1574-6968.1994.tb06744.x. [DOI] [PubMed] [Google Scholar]
- Xu L, Li HL, Vuong C, Vadyvaloo V, Wang JP, Yao YF, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178(19):5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178(3):611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol. 2004;186(16):5267–5280. doi: 10.1128/JB.186.16.5267-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. Activity of the major staphylococcal autolysin Atl. Fems Microbiol Lett. 2006;259(2):260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun. 2006;74(8):4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192(8):2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JFEF, Maniatis T. Molecular Cloning: a laboratory manual. 2. N. Y.: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Mack SRHHL, De Jonge CJ, Anderson RA. The human sperm acrosome reaction does not depend on arachidonic acid metabolism via the cyclooxygenase and lipoxygenase pathways. J Androl. 1992;13:551–559. [PubMed] [Google Scholar]
- Kreiswirth BNLS, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. Fems Microbiol Lett. 1997;151(1):1–8. doi: 10.1016/S0378-1097(97)00116-X. [DOI] [PubMed] [Google Scholar]
- Wieland J, Nitsche AM, Strayle J, Steiner H, Rudolph HK. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na1 pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microb. 2004;70(11):6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, Engelmann S. The influence of agr and sigma(B) in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics. 2004;4(10):3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
- Ji Y, Yu C, Liang X. Transcriptomic analysis of ArlRS two-component signaling regulon, a global regulator, in Staphylococcus aureus. Methods Enzymol. 2007;423:502–513. doi: 10.1016/S0076-6879(07)23024-1. [DOI] [PubMed] [Google Scholar]
- Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol. 2005;187(15):5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187(15):5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde H, Frankenberger S, Zahringer U, Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol. 2010;89(1):103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Yang XM, Li N, Chen JM, Ou YZ, Jin H, Lu HJ, Zhu YL, Qin ZQ, Qu D, Yang PY. Comparative proteomic analysis between the invasive and commensal strains of Staphylococcus epidermidis. Fems Microbiol Lett. 2006;261(1):32–40. doi: 10.1111/j.1574-6968.2006.00327.x. [DOI] [PubMed] [Google Scholar]
- Macintosh RL, Brittan JL, Bhattacharya R, Jenkinson HF, Derrick J, Upton M, Handley PS. The Terminal A Domain of the Fibrillar Accumulation-Associated Protein (Aap) of Staphylococcus epidermidis Mediates Adhesion to Human Corneocytes. J Bacteriol. 2009;191(22):7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential Target Gene Activation by the Staphylococcus aureus Two-Component System saeRS. J Bacteriol. 2010;192(3):613–623. doi: 10.1128/JB.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Growth curves of SE1457ΔsaeRS and the parental strain in aerobic (A) or anaerobic (B) growth conditions. Overnight cultures were diluted 1:200 and incubated at 37°C with shaking at 220 rpm. The OD600 of the cultures was measured at 60 min intervals for 12 h. For anaerobic growth conditions, bacteria were cultured in the Eppendorf tubes that were filled up with the TSB medium and sealed with wax. WT, SE1457; SAE, SE1457ΔsaeRS.
Fig. S2. PIA detection in S. epidermidis biofilms. S. epidermidis strains were grown in 6-well plates under static conditions at 37°C for 24 h. Next, the cells were removed by scraping and collected by centrifugation before being resuspended in 0.5 M EDTA (pH 8.0). After proteinase K treatment (20 mg/mL) for 3 h at 37°C, serial dilutions of the PIA extracts were spotted onto PVDF membranes. Spots corresponding to PIA were quantified using the Quantity-one software. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457saec; 35984, S. epidermidis ATCC35984.
Fig. S3. SE1457ΔsaeRS and wild-type strain 2-DE profiles. SE1457ΔsaeRS and SE1457 were grown in TSB medium at 37°C until the post-exponential growth phase; the bacteria were then separated by centrifugation. Bacteria cell pellets were dissolved in lysis buffer and sonicated on ice. The 2-DE gels were performed using 24 cm immobilized dry strips (IPG, nonlinear, pH 4-7, GE Healthcare) and analyzed by ImageMaster 2D platinum 6.0 software (Amersham Biosciences). Protein spots were identified using a 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, California, USA).
Fig. S4. Detection of Aap expression. Aap in lysostaphin-treated bacterial cells of SE1457ΔsaeRS, SE1457, and SE1457saec was detected by Western blot using an anti-Aap monoclonal antibody (made in our laboratory). Proteins were separated on 7% SDS-PAGE gels and then transferred to polyvinylidene fluoride (PVDF) membranes by electroblotting. Bands corresponding to Aap were quantified using the Quantity-one software. WT, SE1457; SAE, SE1457ΔsaeRS; SAEC, SE1457sae.