Abstract
Previous studies suggest that histones H3 and H4 are posttranslationally modified by binding of the vitamin biotin, catalyzed by holocarboxylase synthetase (HCS). Albeit a rare epigenetic mark, biotinylated histones were repeatedly shown to be enriched in repeat regions and repressed loci, participating in the maintenance of genome stability and gene regulation. Recently, a team of investigators failed to detect biotinylated histones and proposed that biotinylation is not a natural modification of histones, but rather an assay artifact. Here, we describe the results of experiments, including the comparison of various analytical protocols, antibodies, cell lines, classes of histones, and radiotracers. These studies provide unambiguous evidence that biotinylation is a natural, albeit rare, histone modification. Less than 0.001% of human histones H3 and H4 are biotinylated, raising concerns that the abundance might too low to elicit biological effects in vivo. We integrated information from this study, previous studies, and ongoing research efforts to present a new working model in which biological effects are caused by a role of HCS in multiprotein complexes in chromatin. In this model, docking of HCS in chromatin causes the occasional binding of biotin to histones as a tracer for HCS binding sites.
Keywords: Biotin, histones, holocarboxylase synthetase, post-translational modifications
1. Background
Chromatin comprises DNA, histones, and other chromatin proteins [1]. In each nucleosomal core particle, about 146 basepairs of DNA are wrapped around an octamer of core histones (one H3/H3/H4/H4 tetramer and two H2A/H2B dimers). Amino acids in the N-terminal tails of core histones and, to a lesser extent, amino acids in globular domains and C-termini, are exposed at the nucleosomal surface [2]. The N-terminal tails are subject to a multitude of posttranslational modifications, including acetylation and methylation [3]. These modifications play crucial roles in gene regulation. For example, acetylation of lysine (K)-9 in histone H3 (H3K9ac) is associated with transcriptionally active chromatin, whereas dimethylation and trimethylation of K9 (H3K9me2, H3K9me3) are associated with transcriptionally repressed chromatin [3]. Other, less abundant, modifications such as phosphorylation of serine-14 in histone H2B play critical roles in events such as programmed cell death [4].
Hymes et al. suggested that histones are also modified by covalent attachment of the vitamin biotin, mediated by the enzyme biotinidase [5]. While the original report was based on in vitro studies, evidence soon emerged that histone biotinylation is a natural phenomenon, based on probing and tracing biotinylation with streptavidin, anti-biotin, and [3H]biotin in primary human lymphoid cells [6]. While biotinidase undoubtedly has histone biotinyl ligase activity in vitro [5, 7], it is now believed that holocarboxylase synthetase (HCS) is the enzyme responsible for biotinylation of histones in vivo. This conclusion is based on observations that HCS is a nuclear, chromatin-associated protein [8–11]; that phenotypes of HCS knockdown include shortened life span and decreased heat tolerance in Drosophila melanogaster, while phenotypes of biotinidase knockdown are relatively minor [9]; that both human recombinant HCS (rHCS) and its microbial ortholog BirA have enzymatic activity to biotinylate histones and histone-based peptides [12, 13]; and that HCS knockdown causes aberrant gene regulation in human Jurkat lymphoblastoma cells and Drosophila melanogaster [9, 10].
Over the past several years, we have identified the following histone biotinylation sites: K4, K9, K18, and perhaps K23 in histone H3 [13, 14] and K8 and K12 in histone H4 [7, 15]. We provided evidence that K9, K13, K125, K127, and K129 in histone H2A also are targets for biotinylation albeit only in trace amounts [16]. We raised target-specific antibodies to many of these marks and to HCS [7, 14–16]. Using these antibodies, new roles of biotin in gene regulation and genome stability have been discovered. In those studies, K12-biotinylated histone H4 (H4K12bio) was the most prominently featured mark, primarily due to the availability of an antibody of exceptional quality [7]. The anti-H4K12bio used in those studies does not cross-react with non-biotinylated histones, does not cross-react with biotinylated histones other than histone H4, and does not cross-react with biotinylation sites other than K12. Evidence was provided that H4K12bio is enriched at repressed loci and at repeat regions in the human genome [10, 17, 18], and, further, that a low abundance of H4K12bio coincides with de-repression of retrotransposons and increased frequency of chromosomal abnormalities in biotin- and HCS deficient humans, human and murine cell lines, and Drosophila melanogaster [11].
In the early stages of these investigations, we proposed that biotinylation of histones is a rare event; binding of biotin to histones is in the order of only attomoles of biotin incorporated into histones isolated from 106 human lymphocytes [6]. Subsequent studies by independent investigators confirmed that <0.03% of histones are biotinylated in human cell cultures [19]. In a recent report, yet another laboratory failed to detect histone biotinylation marks by using streptavidin, antibodies, and mass spectrometry (MS); based on their negative results those investigators proposed that histone biotinylation is an artifact caused by non-specific probes and purification procedures [20]. Here, we conducted a thorough examination of probe specificity and purification procedures to provide an unambiguous answer to the question of whether biotinylation of histones is a rare event or simply an artifact. In view of our findings that biotin is a natural although rare histone modification, we propose a mechanism to explain the rarity as well as the biological significance.
2. Methods
2.1. Cell culture
Jurkat human lymphoblastoma cells, IMR-90 human fibroblasts, HepG2 human hepatocellular carcinoma cells, and U-937 human monocytic cells were obtained from American Type Culture Collection (Manassas, VA). HeLa human cervical cancer cells and MCF-7 human breast cancer cells were gifts by Drs. Jennifer Wood and Angela Pannier at the University of Nebraska-Lincoln. Cells were cultured using media and conditions as recommended by the American Type Culture Collection. In some experiments, cells were cultured in biotin-defined media (0.025 nM, 0.25 nM, or 10 nM), representing biotin concentrations in plasma from biotin-deficient, biotin-normal, and biotin-supplemented individuals [21, 22]. Biotin-defined media were prepared using customized culture media and biotin-depleted fetal bovine serum as previously described [23]. Where indicated, [3H]biotin (specific activity = 2.0535 TBq/mmol; Perkin Elmer, Boston, MA) or custom-made [14C]biotin (specific activity = 2.2052 GBq/mmol; Moravek, Inc., Brea, CA) were substituted for unlabeled biotin in culture media.
