Abstract
Genetic variation in SIRT1 affects obesity-related phenotypes in several populations. The purpose of this study was to determine whether variation in SIRT1 affects susceptibility to obesity or type 2 diabetes in Pima Indians, a population with very high prevalence and incidence rates of these diseases. Genotypic data from single nucleotide polymorphisms (SNPs) identified by sequencing regions of SIRT1 combined with SNPs in/near SIRT1 from a prior genome-wide association study determined that 4 tag SNPs (rs7895833, rs10509291, rs7896005, and rs4746720) could capture information across this gene and its adjacent 5′ region. The tag SNPs were genotyped in a population-based sample of 3501 Pima Indians (44% had diabetes, 58% female) for association with type 2 diabetes and BMI. Metabolic trait data and adipose biopsies were available on a subset of these subjects. Two tag SNPs, rs10509291 and rs7896005, were nominally associated with type 2 diabetes (P = 0.01, OR = 1.25 95%CI 1.05-1.48, and P = 0.02, OR = 1.17 95%CI 1.02-1.34, respectively; additive P values adjusted for age, sex, birth year, and family membership), but not BMI (adjusted P values 0.52 and 0.45, respectively). Among metabolically characterized subjects with normal glucose tolerance (N = 243), those carrying the diabetes risk allele (T) for rs10509291 and (G) for rs7896005 had a reduced acute insulin response (AIR) to an intravenous glucose bolus (adjusted P = 0.045 and 0.035, respectively). SIRT1 expression in adipose biopsies was negatively correlated with BMI (adjusted P = 0.00001). We conclude that variation in SIRT1 is nominally associated with reduced AIR and increased risk for type 2 diabetes. SIRT1 expression in adipose is correlated with BMI, but it remains unknown whether this is a cause or consequence of obesity.
Keywords: SIRT1, type 2 diabetes, insulin secretion, genetic associations
1. Introduction
Sirtuin 1 (SIRT1) belongs to a group of highly conserved NAD+-dependent protein deacetylases (SIRT1-7) which can be found in the nucleus, cytoplasm, and mitochondria [1]. SIRT1 has been shown to regulate several proteins involved in regulating lipid and glucose metabolism [2]. SIRT1 is widely expressed and its action upon different substrates, including forkhead-box transcription factors (FOXOs), peroxisome proliferator-activated receptor γ (PPARγ), PPARγ-coactivator 1α (PGC1-α), myogenic differentiation 1 (MyoD), and p53 [2,3], results in diverse tissue-specific effects including enhanced glyconeogenesis and repressed glycolysis in the liver [2,4,5], reduction of adipogenesis in adipose tissue [2,6], up-regulation of insulin secretion in pancreatic beta cells [2,7] and increased mitochondrial activity, fatty acid oxidation, and insulin sensitivity in skeletal muscle [2]. Genetic variants in SIRT1 have been shown to be modestly associated with human diabetes and obesity-related phenotypes in several studies [8-11]. The purpose of the present study is to determine whether single nucleotide polymorphisms (SNPs) within the SIRT1 locus have a role in increasing susceptibility to type 2 diabetes or obesity in the Pima Indian population.
