Abstract
Studies have demonstrated that TLR4 and TLR2 expression by monocytes and the blood levels of TLR4 and TLR2 ligand in diabetic patients are significantly incased compared to nondiabetic patients, indicating that more monocytes in diabetic patients may have coactivation of TLR4 and TLR2. Although it has been shown that either TLR4 or TLR2 activation leads to increased expression of proinflammatory cytokines, the effect of coactivation of TLR2 and TLR4 in mononuclear cells on proinflammatory cytokine expression and the underlying molecular mechanisms remain largely unknown. In this study, we found that while TLR1, TLR2, TLR4 and TLR6 were expressed by U937 mononuclear cells, TLR4 was expressed at the highest level. Interestingly, results showed that while activation of either TLR4 or TLR2/6 (TLR2 dimerized with TLR6), but not TLR2/1 (TLR2 dimerized with TLR1), significantly increased IL-6 expression by U937 mononuclear cells, coactivation of TLR4 and TLR2/6, but not TLR4 and TLR2/1, led to a further augmentation on IL-6 expression by increasing IL-6 transcriptional activity, but not mRNA stability. To explore the signaling mechanisms involved in the augmentation, we found that p38 MAPK and NFκB pathways, but not ERK and JNK pathways, were required for the augmentation of IL-6 expression by coactivation of TLR4 and TLR2/6. Furthermore, we found that coactivation of TLR4 and TLR2/6 increased p38 phosphorylation, but not NFkB activity, as compared to activation of TLR4 or TLR2/6 alone. Taken together, this study showed that coactivation of TLR4 and TLR2/6 coordinates an additive augmentation of IL-6 gene transcription via p38 MAPK pathway in U937 mononuclear cells.
Keywords: Toll-like receptor, Interleukin 6, p38 mitogen-activated protein kinase, Inflammation
1. INTRODUCTION
Increasing studies in the recent years have provided strong evidence in supporting an essential role of TLR4 and TLR2 as the receptors for the innate immune response in the pathogenesis of diabetes (Caricilli et al., 2008; Dasu et al.; Devaraj et al., 2009; Devaraj et al., 2008; Radin et al., 2008; Tsukumo et al., 2007). Animal studies have shown that TLR4 deficiency or loss-of-function mutation of TLR4 in mice conferred protection against lipid-induced insulin resistance (Radin et al., 2008; Tsukumo et al., 2007). It has been also reported that inhibition of TLR2 expression with antisense oligonucleotides improved insulin sensitivity and signaling in muscle and white adipose tissue of obese mice (Caricilli et al., 2008). To further explore the relationship between TLR2/TLR4 and diabetes, Devaraj et al. reported that TLR2 and TLR4 expression and signaling triggered by TLR2 and TLR4 activation were increased in monocytes isolated from type 1 diabetic patients (Devaraj et al., 2008). They also showed that patients with type 1 or type 2 diabetes had increased circulating levels of TLR2 and TLR4 ligands such as endotoxin, heat-shock protein 60 (Hsp60), and high-mobility group box1 (HMGB1) (Dasu et al.; Devaraj et al., 2009). Furthermore, it has been shown that high glucose induced TLR2 and TLR4 expression via PKC-alpha and PKC-delta, respectively, by stimulating NADPH oxidase in monocytes (Dasu et al., 2008).
While the role of TLR4 and TLR2 in diabetes was well documented by the above investigations, it was also shown that TLR4 and TLR2 played an essential role in atherosclerosis (den Dekker et al.; Huang and Pope), a major cardiovascular complication of diabetes. Animal studies have shown that the deficiency of TLR4 or TLR2 in mouse models is associated with a significant reduction of atherosclerotic lesions (Bjorkbacka et al., 2004; Liu et al., 2008; Michelsen et al., 2004; Mullick et al., 2005). In addition, studies have shown that TLR4 polymorphism is associated with atherosclerosis (Hernesniemi et al., 2006; Hernesniemi et al., 2008; Lin et al., 2005; Norata et al., 2005). To understand the mechanisms by which TLR4 and TLR2 are involved in atherosclerosis, in vitro studies have demonstrated that the activation of TLR4 or TLR2 led to increased expression of proinflammatory cytokines (Li and Sun, 2007; Tobias and Curtiss, 2005). Since it is well known that inflammation plays a pivotal role in diabetes (Bendtzen et al., 1989) and atherosclerosis (King, 2008; Pankewycz et al., 1995), these findings have elucidated a potential mechanism by which TLR4 and TLR2 activation contributes to diabetes and cardiovascular complication of diabetes.
The findings by recent studies that diabetic patients have increased TLR4 and TLR2 expression by monocytes and increased concentrations of TLR4 and TLR2 ligands in serum (Devaraj et al., 2009; Devaraj et al., 2008) suggest that more monocytes in diabetic patients may have coactivation of both TLR4 and TLR2. Although it is well known that activation of either TLR4 or TLR2 in mononuclear cells increased expression of proinflammatory cytokines, the effect of coactivation of TLR4 and TLR2 on proinflammatory cytokine expression and underlying mechanisms remain largely unknown. In this study, we have tested our hypothesis that coactivation of TLR4 and TLR2 further increases proinflammatory cytokine expression when compared to activation of either TLR4 or TLR2 alone. The results of this study showed that coactivation of TLR4 and TLR2/6 (TLR2 is dimerized with TLR6) coordinated an augmentation on IL-6 transcription in U937 mononuclear cells via p38 mitogen-activated protein kinase (MAPK) signaling pathway.
2. MATERIALS AND METHODS
2.1. Cell Culture and Treatments
U937 mononuclear cells (Sundstrom and Nilsson, 1976) were purchased from American Type Culture Collection (Manassas, VA). The cells were cultured in a 5% CO2 atmosphere in RPMI 1640 medium (GIBCO, Invitrogen Cop. Carlsbad, CA) containing 10% fetal calf serum, 1% MEM non-essential amino acid solution, and 0.6 g/100 ml of HEPES. The medium was changed every 2–3 days. Our previous studies have shown the similarity between U937 cells and human monocytes in cellular response to LPS (Samuvel et al.; Sundararaj et al., 2008; Sundararaj et al., 2009). For cell treatment, LPS from E. coli (Ec) (Sigma, St. Louis, MO) and pam2CSK4 and pam3CSK4 (InvivoGen, San Diego, CA) were used. The LPS was highly purified by phenol extraction and gel filtration chromatography and was cell culture tested. Pam2CSK4 and pam3CSK4 are synthetic diacylated and triacylated lipopeptides, respectively.
2.2. Enzyme-Linked Immunosorbent Assay (ELISA) for Quantification of IL-6 and MMP-1
IL-6 and MMP-1 in conditioned medium were quantified using sandwich ELISA kits according to the protocol provided by the manufacturer (R&D System, Minneapolis, MN). U937 cells were plated into 12-well plate (0.5 × 106/well) and treated with different agonists for 24 h. After the treatment, culture medium was subjected to quantification of IL-6 or MMP-1 using ELISA.
2.3. Real-Time Polymerase Chain Reaction (PCR)
Total RNA was isolated from cells using the RNeasy minikit (Qiagen, Santa Clarita, CA). First-strand complementary DNA (cDNA) was synthesized with the iScript™ cDNA synthesis kit (Bio-Rad, Hercules, CA) using 20 μl of reaction mixture containing 1 μg of total RNA, 4 μl of 5x iScript reaction mixture, and 1 μl of iScript reverse transcriptase. The complete reaction was cycled for 5 minutes at 25 °C, 30 minutes at 42 °C and 5 minutes at 85°C using a PTC-200 DNA Engine (MJ Research, Waltham, MA). The reverse transcription (RT) reaction mixture was then diluted 1:10 with nuclease-free water and used for PCR amplification of cDNA in the presence of the primers. The Beacon designer software (PREMIER Biosoft International, Palo Alto, CA) was used for primer designing (IL-6: 5′ primer sequence, AACAACCTGAACCTTC CAAAGATG; 3′ primer sequence, TCAAACTCCAAAAGACCAGTGATG. MMP-1: 5′ primer sequence, CTGGGAAGCCATCACTTACCTTGC; 3′ primer sequence, GTTTCTAGAGTC GCTGGGAAGCTG). Primers were synthesized (Integrated DNA Technologies, Inc., Coralville, IA) and real-time PCR was performed in duplicate using 25 μl of reaction mixture containing 10 μl of RT mixture, 0.2 μM of both primers, and 12.5 μl of iQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA). Real-time PCR was run in the iCycler™ real-time detection system (Bio-Rad) with a two-step method. The hot-start enzyme was activated (95°C for 2 min) and cDNA was then amplified for 40 cycles consisting of denaturation at 95°C for 10 sec and annealing/extension at 52.5°C for 45 sec. A melt-curve assay was then performed (55°C for 1 min and then temperature was increased by 0.5°C every 10 sec) to detect the formation of primer-derived trimers and dimers. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a control (5′ primer sequence, GAATTTGGCTACAGCAACAGGGTG; 3′ primer sequence, TCTCTTCCTCTTGTGCTCTTGCTG). Data were analyzed with the iCycler iQ™ software. A standard curve was constructed for the conversion of cycle threshold (Ct) to starting quantify (SQ). Quantification was calculated using the SQ of targeted cDNA relative to that of GAPDH cDNA in the same sample.
2.4. IL-6 mRNA Stability Analysis
U937 cells were plated in 12-well plates at a density of 1×106 cells/well and treated with 100 ng/ml LPS, 100 ng/ml pam2CSK4 or LPS plus pam2CSK4 for 4 hours, followed by addition of 10 μg/ml of actinomycin D (Sigma-Aldrich Corporate, St. Louis, MO). Previous studies have shown that actinomycin D at the concentration of 10 μg/ml inhibits transcription effectively in U937 cells (Hayes et al., 2002; Ley et al., 1989; Yamato et al., 1990). U937 cells were harvested at 2 and 4 hours after actinomycin D treatment for quantification of IL-6 mRNA using real-time PCR as described above.