2.2. Purification of histones
Cell nuclei were released by treatment with detergent and collected by gradient centrifugation [6]; histones were extracted overnight with 1 M HCl at 4°C and the pH in the supernatant was adjusted to ~7.0 with 10 M NaOH. For comparison, we used the H2SO4-based extraction protocol by Healy et al. [20]. For some experiments, histones were further purified by high-performance liquid chromatography (HPLC). Briefly, histones were desalted by using PD MidiTrap G-10 columns (GE Healthcare, Piscataway, NJ) and further purified by using a C8 HPLC column (250 mm × 4.6-mm inner diameter, 10 μm particle size; Grace Vydac, Hesperia, CA). Proteins were eluted using the following binary gradient (flow rate = 0.7 mL/min) at room temperature (solvent A = 0.1% trifluoroacetic acid in water; solvent B = 0.1% trifluoroacetic acid in acetonitrile): 0% solvent B for 10 min; linear increase to 30.5% solvent B over 10 min; linear increase to 39% solvent B over 10 min; 39% solvent B held for 5 min; linear increase to 46.7% solvent B over 65 min; linear increase to 100% solvent B over 1 min; 100% solvent B held for 5 min; linear decrease to 0% solvent B over 1 min; and re-equilibration of the column with 0% solvent B for 6 min. Elution of proteins was monitored at 214 nm using a UV/Vis spectrometer, and HPLC fractions were collected at 1-min intervals. The identities of histones in HPLC fractions were confirmed by MS (data not shown) [7].
2.3. Streptavidin blots and western blots of carboxylases and histones
The following biotinylated carboxylases are markers for cellular biotin status [24]: acetyl-CoA carboxylases (ACC) 1 and 2; 3-methylcrotonyl-CoA carboxylase (MCC); propionyl-CoA carboxylase (PCC); and pyruvate carboxylase (PC). Whole cell extracts were prepared as previously described [23], using a buffer that contains protease inhibitors and DNase. If the goal of an experiment was to analyze biotinylated carboxylases (80 – 250 kDa) and histones (11 – 20 kDa) on one single gel, whole cell extracts were resolved using 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA); if the goal of an experiment was to analyze only carboxylases, 3–8% Tris Acetate gels (Invitrogen) were used to improve resolution of individual carboxylases. Transblots of gels were probed with Immuno Pure Streptavidin Horseradish Peroxidase Conjugate (Thermo Scientific, Waltham, MA), diluted 4000-fold in phosphate-buffered saline (PBS) containing 0.05% Tween-20 (TPBS) to detect protein-bound biotin [23]. Total (apo+holo) PCC and PC were probed using an “in-house” rabbit anti-human PCC antibody (antigen = keyhole limpet hemocyanin-conjugated KAGDTVGEGDLLVELE) diluted 250-fold in 0.637 M NaCl/TPBS, and a commercial rabbit anti-human PC antibody (Santa Cruz, Inc., Santa Cruz, CA) diluted 200-fold in 0.537 M NaCl/TPBS before use.
Histones in nuclear extracts were typically resolved using 18% Tris-Glycine gels (Invitrogen) [7]; histones in HPLC fractions were lyophilized and dissolved in distilled water prior to gel electrophoresis. HPLC fractions were resolved using Triton-Acid-Urea gel electrophoresis (TAU-PAGE) and electroblotted using N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) transfer buffer (25 mM CAPS, pH 10.0, and 20% methanol). Transblots were probed with streptavidin peroxidase; polyclonal goat anti-biotin peroxidase conjugate (30,000- to 200,000-fold dilution in TPBS, Sigma); polyclonal rabbit anti-biotin (1000-fold dilution in 0.637 M NaCl/TPBS, Abcam); polyclonal rabbit antibodies to K9-biotinylated histone H3 (H3K9bio), K18-biotinylated histone H3 (H3K18bio) [14], K12-biotinylated histone H4 (H4K12bio) (250-fold dilution in 0.637 M NaCl/TPBS) [7], and commercial rabbit anti-H4K8bio (500-fold dilution in 0.637 M NaCl/TPBS, Abcam); commercial antibodies to the C-termini in histones H3 (17,000-fold dilution in 0.637 M NaCl/TPBS, Santa Cruz, Inc; Santa Cruz, CA) and H4 (5000 fold dilution in 0.637 M NaCl/TPBS, Abcam); anti-acetyl lysine (250-fold dilution in 0.637 M NaCl/TPBS, Abcam); and anti-pan methyl lysine (250-fold dilution in 0.637 M NaCl/TPBS, Abcam). In addition, a new polyclonal antibody to H4K8bio was raised in rabbits as described before [7]. The secondary antibodies used were goat anti-rabbit IgG horseradish peroxidase conjugate (100,000-fold dilution; Sigma), donkey anti-goat IgG horseradish peroxidase conjugate (100,000-fold dilution; Sigma), goat anti-rabbit IgG IRDye800CW conjugate (100,000-fold dilution; LI-COR, Lincoln, NE), and donkey anti-goat IgG IRDye800CW conjugate (100,000-fold dilution; LI-COR) in 0.237M NaCl/TPBS. Controls included pre-immune serum, horseradish peroxidase-conjugated streptavidin (Thermo Scientific), and IRDye 800CW Streptavidin (LI-COR). Bands were visualized using an Odyssey infrared imaging system (LI-COR) or a chemiluminescence-based film developer, depending on the antibodies used.
The target specificities of anti-H3K4bio, anti-H3K9bio, anti-H3K18bio, and anti-H4K8bio were tested as described before [7], using the following antigens: (i) synthetic peptides H3K4bio (denoted N1–13bioK4), H3K9bio (N1–13bioK9), H3K18bio (N13–25bioK18), and a non-biotinylated peptide spanning amino acids 1–25 in histone H3 (N1–25) [14]; (ii) synthetic peptides H4K8bio (N6–15bioK8), H4K12bio (N6–15bioK12), and a non-biotinylated peptide spanning amino acids 1–19 in histone H4 (N1–19) [7]; (iii) synthetic peptides as described before in which acetylation or methylation were substituted for biotinylation (denoted N1–13H3K9ac, N5–14H3K9me2, N13–25 H3K18ac, N6–15H4K8ac, and N6–15H4K12ac); (iv) bulk extracts of nuclear histones from human Jurkat cells [7]; and (v) bulk extracts of nuclear histones from which the biotinylated fraction was removed by using avidin agarose [7]. In addition, competition studies were conducted in which peptides H3K4bio, H3K9bio, H3K18bio, H4K8bio, and H4K12bio were preincubated with anti-sera (0.3 – 12 μg peptide/μL serum) at 4°C for 1 h in 0.637 M NaCl/TPBS prior to probing transblots. Anti-H4K12bio passed these stringent tests in previous studies [7] and was not re-tested, except for testing its reactivity toward acetylation marks. Biotinylated peptides were titrated by using streptavidin [7] and equal loading of all peptides was confirmed with Ponceau stain.