2. Materials and Methods
2.1. Subjects and phenotyping
Subjects are from a longitudinal study examining the etiology of type 2 diabetes and obesity in the Gila River Indian Community [12]. Tag SNPs were genotyped in a population-based sample of full-heritage Pima Indians which included all individuals who had available DNA, measures of diabetes status and body mass index (BMI), and whose heritage was reported as full Pima and/or Tohono O'odham (a closely related tribe). In this sample of 3501 full heritage Pima Indians, the mean BMI was 37.1 +/- 8.4, the mean age was 35.4 +/- 13.0 years, 58% were female and 44% had type 2 diabetes. To assess independent replication in other American Indians, SNPs were further genotyped in a second sample of 3003 individuals who had available DNA and measures of diabetes status and BMI regardless of heritage. Individuals in this second sample were predominately mixed-heritage American Indians, whose reported heritage was, on average, 3/4 American Indian. In this second sample of 3003 “mixed heritage” subjects, the mean BMI was 34.6 +/- 8.7, the mean age was 28.9 +/- 11.8 years, 57% were female and 20% had type 2 diabetes. Diabetes was determined by a 75g oral glucose tolerance test and the results are interpreted according to the criteria of the World Health Organization [13]. BMI was defined as the maximum BMI recorded after the age of 15 years. Metabolic traits were assessed in 424 full-heritage non-diabetic Pima Indians in our Clinical Research Center [14]. A subset of these individuals also underwent muscle and/or adipose biopsies [15]. Insulin action was assessed using the two-step hyperinsulinemic-euglycemic clamp technique, where both basal glucose appearance and insulin-stimulated glucose disappearance (uptake) rates were measured to evaluate insulin sensitivity [14]. Percent body fat was estimated by underwater weighing or dual-energy X-ray absorptiometry (DPX-1; Lunar Radiation) [16]. To determine the acute insulin response (AIR), subjects received a 25g bolus of glucose injected intravenously over 3 minutes and blood samples were collected before infusion, and at 3, 4, 5, 6, 8, and 10 min after the injection for determination of plasma glucose and insulin concentrations. AIR was calculated as the mean increment in plasma insulin concentrations from 3 to 5 min [14]. Analysis of AIR was limited to individuals with normal glucose tolerance (N = 243). All studies were approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases.
2.2. DNA sequencing and genotyping
To identify novel sequence variants, all nine exons, including adjacent exon-intron boundaries extending ∼100 bp into each intron, and 2 kb of the putative promoter region of SIRT1 were sequenced in DNA samples from 24 Pima Indians who were not first degree relatives. Sequencing was performed using Big Dye Terminator (Applied Biosystems) on an automated DNA capillary sequencer (model 3730; Applied Biosystems). SNPs were genotyped by the method of SNPlex (Applied Biosystems) following the manufacturer's protocol.
2.3. Statistical analyses
Statistical analyses were performed using the software of the SAS Institute (Cary, NC). Numeric variables were expressed as means ± SD. The relationship between genotype and continuous variables was assessed by linear regression with adjustment for appropriate covariates (detailed in Tables 2 and 3). The association of SNPs with type 2 diabetes was assessed by logistic regression with adjustments for age and sex. Because many subjects were related, analyses accounted for family membership. For regression modeling in the additive model, homozygotes for the major allele (1/1), heterozygotes (1/2), and homozygotes for the minor allele (2/2) were coded to a numeric variable for genotype (0, 1, and 2). Haploview was used to place SNPs in haplotype blocks and to calculate D' and r2 value. Partial regression plots were used to assess the association of expression and obesity, independent of effects of age and sex. Adjusted values were expressed as residual values plus the mean of the unadjusted levels for both axes. A P value of <0.05 was considered to be nominally significant. Since this gene was analyzed as a biologic candidate gene, we chose to preserve power and did not adjust for multiple testing.
Table 2. Association of SIRT1 tag SNPs with BMI in the population-based sample of 3501 Pima Indians.
| SNP | M/m | Sex | mAF | BMIa | Pb | Pc | ||
|---|---|---|---|---|---|---|---|---|
| M/M | M/m | m/m | ||||||
| rs7895833 | A/G | All | 0.48 | 37.3 ± 8.4 | 37.2 ± 8.5 | 37.0 ± 8.2 | 0.92 | 0.90 |
| Male | 35.9 ± 8.1 | 35.1 ± 7.8 | 35.7 ± 7.8 | |||||
| Female | 38.3 ± 8.5 | 38.5 ± 8.7 | 38.0 ± 8.4 | |||||
| rs10509291 | T/A | All | 0.14 | 36.9 ± 8.2 | 37.2 ± 8.5 | 37.9 ± 9.8 | 0.52 | 0.49 |
| Males | 35.3 ± 7.7 | 35.2 ± 8.2 | 36.2 ± 7.8 | |||||
| Females | 38.0 ± 8.4 | 38.6 ± 8.5 | 39.5 ± 11.3 | |||||
| rs7896005 | A/G | All | 0.30 | 37.0 ± 8.5 | 37.2 ± 8.5 | 37.2 ± 8.2 | 0.45 | 0.60 |
| Male | 35.4 ± 8.0 | 35.1 ± 7.6 | 35.9 ± 8.8 | |||||
| Female | 38.1 ± 8.5 | 38.6 ± 8.7 | 38.3 ± 7.5 | |||||
| rs4746720 | T/C | All | 0.33 | 37.4 ± 8.6 | 37.0 ± 8.5 | 36.7 ± 7.6 | 0.67 | 0.27 |
| Male | 35.3 ± 8.0 | 35.3 ± 8.1 | 35.3 ± 7.5 | |||||
| Female | 38.8 ± 8.7 | 38.2 ± 8.6 | 37.7 ± 7.6 | |||||
Unadjusted mean BMI ± SD by genotypic group is shown.