2.5. IL-6 Promoter Activity Assay
U937 cells grown in 12-well plates at a density of 5×105 cells/well were transiently cotransfected using FuGENE HD transfection reagent (Promega, Madison, WI) with 0.55 μg of p1168huIL6P-luc+ plasmids (Belgian Co-ordinated Collections of Micro-organisms, BCCM™, Brussels, Belgium) and pRL-TK plasmids (Renilla luciferase) (Promega) as an internal control for 24 h. The plasmid p1168huIL6P-luc+ was constructed by inserting the HindIII/XhoI fragment of pBLHIL6CAT containing 1168-bp human IL-6 promoter (hIL6) fragment into the unique HindIII site of pGL3-Basic and has been used in the previous studies on IL-6 promoter activities (Plaisance et al., 1997; Vanden Berghe et al., 1999; Vanden Berghe et al., 1998). The nucleotide sequence of the genomic hIL6 DNA corresponds with the EMBL Nucleotide Sequence Database Accession Number AC073072. After the transfection, the cells were treated with 100 ng/ml LPS, 100 ng/ml pam2CSK4, or LPS plus pam2CSK4 for 24 h. The cells were then rinsed with cold PBS and lysed with reporter lysis buffer (Promega). The lysate was centrifuged at 15,000 g for 5 min at 4°C and the supernatant was used for luciferase activity assay. Both firefly and Renilla luciferase levels were measured in a luminometer using the dual-luciferase reporter assay system according to the instructions from the manufacturer (Promega). The firefly luciferase levels were normalized to the Renilla luciferase levels.
2.6. Immunoblotting of p38, ERK and JNK MAPK
After treatment, cell lysate containing 25–50 μg protein was electrophoresed in a 10% polyacrylamide gel. After transferring proteins to a PVDF membrane, total and phosphorylated MAPK were immunoblotted with anti-total or anti-phosphorylated p38, extracellular regulated kinase (ERK) or c-Jun N-terminal kinases (JNK) MAPK primary antibodies and HRP-conjugated secondary antibody (Cell Signaling Technology, Danvers, MA). Targeted proteins were visualized by incubating the membrane with chemiluminescence reagent (NEN Life Science Products) for 1 min and exposing it to x-ray film for 1–10 min. The X-ray films were scanned using an Epson scanner (Perfection 1200U) and the density of bands on the images was quantified using Adobe Photoshop version 10.0.1. The results were presented as the ratios of phosphorylated MAPK vs. total MAPK.
2.7. Extraction of Nuclear Protein
Nuclear protein was extracted using NE-PER® nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific Inc., Rockfold, IL). The protein concentration in the nuclear fractions was determined using a protein assay kit (Bio-Rad, Hercules, CA).
2.8. Electrophoretic Mobility Shift Assay (EMSA)
Ten μg of nuclear proteins was used for EMSA to determine NFκB DNA-binding activity. DNA–protein binding reactions were performed at room temperature for 20 min in a buffer containing 10 mM Trizma base (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 1 mg poly (dI-dC), and approximately 20 fmole of NFκB oligonucleotide (Integrated DNA Technologies, Inc., Coralville, IA) labeled with biotin using Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific Inc., Rockfold, IL). Protein–DNA complexes were resolved from protein-free DNA in 6% DNA retardation gel (Invitrogen Corporation, Carlsbad, CA) at room temperature in Novex® TBE Running Buffer (0.5X), and electroblotted onto positively charged nylon membranes. The chemiluminescence detection of biotin-labeled probes was conducted by following the instruction provided by the Thermo Fisher Scientific Inc.
2.9. Statistic Analysis
Data were presented as mean ± SD. The ANOVA was applied for statistic analysis to determine the statistical significance of gene expression or luciferase activity among different experimental groups. A value of P< 0.05 was considered significant.
3. RESULTS
3.1. Coactivation of TLR4 and TLR2/6 Increases IL-6 Production
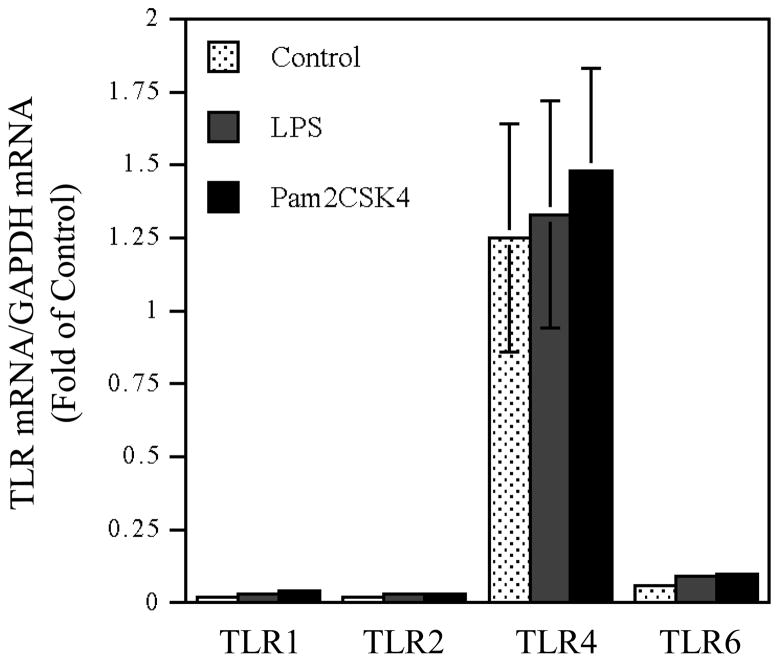
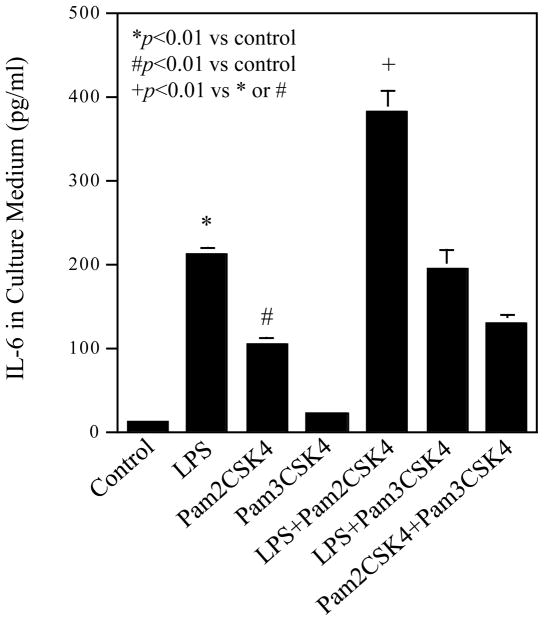
We first determined the expression of TLR1, TLR2, TLR4 and TLR6 by U937 cells. Results showed that while all these TLRs were expressed, TLR4 was expression at the highest level (Fig. 1). Treatment cells with LPS, pam2CSK4 or both LPS and pam2CSK4 did not significantly increase TLR expression. We then studied the effect of coactivation of TLR4 and TLR2 on IL-6 production by U937 cells. LPS was used to activate TLR4 while pam2CSK4 and pam3CSK4 were used to activate TLR2/6 (TLR2 dimerized with TLR6) and TLR2/1 (TLR2 dimerized with TLR1), respectively (Kanczkowski et al., 2007; Nagpal et al., 2009). Results showed that LPS or pam2CSK4, but not pam3CSK4, significantly stimulated IL-6 production markedly (Fig. 2). Interestingly, coactivation of TLR4 and TLR2/6 with LPS and pam2CSK4 had an additive effect on IL-6 production as compared to activation of TLR4 or TLR2/6 alone. In contrast, coactivation of TLR4 and TLR2/1 with LPS and pam3CSK4 or TLR2/6 and TLR2/1 with pam2CSK4 and pam3CSK4 did not further increase IL-6 production as compared to activation of TLR4, TLR2/6 or TLR2/1 alone (Fig. 2).
Figure 1.
TLR expression by U937 cells in the absence or presence of the treatment with LPS, pam2CSK4 or both LPS and pam2CSK4. U937 cells were treated with or without 100 ng/ml of LPS, pam2CSK4 or both LPS and pam2CSK4 for 24 h. After the treatment, cellular mRNA levels of TLR1, TLR2, TLR4 and TLR6 were quantified using real-time PCR and normalized to GAPDH mRNA.
Figure 2.
Stimulation of IL-6 production by TLR4, TLR2 or TLR4 plus TLR2 activation. U937 cells were treated with 100 ng/ml of LPS, 100 ng/ml of pam2CSK4, 100 ng/ml of pam3CSK4, LPS plus pam2CSK4, LPS plus pam3CSK4, or pam2CSK4 plus pam3CSK4 for 24 h. After the treatment, IL-6 released into culture medium was quantified using ELISA. The data presented are the representative of 5 independent experiments with similar results.