Note that it is absolutely critical to adhere to the following steps in the various antibody protocols to ensure target recognition, while, at the same time, minimize non-specific binding. We advise against using modifications proposed elsewhere [20]. First, the concentration of NaCl in regular TPBS is 0.137 M and should not be altered if transblots are probed with streptavidin, anti-biotin peroxidase conjugate, or anti-H4K8ac; the salt concentration should be increased to 0.537 M NaCl for incubations with anti-PC, and to 0.637 M for all other primary antibodies. Second, the salt concentration should be 0.237 M NaCl in TPBS for diluting secondary antibodies. Third, membranes should be incubated with streptavidin and anti-biotin peroxidase conjugate at room temperature for 1 h, and with other primary probes at 4°C overnight; membranes should be incubated with secondary antibodies at room temperature for 1 h. Fourth, membranes should be washed using the same buffers that were used for incubations with primary and secondary antibodies with the following exception. For those primary antibodies that are used at 0.537 M or 0.637 M NaCl, washing steps after secondary antibodies should be conducted in TPBS containing 0.637 M NaCl. Fifth, after transblots are incubated with primary and secondary antibodies, the membranes should be washed three times each for 5 min.
2.4. Radiotracer studies
Healy et al. cultured biotin-depleted HeLa cells in the presence of 8 nM [3H]biotin to trace biotinylation of histones; they observed no meaningful binding of biotin to histones and a weak, yet detectable, binding to carboxylases by using gel electrophoresis and radioimaging [20]. We re-examined their observations using a similar protocol with the following important modifications. First, studies were conducted in both HeLa and Jurkat cells, because we suspected that HeLa cells degrade [3H]biotin by β-oxidation, which would cause a substantial decrease in the amount of tritiated biotin (Online Supplemental Fig. 1). Second, in some experiments custom-made [14C]biotin (Moravek, Inc., Brea, CA) was substituted for [3H]biotin. [14C]Biotin was synthesized following the protocol by Brady et al. [25], yielding a molecule in which the carbonyl-carbon is radiolabeled as opposed to the carboxyl carbon in commercial [14C]biotin; also, the specific radioactivity of the custom-made [14C]biotin (2.2052 GBq/mmol) is substantially greater than what is achieved in non-customized [14C]biotin. Third, gel electrophoresis studies were supplemented with studies in which radiotracer binding was quantified by liquid scintillation counting in order to increase the signal size and to provide a quantitative estimate of the percentage of histones that are biotinylated. Streptavidin, anti-biotin, anti-PCC, and anti-PC were used to demonstrate that the radioactivity in histone extracts is not due to a contamination with carboxylases, contrary to previous claims [20]. Fourth, the percent distribution of biotin between histones and carboxylases was quantified by streptavidin and gel densitometry in human whole cell extracts. Fifth, the concentration of radiotracers was increased 8 nM to 20 nM.
3. Results and Discussion
3.1. Various extents of histone biotinylation in distinct human tissues
Commonly used probes for biotinylated proteins include streptavidin, avidin, and anti-biotin [6, 8, 20, 26]. It has long been recognized that the glycoprotein avidin from egg white does not have the same level of specificity for biotin as the non-glycosylated streptavidin from Streptomyces avidinii [26]. Not surprisingly, recent reports suggest that avidin crossreacts with non-biotinylated histones [19], whereas streptavidin and anti-biotin are specific for biotinylated histones [20]. While none of these three probes can discriminate among histone biotinylation sites (e.g., H4K8bio vs. H4K12bio), both streptavidin and anti-biotin are robust probes for the overall biotinylation status of carboxylases and histones [20]. Note that Bailey et al. report that even streptavidin shows some cross-reactivity with non-biotinylated histones [19].
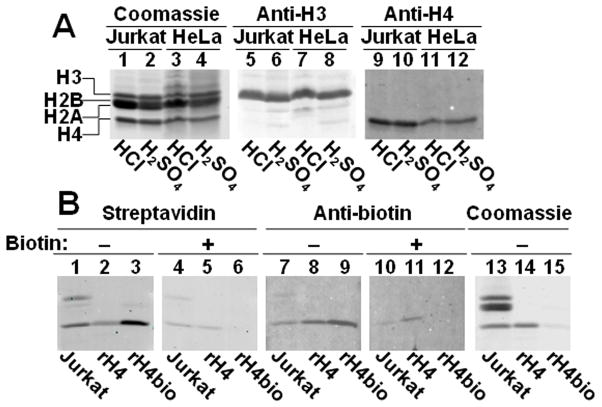
In a first series of experiments, streptavidin and anti-biotin were used to compare the efficiency of HCl- and H2SO4-based procedures for extracting biotinylated histones, and to compare the levels of histone biotinylation in cells from various tissues. When histones from Jurkat and HeLa cells were extracted with HCl or H2SO4, yield and purity of histones were comparable for the two protocols (Fig. 1A), as judged by staining with coomassie blue and probing with anti-H3 and anti-H4. Unless noted otherwise, subsequent experiments were conducted using the widely accepted HCl protocol, which does not require the sequential washing steps (TCA, acetone/HCl, acetone, acetone) of the H2SO4 protocol.
Fig. 1. Comparison of histone extraction protocols, and specificity testing of streptavidin and anti-biotin.
Panel A: Nuclear histones were extracted from Jurkat cells (lanes 1, 5, and 9) or HeLa cells (lanes 3, 7, and 11) by using HCl; for comparison histones were extracted from Jurkat cells (lanes 2, 6, and 10) or HeLa cells (lanes 4, 8, and 12) by using H2SO4+TCA+acetone/HCl+acetone. Histones were probed with coomassie blue (lanes 1–4) and antibodies to the C-termini in histone H3 (lanes 5–8) and H4 (lanes 9–12). Panel B: HCl extracts of Jurkat cell histones (lanes 1, 4, 7, 10, and 13), recombinant human histone H4 (lanes 2, 5, 8, 11, and 14), and chemically biotinylated histone H4 (lanes 3, 6, 9, 12, and 15) were probed with streptavidin without biotin competitor (lanes 1–3) and with 5 mM free biotin (lanes 4–6), and with anti-biotin without biotin competitor (lanes 7–9) and with 5 mM free biotin (lanes 10–12), and with coomassie blue (lanes 13–15).