Additive P-values were adjusted for age, sex, birth year and family membership in the entire sample, and age, birth year and family membership in males and females alone.
P-values for sex interaction.
Table 3. Associations between rs10509291 and rs7896005 and traits that predict type 2 diabetes.
| rs10509291 | rs7896005 | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | |||||||
| TT | TA | AA | P | AA | AG | GG | P | |
| All non-diabetic subjects | ||||||||
| Number (males/females) | 151/117 | 39/28 | 12/5 | 89/78 | 77/59 | 20/14 | ||
| Percent body fatabc | 33.4 ± 8.1 (33.2 ± 6.8) | 32.5 ± 9.5 (33.6 ± 7.0) | 31.2 ± 6.6 (33.1 ± 6.2) | 0.82 | 33.6 ± 8.7 (33.5 ± 6.9) | 33.3 ± 8.2 (33.7 ± 6.8) | 33.5 ± 6.7 (33.4 ± 5.0) | 0.94 |
| Log10 insulin stimulated glucose disposal (mg·kg EMBS-1·min-1)abcd | 0.54 ± 0.11 (0.54 ± 0.09) | 0.56 ± 0.12 (0.55 ± 0.11) | 0.53 ± 0.12 (0.54 ± 0.11) | 0.70 | 0.55 ± 0.12 (0.55 ± 0.10) | 0.54 ± 0.11 (0.54 ± 0.09) | 0.54 ± 0.09 (0.53 ± 0.08) | 0.42 |
| Normal glucose tolerant | ||||||||
| Number (males/females) | 116/74 | 34/13 | 9/2 | 73/45 | 61/36 | 16/12 | ||
| Log10 acute insulin response (μU/ml)abcde | 2.31 ± 0.29 (2.31 ± 0.28) | 2.39 ± 0.24 (2.42 ± 0.23) | 2.40 ± 0.27 (2.36 ± 0.25) | 0.045 | 2.36 ± 0.25 (2.36 ± 0.24) | 2.29 ± 0.32 (2.31 ± 0.31) | 2.29 ± 0.31 (2.27 ± 0.29) | 0.035 |
All subjects are non-diabetic, however, analysis of acute insulin response (AIR) is restricted to individuals with normal glucose tolerance because AIR declines as a consequence of impaired glucose tolerance. Means ± SD for each trait are given as both the raw mean and the adjusted mean (adjusted shown in parenthesis). Covariates for adjusted means and P-values are:
age,
sex,
family membership,
percent body fat, and
glucose disposal rate. EMBS = estimated metabolic body size.
3. Results
Genotypic data from 9 SNPs detected by sequencing (2 in the putative promoter region, 5 in intronic regions, and 2 in the 3′-UTR) and 6 SNPs near/within SIRT1 represented on the Affymetrix Genome-Wide Human SNP Array 6.0 (genotyped in ∼1120 Pima Indians as part of a genome-wide association study, [17]) were used to determine the linkage disequilibrium (LD) pattern across SIRT1 (Fig. 1). Three tag SNPs rs10509291, rs7896005, and rs4746720 (with redundancy defined as r2 >0.90) which capture the known common variation across the SIRT1 locus, and a fourth SNP (rs7895833) approximately 20 kb upstream of SIRT1, were selected for further genotyping in a population-based sample of full-heritage Pima Indians (N = 3501). Neither rs7895833 nor rs4746720 were associated with diabetes (Table 1). In contrast, 2 tag SNPs, rs7896005 which served as a tag for most of the SNPs in SIRT1 (Fig. 1) and rs10509291 were nominally associated with diabetes (Table 1). For rs7896005 the minor (G) allele was the risk allele for diabetes (OR = 1.17, 95%CI 1.02-1.34; P = 0.02 adjusted for age, sex, birth year, and family membership), and for rs10509291, the major (T) allele was associated with increased risk for diabetes (P = 0.01, OR = 1.25 95%CI 1.05-1.48, adjusted for age, sex, birth year, and family membership). There was a modest sex interaction with the association between rs7896005 and diabetes (P = 0.02); therefore, we further analyzed this SNP in men and women separately (58% of the individuals were women). The evidence for association with type 2 diabetes came from the women (OR = 1.37 95% CI 1.14-1.65; P = 0.0008 adjusted for age and birth year) as compared to men (OR = 0.97 95% CI 0.79-1.18; P = 0.74) (Table 1). None of the 4 tag SNPs was associated with BMI in either the entire population-based sample or when men and women were analyzed separately (Table 2).