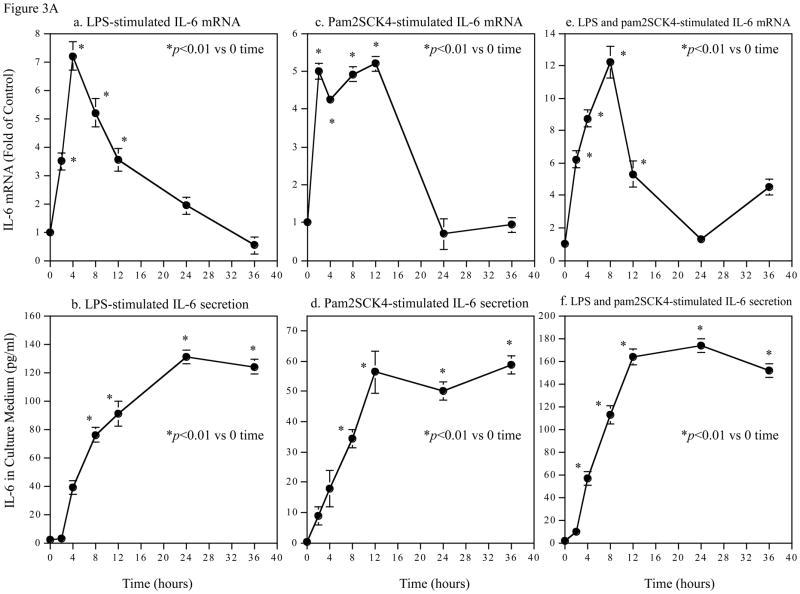
3.2. Time Courses of the Augmentation of IL-6 mRNA Expression and Protein Production Induced by Coactivation of TLR4 and TLR2/6
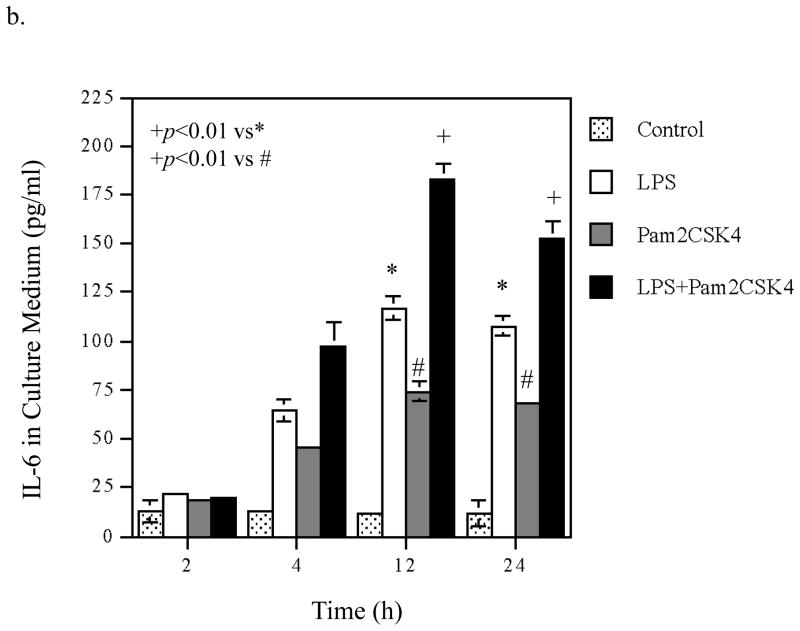
To understand the molecular mechanisms involved in the augmentation of IL-6 production by coactivation of TLR4 and TLR2/6, we first studied the effect of coactivation of TLR4 and TLR2/6 on the kinetics of IL-6 mRNA expression and IL-6 protein production and compared it with that of activation of TLR4 or TLR2/6 alone. Results showed that with LPS stimulation, IL-6 mRNA level was quickly increased, peaked at 4 h and then declined (Fig. 3A, panel a). In consistence with the kinetic of IL-6 mRNA expression, IL-6 protein in culture medium also increased rapidly and reached a plateau at 24 h (Fig. 3A, panel b). With pam2CSK4 stimulation, IL-6 mRNA level was rapidly increased at 2 h, remained at high level for 10 hours, and started dropping rapidly at 12 h (Fig. 3A, panel c). IL-6 protein in culture medium increased rapidly in the first 12 h and then reached a plateau (Fig. 3A, panel d). With stimulation by LPS plus pam2CS4, IL-6 mRNA level peaked at 8 h and then declined (Fig. 3A, panel e). IL-6 protein in culture medium reached a plateau at 12 h (Fig. 3A, panel f).
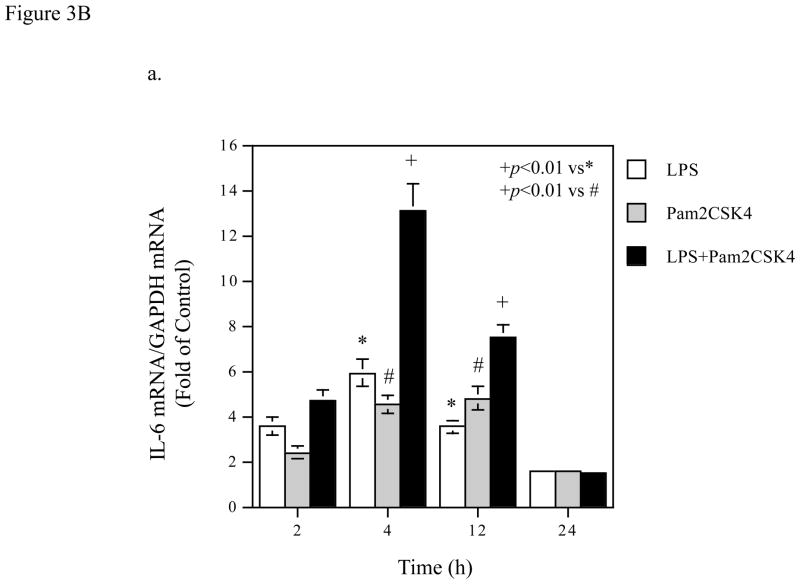
Figure 3.
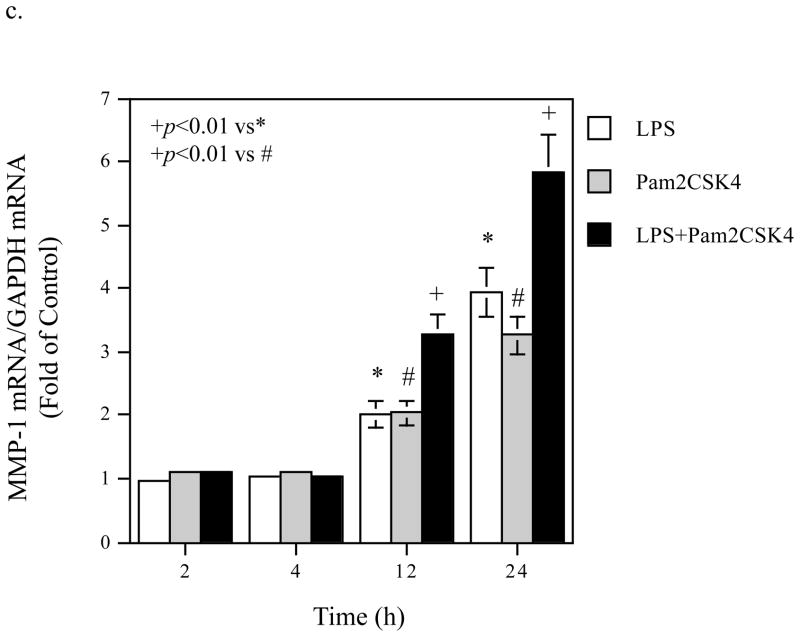
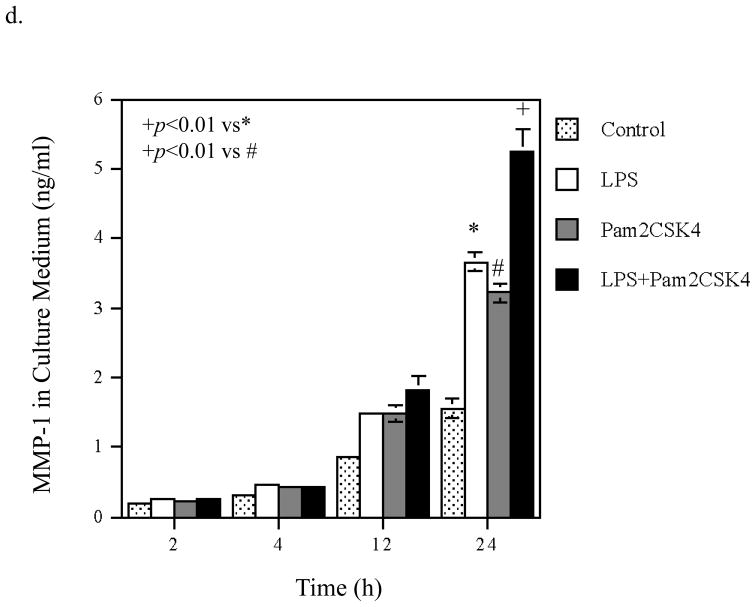
Time Courses of the augmentation of IL-6 mRNA expression and protein production induced by coactivation of TLR4 and TLR2/6. A. Kinetics of IL-6 mRNA expression and protein production in response to LPS (a and b), pam2CSK4 (d and d) or LPS plus pam2CSK4 (e and f). U937 cells were treated with 100 ng/ml of LPS, 100 ng/ml of pam2CSK4 or both for 2, 4, 8, 12, 24, or 36 h. At each time point, RNA was isolated from cells for quantification of IL-6 mRNA using real-time PCR; culture medium was collected for quantification of IL-6 protein production using ELISA. The data presented are the representative of 3 independent experiments with similar results. B. Augmentation of IL-6 mRNA expression (a), IL-6 protein production (b), MMP-1 mRNA expression (c) and MMP-1 protein production (d) by activation of TLR4, TLR2/6, or TLR4 and TLR2/6. U937 cells were treated with 100 ng/ml of LPS, 100 ng/ml of pam2CSK4 or LPS plus pam2CSK4 for 2, 4, 12 or 24 h. Culture medium and cells were collected at each time point. IL-6 and MMP-1 mRNA was quantified using real-time PCR and IL-6 and MMP-1 protein in culture medium was quantified using ELISA. The data presented are the representative of 3 independent experiments with similar results.
Taken together, these data indicate that IL-6 mRNA expression is highly controlled by coactivation of TLR4 and TLR2/6 as well as activation of TLR4 or TLR2/6 alone: IL-6 mRNA expression was subjected to not only a rapid and robust upregulation, but also a fast and vigorous downregulation.