When histones from Jurkat cells were probed with streptavidin and anti-biotin, biotinylation signals were detected for histones H3 and H4 (Fig. 1B, lanes 1 and 7); the signals were substantially decreased if probes were pre-incubated with 5 mM biotin to block biotin-binding site (lanes 4 and 10), suggesting specificity. We did not use recombinant histones as negative controls because of our previous observation that microbial BirA catalyzes biotinylation of histones and that recombinant histones contain detectable amounts of biotin [13]. This observation was confirmed here, using streptavidin and anti-biotin as probes for biotin in recombinant histone H4 (Fig. 1B, lanes 2, 5, 8 and 11). Chemically biotinylated histone H4 was used as positive control; biotinylation signals were detected with both streptavidin and anti-biotin (lanes 3 and 9), and substantially decreased in the presence of 5 mM biotin (lanes 6 and 12). Equal loading of histones from Jurkat cells and recombinant human histone H4 was confirmed by coomassie blue staining (lanes 13 and 14). Chemically biotinylated H4 produced a very strong signal in western blots; hence, we loaded ~1000 times less chemically biotinylated histone H4 than histone extracts from cells (lane 15).
Healy et al. did not detect biotinylated histone H2A in extracts from HeLa cells by using streptavidin and anti-biotin as probes [20]. We attribute the absence of signal in their studies to modifications in analytical protocols as described below.
In a second series of experiments, histone extracts from various cell lineages were probed with streptavidin and anti-biotin. Biotinylation of histones H3 and H4 was detectable in all cells tested, i.e., Jurkat cells, U937 cells, HeLa cells, HepG2 cells, MCF-7 cells, and IMR-90 fibroblasts (Fig. 2A and B). Although the degrees of H3 and H4 biotinylation were moderately different among cell lines, clear biotinylation signals were detectable in histones from each cell. Histone integrity, identity, and equal loading and transfer were confirmed by using coomassie blue (Fig. 2C) and antibodies to the C-termini in histones H3 (Fig. 2D) and H4 (Fig. 2E). In contrast to histones H3 and H4, histones H2A and H2B produced only weak biotinylation signals, suggesting that histone H2A is not a good model to investigate histone biotinylation. This observation is consistent with our previous observations using anti-biotin as a probe [6]. The findings in the Healy et al. paper are largely based on studies with histone H2A [20].
Fig. 2. Biotinylation marks can be detected in bulk extracts from human cells, using streptavidin and anti-biotin as probes.
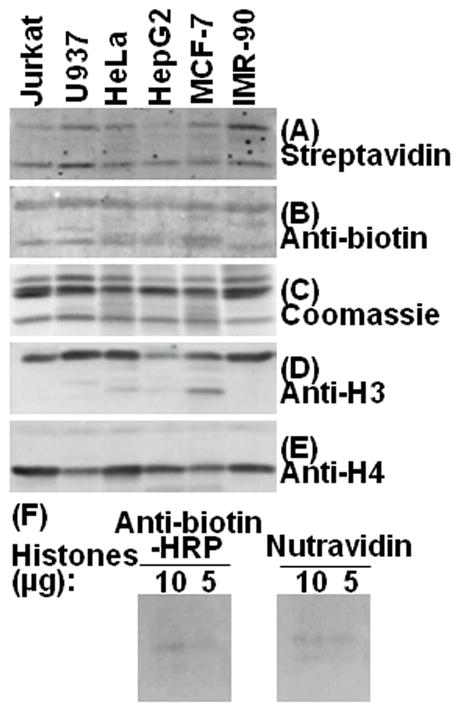
Bulk extracts of histone extracts from various cell lineages were probed with streptavidin (panel A), anti-biotin (panel B), and the loading and transfer controls coomassie blue (panel C), anti-H3 (panel D), and anti-H4 (panel E). Panel F: Histones from HeLa cells were extracted with H2SO4+TCA+acetone/HCl+acetone. Ten or five microgram of histones were loaded per well. Blots were blocked with PBS containing 5% BSA. After probing with horseradish peroxidase-conjugated anti-biotin or Nutravidin, the blots were washed for 6 hrs, and exposed to autoradiography film for 1 min.
Previous studies suggest that HCS interacts directly with histones H3 [12] and H4, but not with histones H2A and H2B (Bao et al., unpublished observation). Evidence suggests that biotinylation of histone H2A depends on the diffusion of the energy-rich intermediate in HCS catalysis, biotinyl-AMP, to histone H2A [27]. Taken together, the proximity of HCS to target histones might explain why histones H3 and H4 are better targets for biotinylation than H2A and H2B. Consistent with this theory, no biotinylation sites have been reported for histone H2B as of today. Based on these observations, we focused our subsequent studies on histones H3 and H4 in this paper.
Next, we isolated and probed histones following the protocol by Healy et al. [20]. That protocol differs from our protocol by the use of H2SO4 for histone extraction, the use of BSA in blocking and washing buffers, and washing membranes for up to 6 h while exposing autoradiography films for only 5–30 s [20]. HeLa cell histones prepared by using the Healy protocol produced a substantially lower biotinylation signal (Fig. 2F) compared to our protocol (Figs. 2A and B). We propose that the extensive wash times of membranes in combination with short exposure of films produced false negative findings in the study by Healy et al. [20]. An independent laboratory also succeeded with detecting biotinylated histones in human embryonic palatal mesenchymal cells by using streptavidin [28].
3.2. Biotinylation site-specific antibodies
In previous studies we generated polyclonal rabbit anti-human H3K4bio, H3K9bio, H3K18bio, H4K8bio, and H4K12bio [7, 14]. Anti-H4K12bio passed a series of stringent specificity tests (see below) and was shown to be target specific; anti-H4K12bio was not retested here, with the exception of re-examining a possible cross-reaction with acetylated H4. The latter test was conducted based on observations by Healy et al. that commercial anti-H4K5bio and anti-H4K8bio (Abcam, Inc.) cross-react with acetylation marks [20]. Because previous batches of antibodies were exhausted, new batches of anti-H3K4bio, anti-H3K9bio, anti-H3K18bio, and anti-H4K8bio were raised in rabbits as described before [7, 14]; their target specificities were tested as follows.