Figure 1.
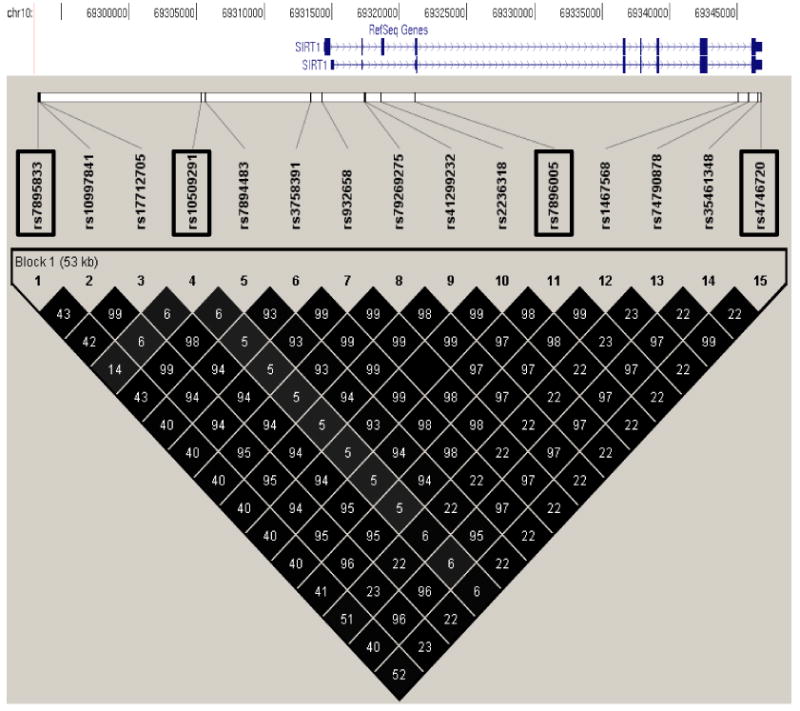
Linkage disequilibrium pattern (D' is indicated by shading and r2 is shown as numbers) for SNPs spanning SIRT1 and the adjacent 5′ (21 kb) region in Pima Indians. Genotypic data from 15 SNPs, identified by sequencing SIRT1 or present on the Affymetrix Genome-Wide Human SNP Array 6.0, were used to determine that all variants could be captured by 4 representative SNPs (denoted by open rectangles). Sequence information for the 3 novel SNPs: SIRT1-1, TGCAATGGCG[C/G]GATCTCGGCT; SIRT1-2, CTCAGCCTCC[C/G]C AGCAGCTGG; SIRT1-3, AATAAAGGCA[G/A] AGCTGGAACC.
Table 1. Association of SIRT1 tag SNPs with type 2 diabetes in the population-based sample of 3501 Pima Indians.