Based on the above kinetic studies, we selected 2, 4, 12 and 24 h as the time points to determine the effect of coactivation of TLR4 and TLR2/6 on IL-6 mRNA expression and protein level in culture medium and compared it with that of activation of TLR4 or TLR2/6 alone. Results showed that while either TLR4 or TLR2/6 activation led to a time-dependent increase in IL-6 mRNA, coactivation of TLR4 and TLR2/6 exerted an additive augmentation on IL-6 mRNA expression at 4 and 12 h (Fig. 3B, panel a). At 24 h, coactivation of TLR4 and TLR2/6 no longer augmented IL-6 mRNA expression. In consistence with the findings on IL-6 mRNA, the augmentation of IL-6 protein level in culture medium by coactivation of TLR4 and TLR2/6 was observed at 12 and 24 h after the treatment (Fig. 3B, panel b).
To show the specificity of the kinetics of IL-6 mRNA expression and protein production regulated by TLR4 and TLR2/6 coactivation, we studied the kinetics of MMP-1 mRNA expression and protein level in culture medium from the same cells. As shown in Fig. 3B, panel c, the augmentation of MMP-1 mRNA expression by coactivation of TLR4 and TLR2/6 was observed at 12 and 24 h. The augmentation of MMP-1 protein level in culture medium by coactivation of TLR4 and TLR2/6 was observed at 24 h (Fig. 3B, panel d).
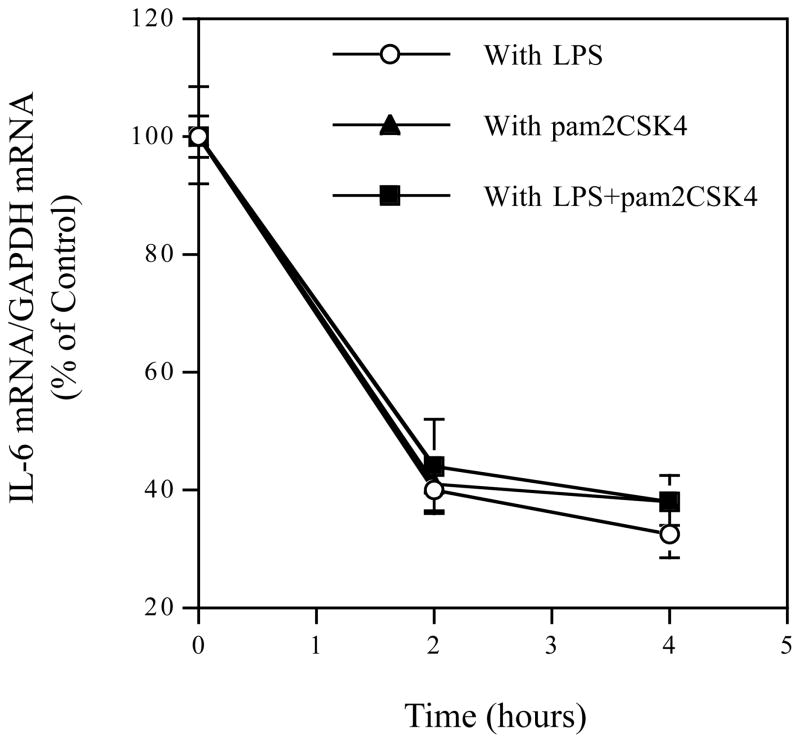
3.4. The Effect of Coactivation of TLR4 and TLR2/6 on IL-6 mRNA Stability
Since it was reported that IL-6 mRNA expression level could be upregulated by enhancing IL-6 mRNA stability (Patil et al., 2004), we determined if coactivation of TLR4 and TLR2/6 augmented IL-6 mRNA expression by increasing IL-6 mRNA stability by using actinomycin D, an inhibitor of transcription (Yamato et al., 1990). The inhibition of IL-6 transcription by actinomycin D was confirmed by the control study that showed that incubation of U937 cells with 10 μg/ml of actinomycin D alone reduced IL-6 mRNA by 77% at 2 h and 80% at 4 h. U937 cells were treated with LPS, pam2CSK4 or LPS plus pam2CSK4 for 4 h and then exposed to 10 μg/ml of actinomycin D for 2 or 4 h. Data from quantitative real-time PCR showed that no significant difference in IL-6 mRNA stability was found in cells treated with LPS, pam2CSK4 or LPS plus pam2CSK4 (Fig. 4), indicating that the augmentation of IL-6 mRNA expression by coactivation of TLR4 and TLR2/6 is not due to increase in IL-6 mRNA stability when compared to activation of TLR4 alone.
Figure 4.
IL-6 mRNA stability in cells treated with LPS, pam2CSK4 or LPS plus pam2CSK4. U937 cells were treated with 100ng/ml LPS, 100ng/ml pam2CSK4 or LPS plus pam2CSK4 for 4 hours, followed by addition of 10 μg/ml of actinomycin D. U937 cells were harvested before and 2 and 4 hours after actinomycin D treatment for quantification of IL-6 mRNA using real-time PCR. The IL-6 mRNA expression was normalized to GAPDH mRNA expression and the data presented are % of the control that was IL-6 mRNA level before the addition of actinomycin D.
3.5. The Effect of Coactivation of TLR4 and TLR2/6 on IL-6 Transcriptional Activity
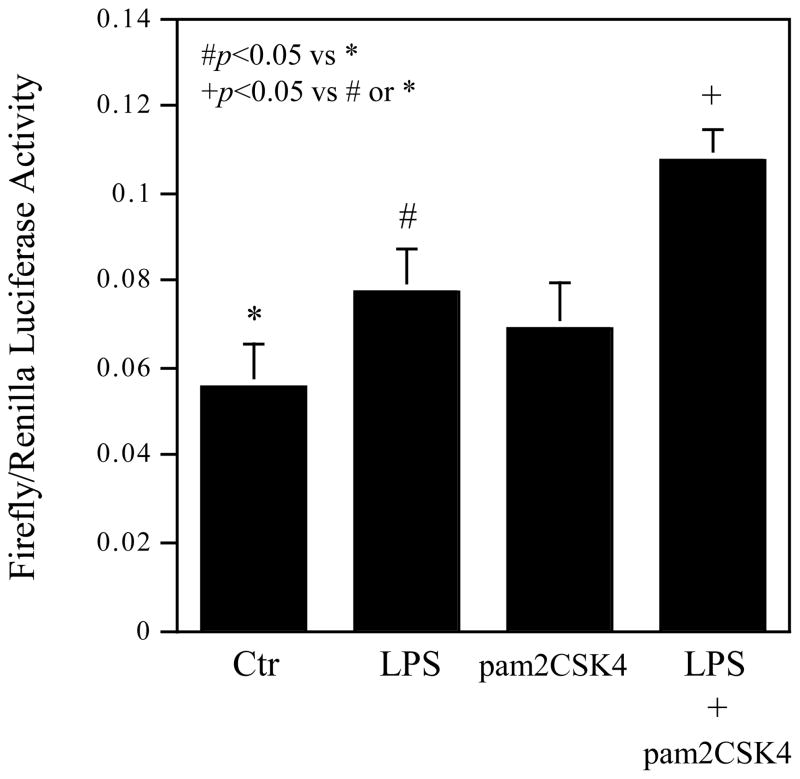
To determine if increase in IL-6 transcription activity is the mechanism involved in the augmentation of IL-6 mRNA expression by coactivation of TLR4 and TLR2/6, we transfected cells with luciferease reporter plasmids, whose expression was driven by an 1168-bp human IL-6 promoter region. Following the transfection, U937 cells were treated with LPS, pam2CSK4 or LPS plus pam2CSK4. Results showed that while LPS significantly stimulated luciferase activity, LPS plus pam2CSK4 increased the luciferase activity to a greater level as compared to LPS alone (Fig. 5), indicating that increased IL-6 transcriptional activity is responsible for the augmentation of IL-6 mRNA expression induced by coactivation of TLR4 and TLR2/6.
Figure 5.
The Effect of coactivation of TLR4 and TLR2/6 on IL-6 transcription. U937 cells were transiently cotransfected using FuGENE HD transfection reagent with 0.55 μg of p1168huIL6P-luc+ plasmids and pRL-TK plasmids (Renilla luciferase) 24 h. The cells were then treated with 100 ng/ml LPS, 100ng/ml pam2CSK4, or LPS plus pam2CSK4 for 24 h. After the treatment, the cells were lysed and the lysate was used for luciferase activity assay. The firefly luciferase levels were normalized to the Renilla luciferase levels. The presented data (mean ± SD) are representative of 3 experiments with similar results.