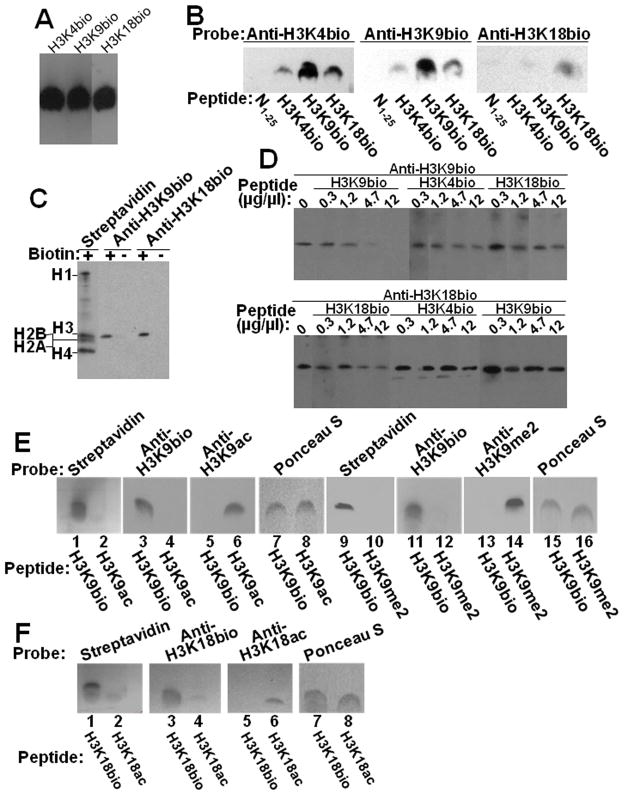
First, we examined the specificities of antibodies to biotinylated histone H3. Synthetic peptides (antigens) H3K4bio, H3K9bio, and H3K18bio [14] were titrated with streptavidin (Fig. 3A) to ensure equal masses of the various synthetic peptides in subsequent specificity tests. Titration of peptide N1–25 (negative control) was not possible due to the absence of biotin [14]; thus, N1–25 was quantified gravimetrically to normalize for loading. Please note that only H4K8bio, but not H4K8ac, produced a signal with streptavidin and anti-biotin (Online Supplemental Figure 2). In a first series of tests, peptides H3K4bio, H3K9bio, H3K18bio, and N1–25 were probed with anti-H3K4bio, anti-H3K9bio, and anti-H3K18bio in all possible combinations; pre-immune sera were included as negative controls. Anti-H3K4bio cross-reacted consistently with peptides H3K9bio and H3K18bio (but not N1–25), and this pan-H3bio antibody was excluded from further testing (Fig. 3B, left panel). Anti-H3K9bio produced a strong signal with peptide H3K9bio; weak signals were produced with peptides H3K4bio and H3K18bio, but not with N1–25 (Fig. 3B, middle panel). Anti-H3K18bio was specific for peptide H3K18bio (Fig. 3B, right panel). None of the pre-immune sera produced a signal with any of the peptides (not shown). Both anti-H3K9bio and anti-H3K18bio were included in additional specificity tests. In a second series of testing, HCl extracts of histones from Jurkat cells were probed with anti-H3K9bio and anti-H3K18bio. Histone extracts contained all five major classes of biotinylated histones, as judged by probing with streptavidin (Fig. 3C). Integrity of proteins was further confirmed by staining with Coomassie blue (not shown). Both anti-H3K9bio and anti-H3K18bio reacted only with histone H3 and did not cross-react with any of the other classes of histones present in the sample (Fig. 3C). When the biotinylated fraction of histones was removed by using avidin beads, streptavidin, anti-H3K9bio, and anti-H3K18bio produced no signal with the remaining fraction of non-biotinylated histones (Fig. 3C). Recombinant histones may contain some biotin [13], and are not good negative controls (see 3.1.). In a third series of testing, extracts of biotinylated human histones were probed with anti-H3K9bio and anti-H3K18bio in the presence of increasing amounts of peptide competitors (Fig. 3D). For anti-H3K9bio, peptide H3K9bio competed with histone H3 for binding to the antibody, whereas competition by peptides H3K4bio and H3K18bio was quantitatively minor. For anti-H3K18bio, only peptide H3K18bio, but not H3K4bio and H3K9bio, competed with histone H3 for binding to the antibody. In a fourth series of testing, we expanded our original array of tests by including acetylated peptides and methylated peptides as potential targets. We focused on modifications that are known to target the same lysine residues as biotinylation, i.e., acetylation of K9 and K18, and dimethylation of K9 [3]. Anti-H3K9bio produced a signal only with peptide H3K9bio, but not with H3K9ac or H3K9me2 (Fig. 3E, lanes 3, 4, 11 and 12); likewise anti-H3K9ac (lanes 5 and 6) and anti-H3K9me2 (lanes 13 and 14) bound only to acetylated and methylated peptides, respectively. Equal loading and transfer of peptides was confirmed by using Ponceau stain (lanes 7, 8, 15, 16). Anti-H3K18bio produced a signal only with peptide H3K18bio, but not with H3K18ac (Fig. 3F, lanes 3 and 4); likewise anti-H3K18ac bound only to H3K18ac (lanes 5 and 6). Equal loading and transfer of peptides was confirmed by using Ponceau stain (lanes 7 and 8). Collectively, these validation experiments suggest that both anti-H3K9bio and anti-H3K18bio are specific for their designated targets, and that both biotinylation marks exist in human cells.
Fig. 3. Specificity of antibodies to H3K4bio, H3K9bio and H3K18bio.
Panel A: Confirmation of equal loading of peptides H3K4bio, H3K9bio, and H3K18bio with streptavidin. Panel B: Transblots of peptides N1–25 (non-biotinylated negative control), H3K4bio, H3K9bio, and H3K18bio were probed with anti-H3K4bio (left), anti-H3K9bio (middle), and anti-H3K18bio (right). Panel C: HCl extracts of histones from Jurkat cells were probed using streptavidin, anti-H3K9bio, and anti-H3K18bio; samples of biotin-free histones (“-”) were generated by using avidin agarose. Panel D: Bulk HCl extracts of histones from Jurkat cells were probed with anti-H3K9bio (top) and anti-H3K18bio (bottom) after pre-incubation of antibodies with increasing amounts of competing peptides H3K4bio, H3K9bio, and H3K18bio; controls (“C”) were prepared without peptide competitors. Note the difference in the order of peptide competitors in the two gels. For some gels, bands from the same analytical runs were electronically re-arranged to facilitate comparisons. Panel E: Peptides H3K9bio, H3K9ac, and H3K9me2 were probed with streptavidin (lanes 1, 2, 9 and 10), anti-H3K9bio (lanes 3, 4, 11 and 12), anti-H3K9ac (lanes 5 and 6), and anti-H3K9me2 (lanes 13 and 14); Ponceau S was used as loading control (lanes 7, 8, 15 and 16). Panel F: Peptides H3K18bio and H3K18ac were probed with streptavidin (lanes 1 and 2), anti-H3K18bio (lanes 3 and 4) and anti-H3K18ac (lanes 5 and 6); Ponceau S was used as loading control (lanes 7 and 8).