| SNPa | M/m | Sex | Risk allele frequency diabetic/non-diabetic | Pb | Risk allele OR(95% CI) | Pc |
|---|---|---|---|---|---|---|
| rs7895833 | A/G | All | 0.52/0.52 | 0.16 | 1.09(0.97-1.23) | 0.49 |
| Males | 0.51/0.52 | 0.87 | 1.01(0.85-1.22) | |||
| Females | 0.52/0.52 | 0.11 | 1.14(0.97-1.33) | |||
| rs10509291 | T/A | All | 0.87/0.85 | 0.01 | 1.25(1.05-1.48) | 0.94 |
| Males | 0.87/0.85 | 0.10 | 1.24(0.96-1.60) | |||
| Females | 0.87/0.85 | 0.06 | 1.24(0.99-1.54) | |||
| rs7896005 | A/G | All | 0.31/0.29 | 0.02 | 1.17(1.02- 1.34) | 0.02 |
| Males | 0.30/0.31 | 0.73 | 0.97(0.79-1.18) | |||
| Females | 0.31/0.28 | 0.0008 | 1.37(1.14-1.65) | |||
| rs4746720 | T/C | All | 0.34/0.32 | 0.79 | 1.02(0.90-1.15) | 0.48 |
| Males | 0.34/0.32 | 0.35 | 1.10(0.90-1.34) | |||
| Females | 0.34/0.32 | 0.60 | 0.96(0.81-1.13) |
Representative SNPs selected from Fig. 1. M, major allele; m, minor allele. The risk allele (defined when analysis is done in All subjects) is underlined.
Additive P-values for association with type 2 diabetes, adjusted for age, sex, birth year, and family membership.
P-value for sex interaction. Subjects for analysis were All (N = 3501, 45% are diabetic); Males (N = 1482, 39% are diabetic); Females (N = 2019, 49% are diabetic).
The nominal associations of rs7896005 and rs10509291with type 2 diabetes led us to evaluate whether these variants were further associated with metabolic traits predictive of type 2 diabetes, namely percent body fat, insulin resistance, and glucose-stimulated acute insulin response. Neither SNP was associated with percent body fat or insulin resistance as assessed by insulin-stimulated glucose disposal rate during a clamp (Table 3). However, both SNPs were associated with acute insulin response where subjects with normal glucose tolerance carrying the diabetes risk alleles (rs10509291, T and rs7896005, G) had reduced insulin secretion (P = 0.048 and 0.035 respectively, after adjusting for age, sex, percent body fat, and insulin-stimulated glucose disposal rate; Table 3). There was not a statistically significant sex interaction for rs7896005 and acute insulin response, although this analysis may be less reliable due to the small sample size of subjects with normal glucose tolerance and the fact that only 38% of these subjects are female (Table 3). It is also possible that the positive associations are false; the P values in this study are of nominal significance and none would have remained significant if adjusted for multiple testing.
To assess replication of the type 2 diabetes association in other Native Americans, rs7896005 was genotyped in a second population-based sample of 3003 mixed-heritage American Indians from the same longitudinal study. In addition to adjusting for age, sex, and family membership, association data in this second sample was adjusted for percentage of Indian heritage, since prevalence rates of type 2 diabetes are highly associated with ethnicity. Association analysis of this SNP in individuals of mixed-heritage is confounded by the fact that the major and minor alleles are exactly flipped in full-heritage Pima Indians as compared to Caucasians, thus differences in allele frequencies between diabetes cases and controls may be masked by striking differences in allele frequencies based on ethnicity. Indeed, after adjustment for fraction Indian heritage, the association of rs7896005 with type 2 diabetes in this second sample was non-significant (P = 0.45, OR = 1.06 95% CI 0.91-1.23, adjusted for age, sex, birth year, family membership, and fraction Indian heritage; Table 4). Since both odds ratios go in the same direction, the combined sample remains nominally significant (combined P = 0.03; OR = 1.12 95% CI 1.01-1.24; Table 4).
Table 4. Association results of the rs7896005 SNP with type 2 diabetes in the full-heritage Pima Indians, mixed-heritage American Indians, and combined samples.
| Sample | Diabetic | Non-diabetic | Pb | OR(95% CI) | |||||
| mAF | M/M | M/m | m/m | M/M | M/m | m/m | |||
| Full-heritage Pima Indians | 0.30 | 653 | 614 | 119 | 869 | 710 | 148 | 0.02 | 1.17(1.02-1.34) |
| Mixed-heritage American Indians | 0.35 | 279 | 353 | 95 | 1249 | 1271 | 357 | 0.45 | 1.06(0.91-1.23) |
| Combineda | 0.33 | 932 | 967 | 214 | 2118 | 1981 | 505 | 0.03 | 1.12(1.01-1.24) |
mAF, minor allele frequency. M, major allele; m, minor allele.
Combined = full-heritage Pima Indians + mixed-heritage American Indians.