3.6. Requirement of p38 MAPK and NFκB Pathways for Augmentation of IL-6 Expression by Coactivation of TLR4 and TLR2/6
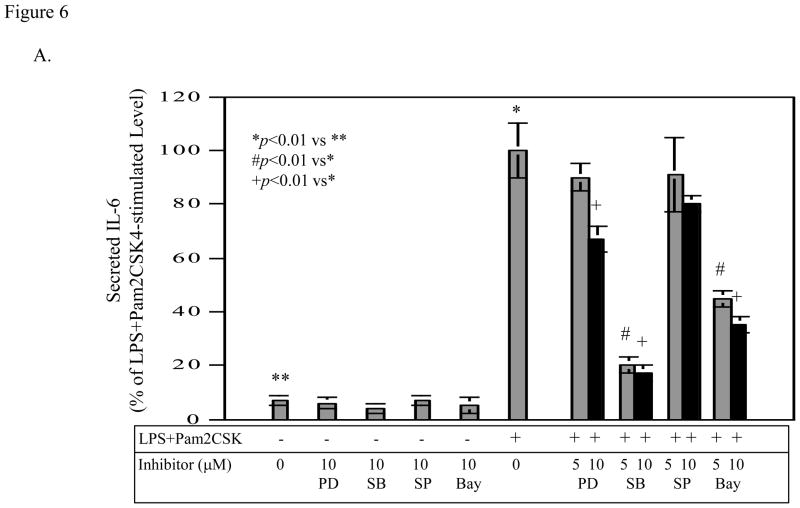
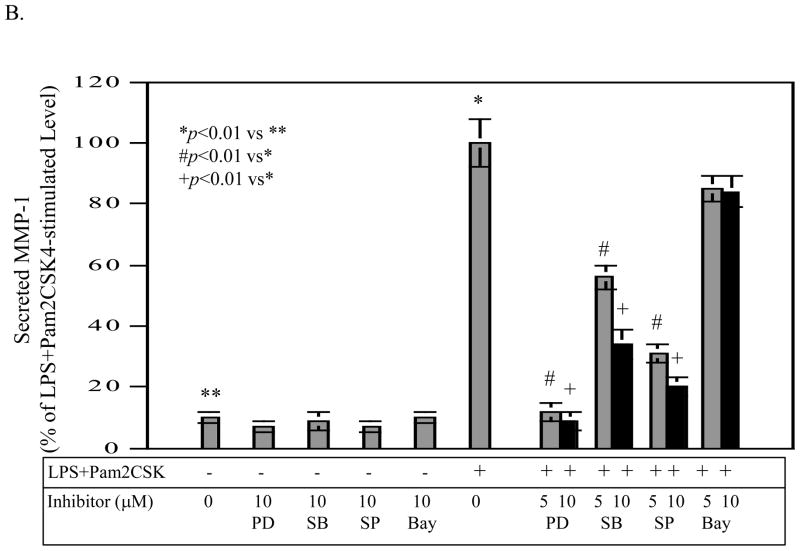
To further understand how coactivation of TLR4 and TLR2/6 augmented IL-6 expression as compared to activation of TLR4 or TLR2/6 alone, we sought to identify the signaling pathways required for the IL-6 upregulation using specific inhibitors for different signaling pathways. Results showed that SB203580 and Bay117085 were effective in inhibition of IL-6 production induced by coactivation of TLR4 and TLR2/6 (Fig. 6A). In contrast, PD98059 and SP600125 at the same concentrations had insignificant effect. These results suggest that p38 MAPK and NFκB pathways are required for IL-6 upregulation induced by coactivation of TLR4 and TLR2/6.
Figure 6.
The effect of PD98059, SB203580, SP600125 and Bay117085 on the stimulated IL-6 (A) and MMP-1 production (B) by LPS plus pam2CSK4. U937 cells were treated with 100 ng/ml of LPS plus 100 ng/ml of pam2CSK4 in the absence or presence of 5 or 10 μM of PD98059, SB203580, SP600125 or Bay117085 for 24 h. For control, U937 cells were treated with 10 μM of PD98059, SB203580, SP600125 or Bay117085 alone. After the treatment, IL-6 and MMP-1 secreted in medium were quantified using ELISA. The data were presented as % of LPS plus pam2CSK4-stimulated IL-6 or MMP-1 production. The data presented are the representative of 2 independent experiments with similar results.
To show the specificity of the requirement of p38 MAPK and NFκB pathways in IL-6 production, we determined the effects of these inhibitors on MMP-1 production by U937 cells. As shown in Fig. 6B, PD98059, SB203580, and SP600125 effectively inhibited MMP-1 production, indicating that the ERK, p38 MAPK, and JNK pathways are required for the stimulation of MMP-1 expression by coactivation of TLR4 and TLR2/6.
3.7. P38 MAPK Signaling Was Increased by Coactivation of TLR4 and TLR2/4
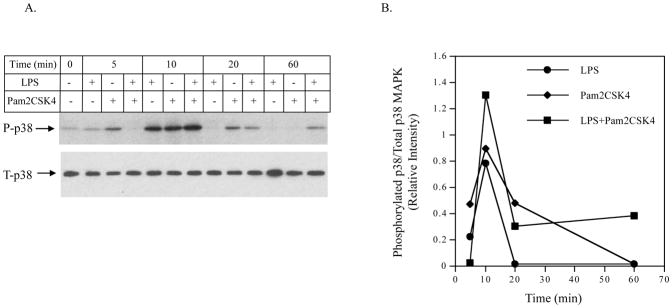
The findings from the above studies suggest that the p38 MAPK and NFκB pathways may mediate the augmentation of IL-6 expression by coactivation of TLR4 and TLR2/6. Thus, we determined if coactivation of TLR4 and TLR2/6 increased p38 MAPK and NFκB pathway signaling as compared to activation of TLR4 or TLR2/6 alone. Since our previous studies have shown that p38 MAPK is activated rapidly (Sundararaj et al., 2008), we determined the phosphorylation of p38 MAPK within 1 h.
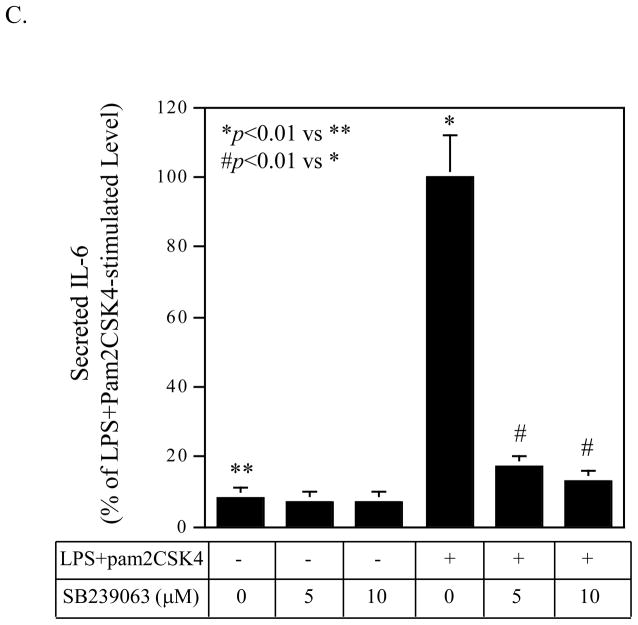
Interestingly, results showed that the kinetics of p38 MAPK phosphorylation in response to LPS, pam2CSK4 or LPS plus pam2CSK4 were different. As shown in Fig. 7A and 7B, LPS, pam2CSK4 or LPS plus pam2CSK4 stimulated the p38 MAPK phosphorylation in a time-dependent manner and the stimulation peaked at 10 min. At 10 min, the combination of LPS and pam2CSK4 was more potent than LPS or pam2CSK4 alone in the stimulation of p38 MAPK phosphorylation. Furthermore, the phosphorylation of p38 MAPK at 60 min was only observed in cells treated with LPS plus pam2CSK4, but not LPS or pam2CSK4 alone. Thus, these data showed that coactivation of TLR4 and TLR2/6 triggered a stronger and prolonged p38 MAPK signaling. To confirm the role of p38 MAPK pathway in the augmentation of IL-6 expression by coactivaiton of TLR4 and TLR2/6, we used another p38 MAPK inhibitor SB239063 (Strassburger et al., 2008) to block p38 MAPK signaling. Results showed that 10 μM of SB239063 inhibited IL-6 secretion stimulated by LPS plus pam2CSK4 by 88% (Fig. 7C).
Figure 7.