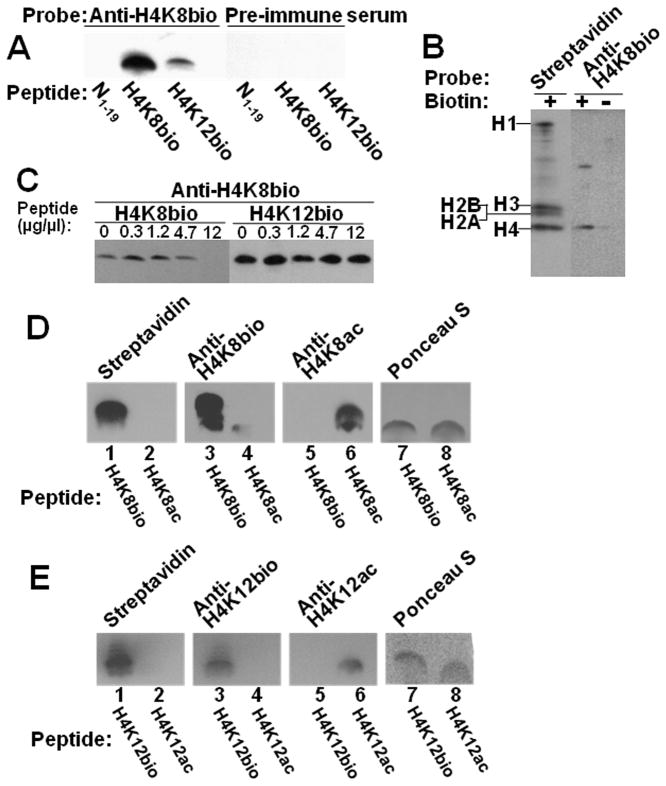
Next, we examined the specificity of the new preparation of anti-H4K8bio raised in this study. The new preparation of anti-H4K8bio was target specific. In a first series of testing, peptides H4K8bio, H4K12bio, and N1–19 were probed with anti-H4K8bio; pre-immune sera were included as negative controls. Anti-H4K8bio produced a strong signal with peptide H4K8bio, although peptide H4K12bio also produced a weak signal; no signal was detectable with peptide N1–19 (Fig. 4A). None of the pre-immune sera produced a signal with any of the peptides (Fig. 4A). In a second series of testing, HCl extracts of histones from Jurkat cells were probed with anti-H4K8bio. Histone extracts contained all five major classes of biotinylated histones, as judged by probing with streptavidin (Fig. 4B). Integrity of proteins was further confirmed by staining with Coomassie blue (not shown). Anti-H4K8bio reacted only with histone H4 (Fig. 4B). If the biotinylated fraction of histones was removed by using avidin beads prior to western blot analysis, anti-H4K8bio produced no signal with the remaining fraction of non-biotinylated histones (Fig. 4B). In a third series of testing, HCl extracts of histones from Jurkat cells were probed with anti-H4K8bio in the presence of increasing amounts of peptide competitors. For anti-H4K8bio, peptide H4K8bio competed with histone H4 for binding to the antibody, whereas competition by peptide H4K12bio was quantitatively minor (Fig. 4C). Finally, we determined whether H4K8bio and H4K12bio are specific for the biotin mark or if they cross-react with acetylation marks at K8 and K12. Anti-H4K8bio produced a signal only with peptide H4K8bio, but not with H4K8ac (Fig. 4D, lanes 3 and 4); likewise, anti-H4K8ac bound only to H4K8ac (lanes 5 and 6). Equal loading and transfer of peptides was confirmed by using Ponceau stain (lanes 7 and 8). Anti-H4K12bio produced a signal only with peptide H4K12bio, but not with H4K12ac (Fig. 4E, lanes 3 and 4); likewise, anti-H4K12ac bound only to H4K12ac (lanes 5 and 6). Equal loading and transfer of peptides was confirmed by using Ponceau stain (lanes 7 and 8). Collectively, these validation experiments and previous studies [7] suggest that both anti-H4K8bio and anti-H4K12bio are specific for their designated targets, and that both biotinylation marks exist in human cells. Finally, we re-examined whether commercial anti-H4K8bio (Abcam) cross-reacts with acetylated histones, as suggested by previous studies [20]. We could not reproduce the findings from those previous studies and observed that the commercial anti-H4K8bio is specific for biotin (data not shown). We attribute these apparently conflicting results to the procedures used for preparing targets for specificity testing. Healy et al. conducted acetylation and biotinylation of recombinant histone H4 by using recombinant histone acetyl transferase and recombinant BirA, respectively, to produce targets for anti-H4K8bio [20]. In contrast, in this study we used chemically defined acetylated and biotinylated peptides to test for possible cross-reactivities of antibodies to biotinylated histones with acetylated histones. Only the use of synthetic peptides permits to control for the extent and site of acetylation and biotinylation, whereas the enzymatic modification of recombinant histones by Healy et al. produces a poorly characterized mix of various compounds that are acetylated or biotinylated at unidentified lysine residues. In addition, in previous studies we reported that the biotinylation of histones by recombinant BirA in vitro is an extremely slow process [13] whereas it is conceivable that acetylation by recombinant acetyl transferase proceeds at a much faster rate.
Fig. 4. Specificity of antibodies to H4K8bio and H4K12bio.
Panel A: Transblots of peptides N1–19 (non-biotinylated negative control), H4K8bio, and H4K12bio were probed with anti-H4K8bio; pre-immune serum was used as negative control. Panel B: HCl extracts of histones from Jurkat cells were probed using streptavidin and anti-H4K8bio; samples of biotin-free histones (“-”) were generated by using avidin agarose. Panel C: HCl extracts of histones from Jurkat cells were probed with anti-H4K8bio after pre-incubation of antibodies with increasing amounts of competing peptides H4K8bio and H4K12bio; controls (“C”) were prepared without peptide competitors. For some gels, bands from the same analytical runs were electronically rearranged to facilitate comparisons. Panel D: Peptides H4K8bio and H4K8ac were probed with streptavidin (lanes 1 and 2), anti-H4K8bio (lanes 3 and 4) and anti-H4K8ac (lanes 5 and 6); Ponceau S was used as loading control (lanes 7 and 8). Panel E: Peptides H4K12bio and H4K12ac were probed with streptavidin (lanes 1 and 2), anti-H4K12bio (lanes 3 and 4) and anti-H4K12ac (lanes 5 and 6); Ponceau S was used as loading control (lanes 7 and 8).