P-values were adjusted for age, sex, birth year, family membership, and fraction Indian heritage.
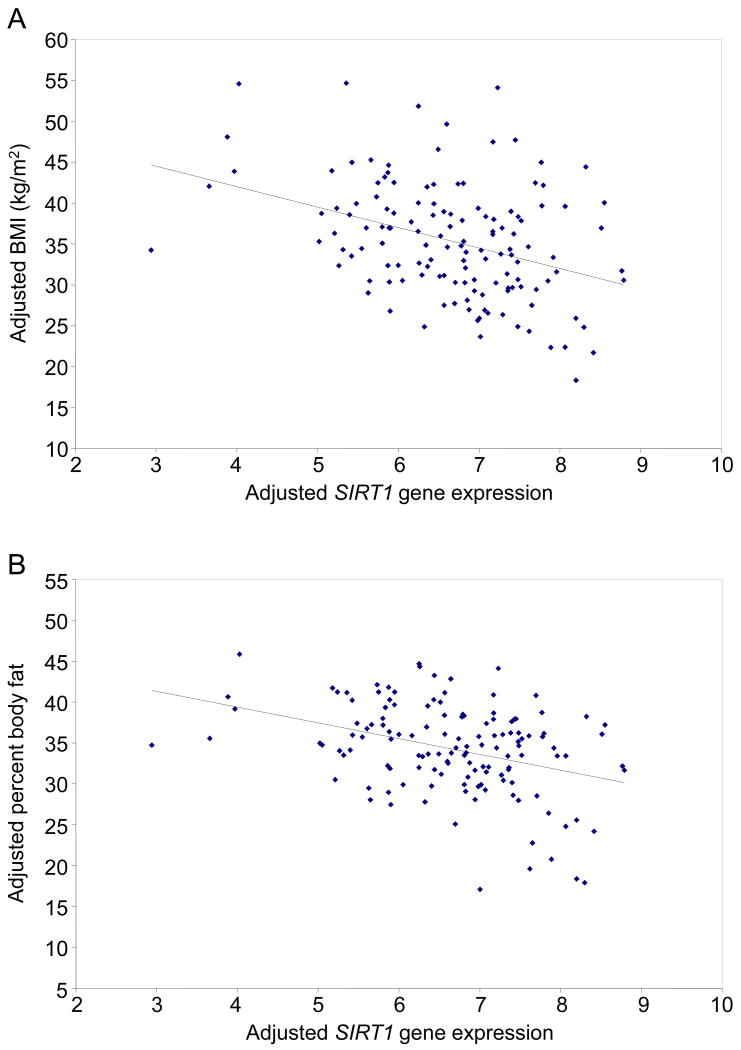
Although none of the tag SNPs were associated with either BMI in the population-based sample or percent body fat in the metabolically characterized subset, we observed that SIRT1 expression levels in adipose biopsies (N = 135) were strongly associated with the donors' BMI (P = 0.00001 adjusted for age and sex, P = 0.0005 and 0.006 in men and women, respectively; Fig. 2A) and percent body fat (0.00001 adjusted for age and sex, P = 0.00008 and 0.03 in men and women, respectively; Fig. 2B).
Figure 2.
Correlation of SIRT1 gene expression levels in adipose tissue isolated from 135 Pima Indian subjects with (A) BMI and (B) percent body fat after adjusting both axes for age and sex.
4. Discussion
SIRT1 is considered a master regulator because it is involved in several energy homeostasis pathways [18]. For example, SIRT1 has been shown to regulate gluconeogenesis in the liver, decrease lipogenesis and inhibit adipogenesis, repress inflammation in macrophages [18], and stimulate mitochondrial biogenesis and removal of damaged mitochondria [19]. Most of the variation in SIRT1 falls into a single LD block (fig. 1) which could suggest that conservation of this master regulator is essential for survival. A common variant (rs7896005) which serves as the representative SNP for almost all of the variation across the SIRT1 gene was nominally associated with type 2 diabetes in Pima Indians. The effect of this SNP on diabetes risk was greater in women than in men. Pre-diabetic trait analyses suggest that SNPs in SIRT1 increase risk for diabetes primarily by reduction of glucose-stimulated insulin secretion. We did not observe an effect on insulin sensitivity of peripheral tissues or an effect on adiposity. The statistical evidence for a sex interaction in diabetes risk is modest, and these findings will ultimately need to be confirmed in other populations. Rs7896005 tags the SIRT1 promoter SNP rs3758391 (fig. 1) located within a putative p53 binding site [20]. Naqvi et al. showed that the rs3758391 C allele (the Pima Indian risk allele) had lower binding affinity for p53 compared to the T allele suggesting that this variant may be functional [20]. Rs3758391 has previously been reported to be nominally associated (P = 0.03) with type 2 diabetes in cases (N = 519, 71.4% female) and controls (N = 547, 26.8% female) from Mexico City but the risk allele in Mexicans (T) would be predicted to be the non-risk allele in Pima Indians. Sex interaction was not assessed in the Mexican study, but it is noteworthy that the frequency of females in the cases and controls was statistically different (71.4% vs. 28.6%, P <0.05) which could potentially confound associations [8].