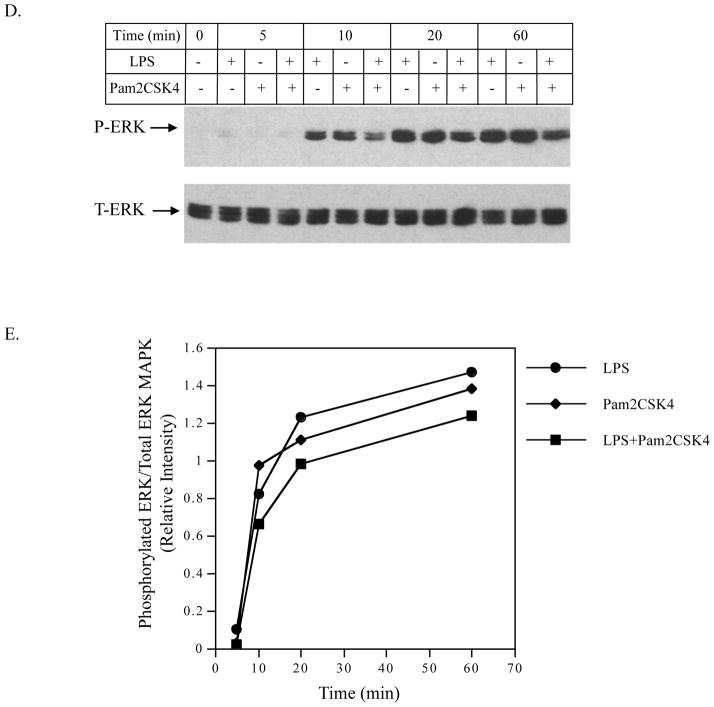
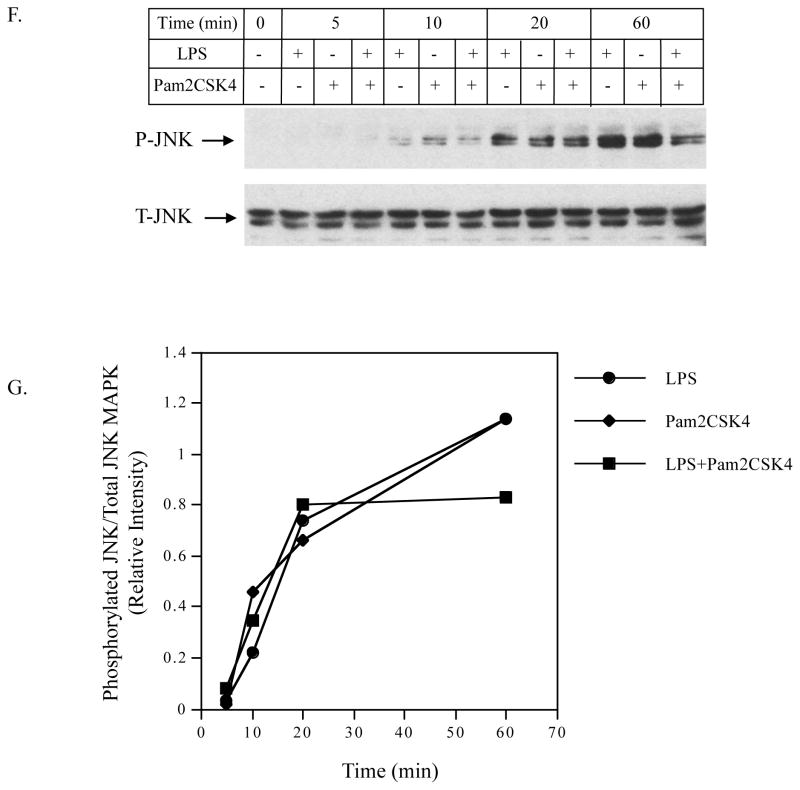
A and B: The effect of LPS, pam2CSK4 or LPS plus pam2CSK4 on phosphorylation of p38 MAPK. U937 cells were treated with LPS, pam2CSK4 or both for 0, 5, 10, 20 or 60 min. At each time point, cells were lysed and the cell lysate was subjected to immunoblotting to detect phosphorylated and total p38 MAPK. C: The effect of p38 MAPK inhibitor SB239063 on IL-6 secretion in response to LPS plus pam2CSK4. U937 cells were treated with or without 100 ng/ml of LPS plus pam2CSK4 in the absence or presence of 5 or 10 μM of SB239063 for 24 h. After the treatment, IL-6 in culture medium was quantified using ELISA. D to G: The effect of LPS, pam2CSK4 or LPS plus pam2CSK4 on ERK MAPK, and JNK MAPK. Similar studies as described in A and B were conducted to determine phosphorylated and total ERK and JNK MAPK. The bands for phosphorylated p38, ERK and JNK MAPK in the images were quantified using densitometric scanning and normalized to total p38, ERK and JNK MAPK (B, E and G). The data presented are the representative of 3 independent experiments with similar results. P-p38, P-ERK, P-JNK: phosphorylated p38, phosphorylated ERK, phosphorylated JNK. T-p38, T-ERK, T-JNK: total p38, total ERK, total JNK.
In contrast to the phosphorylation of p38 MAPK, no enhancement of the phosphorylation of ERK (Fig. 7D and 7E) and JNK (Fig. 7F and 7G) was observed in cells treated with LPS plus pam2CSK4.
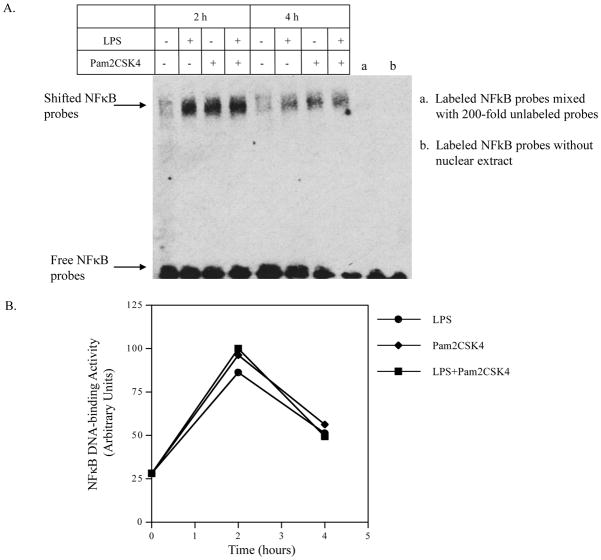
To determine if NFκB signaling is also enhanced by coactivation of TLR4 and TLR2/6, electrophoretic mobility shift assay was performed to show the nuclear NFκB DNA-binding activity. Results showed that while LPS and pam2CSK alone markedly induced NFκB DNA-binding activity, the combination of LPS and pam2CSK4 did not further enhance NFκB activity (Fig. 8A and 8B).
Figure 8.
The effect of LPS, pam2CSK4 or both on NFκB DNA-binding activity. U937 cells were treated with 100 ng/ml of LPS, 100 ng/ml of pam2CSK4 or both for 2 or 4 h. At each time point, nuclear protein was extracted and subjected to electrophoretic motility shift assay to determine NFκB DNA-binding activity as described in Methods. The data presented are the representative of 2 independent experiments with similar results.
4. DISCUSSION
In this study, we demonstrated that coactivation of TLR4 and TLR2/6 coordinates an additive augmentation of IL-6 expression by mononuclear U937 cells. Given the crucial role of IL-6 in diabetes and diabetic complications, this study has revealed an important linkage between the TLR4/TLR2 expression by mononuclear cells and the pathogenesis of diabetes and diabetic complications.
We focused on IL-6 in this study because IL-6 is a multifunctional cytokine involved in the acute phase response, immunity, hematopoiesis, and inflammation (Ishihara and Hirano, 2002; Kishimoto, 2006). IL-6 is a cytokine produced by a broad array of cell types including T cells, B cells, monocytes, macrophages, fibroblasts, endothelial cells, adipocytes, smooth muscle cells and synovial cells. IL-6 is a marker for cardiovascular disease (Kristiansen and Mandrup-Poulsen, 2005; Rattazzi et al., 2003), which is considered as an inflammatory disease and a major complication of diabetes. Studies have well documented that the plasma levels of IL-6 and C-reactive protein are strong independent predictors of risk of future cardiovascular events, both in patients with a history of coronary heart disease and in apparently healthy subjects (Rattazzi et al., 2003). In addition, IL-6 plays an essential role in many other chronic inflammatory diseases such as rheumatoid arthritis, systemic-onset juvenile chronic arthritis, osteoporosis, psoriasis, and autoimmune diseases such as antigen-induced arthritis and experimental allergic encephalomyelitis (Ishihara and Hirano, 2002). IL-6 is also important in the pathogenesis of infectious diseases such as periodontal disease (Geivelis et al., 1993; Rusconi et al., 1991). Furthermore, numerous studies have further demonstrated that IL-6 is not just a marker for inflammation-associated diseases; it is a major player involved in the initiation and progression of the diseases (Lowe, 2001; Moutsopoulos and Madianos, 2006; Rattazzi et al., 2003).
IL-6 plays an important role in the acute phase response to inflammation (Akira and Kishimoto, 1992). In consistence with this notion, our study showed that coactivation of TLR4 and TLR2/6 as well as activation of TLR4 or TLR2/6 alone led to a rapid and transient increase in IL-6 mRNA expression. The IL-6 mRNA expression was robustly stimulated to the peak in 2–8 hours and then vigorously suppressed in the next 12–18 hours. In consistence with the kinetic of IL-6 mRNA expression, although there was a rapid increase in IL-6 protein production, it reached a plateau at 12–24 hours, indicating that IL-6 protein secretion was nearly halted at that time. These findings clearly indicate that IL-6 expression is highly and critically controlled at the mRNA expression level.
The IL-6 mRNA expression kinetic pattern in response to TLR2/6 activation is different from that to TLR4 activation. As shown in Fig. 3A, IL-6 mRNA expression was kept at high level for 10 h after stimulation with pam2CSK4. In contrast, it dropped quickly after reaching the peak at 4 h after stimulation with LPS. Furthermore, the protein level in culture medium for cells treated with pam2CSK4 reached plateau at 12 h while the protein level in culture medium for cells treated with LPS reached plateau at 24 h. Interestingly, when both TLR4 and TLR2/6 were activated by LPS and pam2CSK4, IL-6 mRNA expression kinetic pattern was similar to that induced by LPS, but IL-6 protein secretion pattern was similar to that induced by pam2CSK4 (Fig. 3B), indication a crosstalk between TLR4 and TLR2/6 as the results of TLR4 and TLR2/6 coactivation that regulates IL-6 expression.
In response to TLR4, TLR2/6 or both TLR4 and TLR2/6 activation, the kinetic pattern of IL-6 mRNA expression is different from that of MMP-1 mRNA expression (Fig. 3B, a and c panels). The former showed a quick and transient upregulation while the latter showed a slow and graduate upregulation. At 24 h, IL-6 expression reached below the baseline, but MMP-1 expression was still in rising. We have shown that IL-6 is a potent stimulator for MMP-1 expression and IL-6 has synergistic effect with LPS on MMP-1 expression (Li et al., 2010), the early expression of IL-6 may further increase LPS-induced MMP-1 expression.
The crosstalk between TLR4 and TLR2/6 led to an additive augmentation on IL-6 mRNA expression. Augmentation of mRNA expression level of a particular gene can result from increased transcriptional activity of the gene or/and increased mRNA stability (or decreased mRNA turnover). Since our studies presented in Fig. 2 indicate that IL-6 mRNA was degraded rapidly after reaching the peak and the previous studies have shown that IL-6 mRNA expression level is increased by enhancing IL-6 mRNA stability (Patil et al., 2004), we determined if IL-6 mRNA stability is increased by coactivation of TLR4 and TLR2/6. The studies that quantified the cellular IL-6 mRNA after transcription was inhibited by actinomycin D excluded the possibility. Without enhancement of mRNA stability, increase IL-6 transcriptional activity by coactivation of TLR4 and TLR2/6 is the only possible mechanism involved in the augmentation of IL-6 mRNA expression. Indeed, our studies analyzing IL-6 promoter activity in cells transfected with plasmids containing 1168-bp of IL-6 promoter and the luciferase reporter showed that coactivation of TLR4 and TLR2/6 stimulated a higher IL-6 promoter activity than activation of TLR4 alone.