Additional experiments were conducted using HPLC-purified histone H4 and TAU-PAGE to further corroborate that anti-H4K8bio and anti-H4K12bio are specific for biotin and do not cross-react with acetyl and methyl residues. When H4 was probed with an antibody to its C-terminus, four distinct bands were detectable in addition to a few faint bands (Fig. 5). When the same extract was probed with anti-H4K8bio and anti-H4K12bio, two and four bands, respectively, were detectable that were distinct from those observed with anti-acetyl lysine. There was some overlap among bands probed with anti-H4K12bio and anti-pan methyl lysine.
Fig. 5. Detection of biotinylated histone H4 by TAU-PAGE.
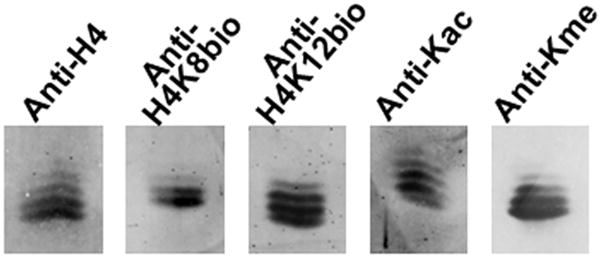
Histone H4 fraction from Jurkat cells was separated by TAU-PAGE. Transblots were probed with anti-H4, anti-H4K8bio, anti-H4K12bio, anti-acetyl Lysine (anti-Kac), and anti-pan methyl Lysine (anti-Kme).
Finally, we probed histone extracts from various cells with anti-H3K9bio, anti-H3K18bio, anti-H4K8bio, and anti-H4K12bio. All four biotinylation marks were detectable in all cell lines, although the abundance of individual biotinylation marks varied among cell lines (Fig. 6). Samples were also probed with coomassie blue, and antibodies to the C-termini in histones H3 and H4 to permit evaluation of sample integrity, loading and transfer, and purity.
Fig. 6. Detection of biotinylated histones in human cells by immunoblots.
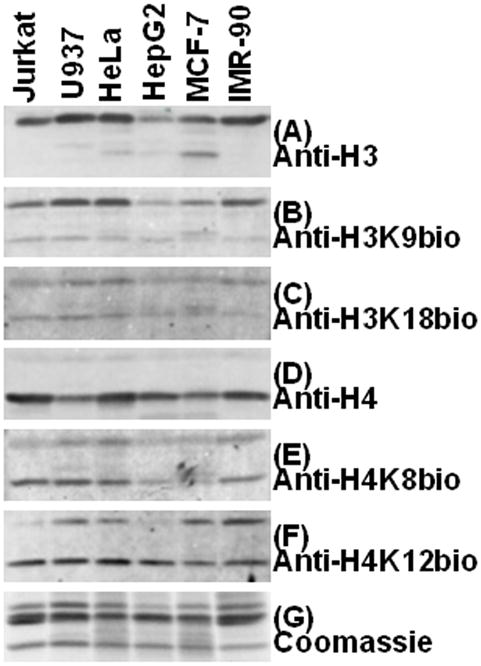
Histones were extracted from Jurkat, U937, HeLa, HepG2, MCF-7 and IMR-90 cells by using HCl. Transblots were probed with anti-H3 (panel A), anti-H3K9bio (panel B), anti-H3K18bio (panel C), anti-H4 (panel D), anti-H4K8bio (panel E) and anti-H4K12bio (panel F). Sample integrity, and equal loading of histones were confirmed using coomassie blue staining (panel G).
3.3. Radiotracer studies
In previous studies, radiolabeled biotin was used to trace histone biotinylation; these studies produced controversial results. In one study, we cultured human lymphocytes ex vivo with [3H]biotin and detected a signal that clearly exceeded background noise [6]. In a second study, Healy did not detect binding of [3H]biotin to histones in biotin-depleted HeLa cells in culture [20]. In a third study, commercial carboxyl-labeled [14C]biotin was used in an attempt at quantifying histone biotinylation; the binding of [14C]biotin to histones was low [19]. Commercial carboxyl-labeled [14C]biotin (as opposed to carbonyl-labeled [14C]biotin) is not an appropriate radiotracer for histone biotinylation due to the possible loss of radiolabel in the β-oxidation of biotin in some cell lines (see below).
A series of radiotracer studies was conducted to determine whether radiolabeled biotin is bound to histones in Jurkat lymphoid cells and HeLa adenocarcinoma cells. In a first step, both cell lines were biotin-depleted using a protocol similar to that by Healy [20] with the following modifications. For depletion, cells were maintained in media containing 0.025 nM biotin rather than the biotin-free medium. Our rationale was that we did not want to abolish essential metabolic pathways by using biotin-free medium, and we intended to maintain a near-normal expression of HCS, which is known to depend on biotin [10, 28, 29]. Also, we extended the depletion period to two weeks to account for a possible slow turnover of biotinylated histones. After the 2-wk depletion period, the abundance of biotinylated carboxylases was substantially lower in Jurkat cells compared with cells cultured in media containing a physiological concentration (0.25 nM) of biotin (Fig. 7, lane 1 vs. 2); when cell cultures were continued in medium containing 10 nM biotin for a 1-wk repletion period, the abundance of biotinylated carboxylases increased to levels similar to those observed in physiological medium (lane 1 vs. 3). The changes in holocarboxylase abundance were due to decreased availability of biotin, not to decreased expression of carboxylases, as judged by probing with anti-PC and anti-PCC. Results were similar in HeLa cells (Fig. 7, lanes 4–6), confirming the observations by Healy et al. [20].
Fig. 7. Validation of the biotin depletion and repletion protocol.