The finding of a role of SIRT1 in glucose-stimulated insulin secretion is consistent with prior studies in mice. Bordone et al., found that glucose-stimulated insulin secretion was reduced in Sirt1-/- mice as compared to wild-type mice [7]. Conversely, transgenic (BESTO) mice with high expression of Sirt1 in islet beta cells had significantly improved glucose tolerance and enhanced insulin response to glucose and KCl compared to their non-transgenic siblings [21], suggesting that an increased dosage of Sirt1 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. In a follow-up study using the same BESTO mice to analyze beta cell function of aged mice (18-24 months of age), Ramsey et al. observed that older BESTO mice no longer had the improved glucose tolerance and glucose-stimulated insulin secretion that was observed in the young BESTO mice (3-8 months of age) [22]. However, administration of nicotinamide mononucleotide (NMN) to the aged mice restored these two phenotypes in a sex-dependent manner (female mice only) [22]. Our observation that the effect of SIRT1 on type 2 diabetes is more prominent in females along with the gender differences observed by Ramsey et al. suggests a potential sex-specific mechanism. Sirt1 regulates ERalpha expression in some tissues [23], and estrogens and their receptors have been shown to affect islet physiology. In the beta cell, ERalpha enhances islet survival and improves islet lipid homeostasis and insulin biosynthesis [24]. Consequently, a reduction in SIRT1 deacetylase activity could potentially reduce ERalpha expression and thereby impair insulin secretion in the beta cell. Since the SIRT1 promoter SNP rs3758391 (tagged by rs7896005) has been shown to lower the binding affinity for p53 [20], further functional studies could include analyzing the in vitro effects of this SNP on transcriptional activity in a pancreatic beta cell line.
Prior studies in European Caucasians have reported associations between SIRT1 SNPs and obesity [9-11] and it has been shown that Sirt1 attenuates adipogenesis and induces lipolysis [6]. We did not observe an association between the SIRT1 SNPs and BMI or percent body fat in Pima Indians, which could be due to different lifestyle/environmental influences interacting with genotype, or due to the fact that Pima Indians are on average much more obese than European Caucasians. However, we did observe a strong negative correlation of SIRT1 expression in adipose with BMI and percent body. A similar result was seen with Swedish non-obese and obese siblings where SIRT1 expression levels in adipose were also lower in the obese siblings compared to the lean siblings [25]. The negative correlation of SIRT1 expression suggests that the SIRT1 pathway may indeed have a role, either primary or secondary, in body composition in Pima Indians. It is also possible that for Pima Indians SIRT1 expression levels in adipose are impacted by inherited variation in a tissue specific upstream regulatory factor. Alternatively, a correlation between SIRT1 expression and BMI and percent body fat could occur as a consequence of obesity.
In summary, common variation in and near SIRT1 is associated with reduced glucose-stimulated insulin secretion in pancreatic beta cells which may increase risk for type 2 diabetes in Pima Indians. However, variation within the SIRT1 gene is not a major risk factor for either type 2 diabetes or obesity in Pima Indians.