It has been shown that ligation of TLR4 by LPS and TLR2/1 by pam3CSK4 synergistically stimulated anti-inflammatory cytokine IL-10, but not proinflammatory cytokine TNFα, in murine dendritic cells (Hirata et al., 2008). Further investigations in this study showed that both p38 MAPK and JNK pathways were involved in the selective synergy in anti-inflammatory cytokine production. It is noteworthy that the synergy for anti-inflammatory cytokine production in this study was generated by coactivation of TLR4 and TLR2/1, but not TLR2/6. In contrast, our current study showed that coactivation of TLR4 and TLR2/6 led to an augmentation of proinflammatory cytokine IL-6 expression in mononuclear cells via p38 MAPK pathway, but not JNK pathway. It is possible that the coactivation of TLR4 with TLR2/6 or that of TLR4 with TLR2/6 may affect the balance between proinflammatory cytokine and anti-inflammatory cytokine expressions. These findings warrant further investigations.
Our further studies on signaling pathways showed that both p38 MAPK and NFκB pathways were essential for the augmentation of IL-6 expression induced by coactivation of TLR4 and TLR2/6. Interestingly, it was found that coactivation of TLR4 and TLR2/6 did not increase NFκB activity, although NFkB pathway was required for the augmentation of IL-6 expression. In contrast, p38 MAPK signaling was found to be enhanced and prolonged by coactivation of TLR4 and TLR2/6 as compared to activation of TLR4 or TLR2/6 alone, indicating that p38 MAPK is likely to play an essential role in the augmentation of IL-6 expression by coactivation of TLR4 and TLR2/6.
p38 MAPK pathway has been shown to mediate IL-6 expression in different types of cells in response to LPS (Thirunavukkarasu et al., 2006), IL-1β (Chae et al., 2005), prostaglandin E2 (Williams et al., 2000), and mechanical stress (Zampetaki et al., 2005). Thirunavukkarasu et al. reported that p38 MAPK pathway inhibitor SB203580 and NFκB pathway inhibitor pyrrolidine dithiocarbamate (PDTC), but not ERK pathway inhibitor PD98059 and JNK pathway inhibitor SP600125, inhibited IL-6 expression in hepatic stellate cells stimulated with LPS (Thirunavukkarasu et al., 2006), which is in agreement with our findings in this study. Although it was proposed that p38 MAPK interacted with NFκB pathway for IL-6 expression in smooth muscle cells by mechanical stress (Zampetaki et al., 2005), Chae et al. reported that p38 MAPK and NFκB pathways acted independently in IL-1β-stimulated human gingival fibroblasts for IL-6 expression (Chae et al., 2005). In our current study, we found that p38 MAPK, but not NFκB pathway, was enhanced by coactivation of TLR4 and TLR2/6, suggesting that p38 MAPK pathway is not linked upstream to the NFκB signaling pathway for IL-6 expression.
The present study showed that upon their activation, TLR4 and TLR2/6 shared many similar biological activities related to IL-6 expression. First, activation of either TLR4 or TLR2/6 led to rapid ERK, JNK, p38 MAPK and NFκB activation. Second, a fast and transient increase in IL-6 mRNA expression was observed in cells with either TLR4 or TLR2/6 activation. Third, a similar action between TLR4 and TLR2/6 activation on IL-6 protein production was observed. Besides these similarities, our study also showed some subtle differences between TLR4 and TLR2/6. First, as compared to TLR2/6 activation, TLR4 activation triggered a faster and more transient upregulation of IL-6 mRNA expression. Second, TLR4 activation leads to a more potent upregulation of IL-6 protein production than TLR2/6 activation. Third, the time period for p38 MAPK phosphorylation induced by TLR2/6 activation was longer than that by TLR4 activation. These differences may be related to the different structures of TLR4 and TLR2/6 signaling pathways. Interestingly, Re et al. have reported that TLR2 or TLR4 activation in dendritic cells leads to the transcriptional activation of different cytokine and chemokine genes (Re and Strominger, 2001), indicating a difference in genes targeted by TLR2 and TLR4 activation.
In conclusion, the present study showed for the first time that coactivation of TLR4 and TLR2/6 coordinated an additive augmentation on IL-6 gene transcription via p38 MAPK pathway. These findings suggest that p38 MAPK signaling pathway is the potential target for inhibition of IL-6 expression by mononuclear cells induced by coactivation of TLR4 and TLR2/6.
Acknowledgments
This work was supported by NIH grant DE016353 and a Merit Review Grant from Department of Veterans Affairs (to Y.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Kishimoto T. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunological reviews. 1992;127:25–50. doi: 10.1111/j.1600-065x.1992.tb01407.x. [DOI] [PubMed] [Google Scholar]
- Bendtzen K, Buschard K, Diamant M, Horn T, Svenson M. Possible role of IL-1, TNF-alpha, and IL-6 in insulin-dependent diabetes mellitus and autoimmune thyroid disease. Thyroid Cell Group. Lymphokine Res. 1989;8:335–40. [PubMed] [Google Scholar]
- Bjorkbacka H, Kunjathoor VV, Moore KJ, Koehn S, Ordija CM, Lee MA, Means T, Halmen K, Luster AD, Golenbock DT, Freeman MW. Reduced atherosclerosis in MyD88-null mice links elevated serum cholesterol levels to activation of innate immunity signaling pathways. Nat Med. 2004;10:416–21. doi: 10.1038/nm1008. [DOI] [PubMed] [Google Scholar]
- Caricilli AM, Nascimento PH, Pauli JR, Tsukumo DM, Velloso LA, Carvalheira JB, Saad MJ. Inhibition of toll-like receptor 2 expression improves insulin sensitivity and signaling in muscle and white adipose tissue of mice fed a high-fat diet. J Endocrinol. 2008;199:399–406. doi: 10.1677/JOE-08-0354. [DOI] [PubMed] [Google Scholar]
- Chae HJ, Byun JO, Chae SW, Kim HM, Choi HI, Pae HO, Chung HT, Kim HR. p38 MAPK and NF-kappaB on IL-6 release in human gingival fibroblasts. Immunopharmacol Immunotoxicol. 2005;27:631–46. doi: 10.1080/08923970500418851. [DOI] [PubMed] [Google Scholar]
- Dasu MR, Devaraj S, Park S, Jialal I. Increased Toll-like Receptor activation and TLR ligands in Recently Diagnosed Type 2 diabetes Subjects. Diabetes Care. 2010;33:861–8. doi: 10.2337/dc09-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasu MR, Devaraj S, Zhao L, Hwang DH, Jialal I. High glucose induces toll-like receptor expression in human monocytes: mechanism of activation. Diabetes. 2008;57:3090–8. doi: 10.2337/db08-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dekker WK, Cheng C, Pasterkamp G, Duckers HJ. Toll like receptor 4 in atherosclerosis and plaque destabilization. Atherosclerosis. 2010;209:314–20. doi: 10.1016/j.atherosclerosis.2009.09.075. [DOI] [PubMed] [Google Scholar]
- Devaraj S, Dasu MR, Park SH, Jialal I. Increased levels of ligands of Toll-like receptors 2 and 4 in type 1 diabetes. Diabetologia. 2009;52:1665–8. doi: 10.1007/s00125-009-1394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S, Dasu MR, Rockwood J, Winter W, Griffen SC, Jialal I. Increased toll-like receptor (TLR) 2 and TLR4 expression in monocytes from patients with type 1 diabetes: further evidence of a proinflammatory state. J Clin Endocrinol Metab. 2008;93:578–83. doi: 10.1210/jc.2007-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geivelis M, Turner DW, Pederson ED, Lamberts BL. Measurements of interleukin-6 in gingival crevicular fluid from adults with destructive periodontal disease. J Periodontol. 1993;64:980–3. doi: 10.1902/jop.1993.64.10.980. [DOI] [PubMed] [Google Scholar]
- Hayes MM, Lane BR, King SR, Markovitz DM, Coffey MJ. Peroxisome proliferator-activated receptor gamma agonists inhibit HIV-1 replication in macrophages by transcriptional and post-transcriptional effects. J Biol Chem. 2002;277:16913–9. doi: 10.1074/jbc.M200875200. [DOI] [PubMed] [Google Scholar]
- Hernesniemi J, Lehtimaki T, Rontu R, Islam MS, Eklund C, Mikkelsson J, Ilveskoski E, Kajander O, Goebeler S, Viiri LE, Hurme M, Karhunen PJ. Toll-like receptor 4 polymorphism is associated with coronary stenosis but not with the occurrence of acute or old myocardial infarctions. Scand J Clin Lab Invest. 2006;66:667–75. doi: 10.1080/00365510600933011. [DOI] [PubMed] [Google Scholar]
- Hernesniemi JA, Raitakari OT, Kahonen M, Juonala M, Hutri-Kahonen N, Marniemi J, Viikari J, Lehtimaki T. Toll-like receptor 4 gene (Asp299Gly) polymorphism associates with carotid artery elasticity. The cardiovascular risk in young Finns study. Atherosclerosis. 2008;198:152–9. doi: 10.1016/j.atherosclerosis.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Hirata N, Yanagawa Y, Ebihara T, Seya T, Uematsu S, Akira S, Hayashi F, Iwabuchi K, Onoe K. Selective synergy in anti-inflammatory cytokine production upon cooperated signaling via TLR4 and TLR2 in murine conventional dendritic cells. Molecular immunology. 2008;45:2734–42. doi: 10.1016/j.molimm.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Huang Q, Pope RM. Toll-like receptor signaling: a potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J Leukoc Biol. 2010;88:253–62. doi: 10.1189/jlb.0310126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–68. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Kanczkowski W, Morawietz H, Ziegler CG, Funk RH, Schmitz G, Zacharowski K, Mohn CE, Ehrhart-Bornstein M, Bornstein SR. Pam3CSK4 and LTA-TLRs ligands associated with microdomains induce IL8 production in human adrenocortical cancer cells. Horm Metab Res. 2007;39:457–60. doi: 10.1055/s-2007-980189. [DOI] [PubMed] [Google Scholar]