Acknowledgments
We thank the many persons who volunteered for these studies. This work was funded by the Intramural Program of NIDDK, NIH
Footnotes
Disclosure: The authors declare that there is no conflict of interest associated with this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelly G. A review of the sirtuin system, its clinical implications, and the potential role of dietary activators like resveratrol: part 1. Altern Med Rev. 2010;15:245–263. [PubMed] [Google Scholar]
- 2.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci. 2009;1173 1:E10–9. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol Metab. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 5.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 6.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006:4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz M, Valladares-Salgado A, Garcia-Mena J, Ross K, Edwards M, Angeles-Martinez J, Ortega-Camarillo C, de la Peña JE, Burguete-Garcia AI, Wacher-Rodarte N, Ambriz R, Rivera R, D'artote AL, Peralta J, Parra EJ, Kumate J. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev. 2010;26:261–270. doi: 10.1002/dmrr.1082. [DOI] [PubMed] [Google Scholar]
- 9.Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, Van Hul W, Van Gaal LF. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008;124:431–436. doi: 10.1007/s00439-008-0567-8. [DOI] [PubMed] [Google Scholar]
- 10.van den Berg SW, Dolle ME, Imholz S, van der A DL, van't Slot R, Wijmenga C, Verschuren WM, Strien C, Siezen CL, Hoebee B, Feskens EJ, Boer JM. Genetic variations in regulatory pathways of fatty acid and glucose metabolism are associated with obesity phenotypes: a population-based cohort study. Int J Obes (Lond) 2009;33:1143–1152. doi: 10.1038/ijo.2009.152. [DOI] [PubMed] [Google Scholar]
- 11.Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, Sijbrands EJ, Witteman JC, Pols HA, van Duijn CM, Uitterlinden AG. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978;108:497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. Diabetes Mellitus: Report of a WHO Study Group. Geneva: World Health Org; 1985. Tech. Rep. Ser., no. 727. [PubMed] [Google Scholar]
- 14.Lillioja S, Mott DM, Spraul M, Ferraro R, Foley JE, Ravussin E, Knowler WC, Bennett PH, Bogardus C. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 15.Bian L, Hanson RL, Ossowski V, Wiedrich K, Mason CC, Traurig M, Muller YL, Kobes S, Knowler WC, Baier LJ, Bogardus C. Variants in ASK1 are associated with skeletal muscle mRNA expression, in vivo insulin resistance, and type 2 diabetes in Pima Indians. Diabetes. 2010;59:1276–1282. doi: 10.2337/db09-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norman RA, Tataranni PA, Pratley R, Thompson DB, Hanson RL, Prochazka M, Baier L, Ehm MG, Sakul H, Foroud T, Garvey WT, Burns D, Knowler WC, Bennett PH, Bogardus C, Ravussin E. Autosomal genomic scan for loci linked to obesity and energy metabolism in Pima Indians. Am J Hum Genet. 1998;62:659–668. doi: 10.1086/301758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malhotra A, Kobes S, Knowler WC, Baier L, Bogardus C, Hanson RL. A genome-wide association study of body mass index in American Indians. Obesity. doi: 10.1038/oby.2011.178. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menzies KJ, Hood DA. The role of SirT1 in muscle mitochondrial turnover. Mitochondrion. 2011 doi: 10.1016/j.mito.2011.03.001. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 20.Naqvi A, Hoffman TA, DeRicco J, Kumar A, Kim CS, Jung SB, Yamamori T, Kim YR, Mehdi F, Kumar S, Rankinen T, Ravussin E, Irani K. A single-nucleotide variation in a p53-binding site affects nutrient-sensitive human SIRT1 expression. Hum Mol Genet. 2010;19:4123–4133. doi: 10.1093/hmg/ddq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Méneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Y, Li H, Gu Y, Davidson NE, Zhou Q. Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis. 2010;31:382–387. doi: 10.1093/carcin/bgp308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, Mauvais-Jarvis F. Minireview: Estrogenic protection of beta-cell failure in metabolic diseases. Endocrinology. 2010;151:859–864. doi: 10.1210/en.2009-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark SJ, Falchi M, Olsson B, Jacobson P, Cauchi S, Balkau B, Marre M, Lantieri O, Andersson JC, Jernås M, Aitman TJ, Richardson S, Sjöström L, Wong HY, Carlsson LM, Froguel P, Walley AJ. Association of Sirtuin 1 (SIRT1) Gene SNPs and Transcript Expression Levels With Severe Obesity. Obesity. 2011 doi: 10.1038/oby.2011.200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]