- King GL. The role of inflammatory cytokines in diabetes and its complications. J Periodontol. 2008;79:1527–34. doi: 10.1902/jop.2008.080246. [DOI] [PubMed] [Google Scholar]
- Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res Ther. 2006;8(Suppl 2):S2. doi: 10.1186/ar1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen OP, Mandrup-Poulsen T. Interleukin-6 and diabetes: the good, the bad, or the indifferent? Diabetes. 2005;54(Suppl 2):S114–24. doi: 10.2337/diabetes.54.suppl_2.s114. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Connolly NL, Katamine S, Cheah MS, Senior RM, Robbins KC. Tissue-specific expression and developmental regulation of the human fgr proto-oncogene. Mol Cell Biol. 1989;9:92–9. doi: 10.1128/mcb.9.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sun B. Toll-like receptor 4 in atherosclerosis. J Cell Mol Med. 2007;11:88–95. doi: 10.1111/j.1582-4934.2007.00011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Samuvel DJ, Sundararaj KP, Lopes-Virella MF, Huang Y. IL-6 and high glucose synergistically upregulate MMP-1 expression by U937 mononuclear phagocytes via ERK1/2 and JNK pathways and c-Jun. Journal of cellular biochemistry. 2010;110:248–59. doi: 10.1002/jcb.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chang YM, Yu JM, Yen JH, Chang JG, Hu CJ. Toll-like receptor 4 gene C119A but not Asp299Gly polymorphism is associated with ischemic stroke among ethnic Chinese in Taiwan. Atherosclerosis. 2005;180:305–9. doi: 10.1016/j.atherosclerosis.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Liu X, Ukai T, Yumoto H, Davey M, Goswami S, Gibson FC, 3rd, Genco CA. Toll-like receptor 2 plays a critical role in the progression of atherosclerosis that is independent of dietary lipids. Atherosclerosis. 2008;196:146–54. doi: 10.1016/j.atherosclerosis.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe GD. The relationship between infection, inflammation, and cardiovascular disease: an overview. Ann Periodontol. 2001;6:1–8. doi: 10.1902/annals.2001.6.1.1. [DOI] [PubMed] [Google Scholar]
- Michelsen KS, Wong MH, Shah PK, Zhang W, Yano J, Doherty TM, Akira S, Rajavashisth TB, Arditi M. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc Natl Acad Sci U S A. 2004;101:10679–84. doi: 10.1073/pnas.0403249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos NM, Madianos PN. Low-grade inflammation in chronic infectious diseases: paradigm of periodontal infections. Ann N Y Acad Sci. 2006;1088:251–64. doi: 10.1196/annals.1366.032. [DOI] [PubMed] [Google Scholar]
- Mullick AE, Tobias PS, Curtiss LK. Modulation of atherosclerosis in mice by Toll-like receptor 2. J Clin Invest. 2005;115:3149–56. doi: 10.1172/JCI25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal K, Plantinga TS, Wong J, Monks BG, Gay NJ, Netea MG, Fitzgerald KA, Golenbock DT. A TIR domain variant of MyD88 adapter-like (Mal)/TIRAP results in loss of MyD88 binding and reduced TLR2/TLR4 signaling. J Biol Chem. 2009;284:25742–8. doi: 10.1074/jbc.M109.014886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norata GD, Garlaschelli K, Ongari M, Raselli S, Grigore L, Benvenuto F, Maggi FM, Catapano AL. Effect of the Toll-like receptor 4 (TLR-4) variants on intima-media thickness and monocyte-derived macrophage response to LPS. J Intern Med. 2005;258:21–7. doi: 10.1111/j.1365-2796.2005.01509.x. [DOI] [PubMed] [Google Scholar]
- Pankewycz OG, Guan JX, Benedict JF. Cytokines as mediators of autoimmune diabetes and diabetic complications. Endocr Rev. 1995;16:164–76. doi: 10.1210/edrv-16-2-164. [DOI] [PubMed] [Google Scholar]
- Patil C, Zhu X, Rossa C, Jr, Kim YJ, Kirkwood KL. p38 MAPK regulates IL-1beta induced IL-6 expression through mRNA stability in osteoblasts. Immunological investigations. 2004;33:213–33. doi: 10.1081/imm-120034231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol. 1997;17:3733–43. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin MS, Sinha S, Bhatt BA, Dedousis N, O’Doherty RM. Inhibition or deletion of the lipopolysaccharide receptor Toll-like receptor-4 confers partial protection against lipid-induced insulin resistance in rodent skeletal muscle. Diabetologia. 2008;51:336–46. doi: 10.1007/s00125-007-0861-3. [DOI] [PubMed] [Google Scholar]
- Rattazzi M, Puato M, Faggin E, Bertipaglia B, Zambon A, Pauletto P. C-reactive protein and interleukin-6 in vascular disease: culprits or passive bystanders? J Hypertens. 2003;21:1787–803. doi: 10.1097/00004872-200310000-00002. [DOI] [PubMed] [Google Scholar]
- Re F, Strominger JL. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human dendritic cells. J Biol Chem. 2001;276:37692–9. doi: 10.1074/jbc.M105927200. [DOI] [PubMed] [Google Scholar]
- Rusconi F, Parizzi F, Garlaschi L, Assael BM, Sironi M, Ghezzi P, Mantovani A. Interleukin 6 activity in infants and children with bacterial meningitis. The Collaborative Study on Meningitis. Pediatr Infect Dis J. 1991;10:117–21. doi: 10.1097/00006454-199102000-00008. [DOI] [PubMed] [Google Scholar]
- Samuvel DJ, Sundararaj KP, Li Y, Lopes-Virella MF, Huang Y. Adipocyte-mononuclear cell interaction, Toll-like receptor 4 activation, and high glucose synergistically up-regulate osteopontin expression via an interleukin 6-mediated mechanism. J Biol Chem. 2010;285:3916–27. doi: 10.1074/jbc.M109.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburger M, Braun H, Reymann KG. Anti-inflammatory treatment with the p38 mitogen-activated protein kinase inhibitor SB239063 is neuroprotective, decreases the number of activated microglia and facilitates neurogenesis in oxygen-glucose-deprived hippocampal slice cultures. European journal of pharmacology. 2008;592:55–61. doi: 10.1016/j.ejphar.2008.06.099. [DOI] [PubMed] [Google Scholar]
- Sundararaj KP, Samuvel DJ, Li Y, Nareika A, Slate EH, Sanders JJ, Lopes-Virella MF, Huang Y. Simvastatin suppresses LPS-induced MMP-1 expression in U937 mononuclear cells by inhibiting protein isoprenylation-mediated ERK activation. J Leukoc Biol. 2008;84:1120–9. doi: 10.1189/jlb.0108064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaj KP, Samuvel DJ, Li Y, Sanders JJ, Lopes-Virella MF, Huang Y. Interleukin-6 released from fibroblasts is essential for up-regulation of matrix metalloproteinase-1 expression by U937 macrophages in coculture: cross-talking between fibroblasts and U937 macrophages exposed to high glucose. J Biol Chem. 2009;284:13714–24. doi: 10.1074/jbc.M806573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;17:565–77. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu C, Watkins SC, Gandhi CR. Mechanisms of endotoxin-induced NO, IL-6, and TNF-alpha production in activated rat hepatic stellate cells: role of p38 MAPK. Hepatology. 2006;44:389–98. doi: 10.1002/hep.21254. [DOI] [PubMed] [Google Scholar]
- Tobias P, Curtiss LK. Thematic review series: The immune system and atherogenesis. Paying the price for pathogen protection: toll receptors in atherogenesis. J Lipid Res. 2005;46:404–11. doi: 10.1194/jlr.R400015-JLR200. [DOI] [PubMed] [Google Scholar]
- Tsukumo DM, Carvalho-Filho MA, Carvalheira JB, Prada PO, Hirabara SM, Schenka AA, Araujo EP, Vassallo J, Curi R, Velloso LA, Saad MJ. Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes. 2007;56:1986–98. doi: 10.2337/db06-1595. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W, De Bosscher K, Boone E, Plaisance S, Haegeman G. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–8. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- Vanden Berghe W, Plaisance S, Boone E, De Bosscher K, Schmitz ML, Fiers W, Haegeman G. p38 and extracellular signal-regulated kinase mitogen-activated protein kinase pathways are required for nuclear factor-kappaB p65 transactivation mediated by tumor necrosis factor. J Biol Chem. 1998;273:3285–90. doi: 10.1074/jbc.273.6.3285. [DOI] [PubMed] [Google Scholar]
- Williams JA, Pontzer CH, Shacter E. Regulation of macrophage interleukin-6 (IL-6) and IL-10 expression by prostaglandin E2: the role of p38 mitogen-activated protein kinase. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research. 2000;20:291–8. doi: 10.1089/107999000312423. [DOI] [PubMed] [Google Scholar]
- Yamato K, el-Hajjaoui Z, Simon K, Koeffler HP. Modulation of interleukin-1 beta RNA in monocytic cells infected with human immunodeficiency virus-1. J Clin Invest. 1990;86:1109–14. doi: 10.1172/JCI114815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Zhang Z, Hu Y, Xu Q. Biomechanical stress induces IL-6 expression in smooth muscle cells via Ras/Rac1-p38 MAPK-NF-kappaB signaling pathways. Am J Physiol Heart Circ Physiol. 2005;288:H2946–54. doi: 10.1152/ajpheart.00919.2004. [DOI] [PubMed] [Google Scholar]