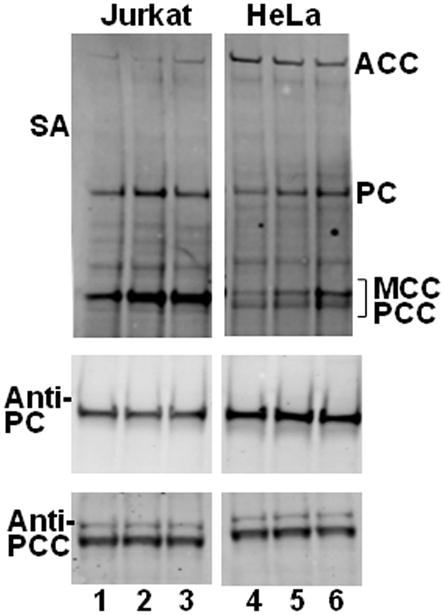
Panel A: Jurkat cells after a 2-wk depletion in biotin-deficient medium (0.025 nM, lane 1) compared with cells cultured in medium containing a physiological concentration of biotin (0.25 nM) (lane 2), and cells after a 1-wk repletion in medium containing a pharmacological concentration of biotin (10 nM) (lane 3). Biotinylated carboxylases were probed using streptavidin (SA). Equal expression, loading, and transfer of carboxylases was confirmed using anti-PC and anti-PCC. Panel B: As described for panel A, but HeLa cells (lanes 4–6) were substituted for Jurkat cells.
We then asked why Healy et al. detected a faint [3H]biotinylation signal in carboxylases but no signal in histones [20]. We hypothesized that biotinylated carboxylases are more abundant than biotinylated histones in human cells, and that there was enough intact [3H]biotin to trace carboxylases but not histones. To test this theory, cells were cultured in media containing 10 nM of unlabeled biotin; whole cell extracts of proteins were prepared using our routine protocol [23] or, where indicated, by following the protocol by Healy et al. [20]. Carboxylases and histones have very different molecular weights (80 – 250 kDa and 11 – 20 kDa, respectively) and do not separate well if run on a single electrophoresis gel. However, it is very clear in Jurkat cells that the amount of carboxylase-bound biotin exceeds histone-bound biotin substantially, using streptavidin as probe for whole cell extracts (Fig. 8A, lane 1); the total carboxylase signal was about 14 times the total histone signal, as judged by gel densitometry. If whole cell proteins were extracted following the protocol by Healy et al. [20], the histone biotinylation signal was lost (lane 2). Importantly, acid extracts of Jurkat cell histones revealed no meaningful contamination with carboxylases, if run on 4–12% Bis-Tris gels and using streptavidin (lane 3) and anti-PC (lane 6) as probes. The identities of carboxylases and histones in these blots were confirmed using antibodies to PC, histone H3, and histone H4 (lanes 4–12). Taken together, we conclude that biotinylated carboxylases are more abundant than biotinylated histones and that acid extracts of Jurkat cell histones are free of contamination with carboxylases. This is not consistent with the proposal by Healy et al. that binding of radiolabeled biotin to histones and quantitation by liquid scintillation counting is an artifact due to contamination with carboxylases [20].
Fig. 8. Binding of radiolabeled biotin to histones.
Panel A: Whole cell extracts prepared by using our protocol (lanes 1, 4, 7 and 10), prepared according to Healy’s protocol (lanes 2, 5, 8 and 11), and acid histones extracts (lanes 3, 6, 9 and 12) from Jurkat cells were resolved using 4–12% Bis-Tris gels and probed with streptavidin (SA). Protein identities were verified by using antibodies to PC, histone H3, and histone H4. Panel B: As described for panel B, but HeLa cells were substituted for Jurkat cells. Lanes loaded with acid extracts were electronically combined with those loaded with whole cell extracts.
Next, we tested the hypothesis that the cell model used by Healy et al. (HeLa cells) contains untypically high concentrations of carboxylases, and that contamination of histone extracts with carboxylases are specific for this cell line. Our experiments in HeLa cells produced results similar to those reported by Healy et al., i.e., a faint carboxylase signal was detectable in acid extracts (Fig. 8B, lane 3), while no histone signal was detectable in whole cell extracts (lane 1) and carboxylase extracts (lane 2). Identities of proteins were confirmed as described for above (Fig. 8B, lanes 4–12). Importantly, the abundance of PC in HeLa cells greatly exceeds that in Jurkat cells (compare lanes 4 and 5 in panel A to panel B). This observation is consistent with our hypothesis that the great abundance of holo-carboxylases in HeLa cells is specific for this cell type but does not apply to studies conducted in Jurkat cells.
Given that acid histone extracts from Jurkat cells are essentially free of carboxylases, we proceeded with the quantitation of radiolabeled histones by liquid scintillation counting. Biotin-depleted Jurkat and HeLa cells were repleted with 20 nM [3H]biotin and histones were collected by HCl extraction. To exclude firmly the contamination of carboxylases, histones (11–20 kDa) were purified by gel electrophoresis and electroelution. Eluted proteins were lyophilized and dissolved with 1 mL pure water, mixed with 5 mL liquid scintillation fluid and counted for 5 min. In Jurkat cells, 5.7 ± 0.4 Bq was bound per 1 mg protein, suggesting that 2.6 ± 0.2 fmoles of biotin were bound per mg of histones. In the case of HeLa cells, 230 ± 27 Bq was bound per 1 mg of proteins, suggesting that 104 ± 12 fmoles of biotin were bound per mg of histones.
We used the following calculation to estimate the percentage of histones that is biotinylated in cells. Each nucleosome contains one molecule of H1 (20 kDa), and two molecules each of H2A (14 kDa), H2B (14 kDa), H3 (15 kDa), and H4 (11 kDa). Thus, 1 mg of total histones equals about 7.8 nmoles of histones. We made the following assumptions: (i) Jurkat cells contain ~2.6 fmol [3H]biotin/1 mg (7.8 nmol) histone; (ii) [3H]biotin binds to H3 and H4, but not to H1, H2A, H2B; and (iii) the molar ratio of H3 and H4 equals 1 in histone extracts. Based on these assumptions, about 0.0000082% of histones H3 and H4 are biotinylated. In case of HeLa cells, we estimate that 0.0003% of histones H3 and H4 are biotinylated. Clearly, this is consistent with previous reports that histone biotinylation is a rare epigenetic mark [6, 19].
Previous studies suggest that cell from the lymphoid lineage, e.g., Jurkat cells, do not catabolize biotin by β-oxidation in meaningful quantities [30]; no information is available for HeLa cells. Hence, in some studies, we substituted [14C]biotin for [3H]biotin. The binding of [14C]biotin to histones was similar to that described for [3H]biotin, judged by liquid scintillation counting (data not shown). This observation suggests that biotin catabolism is not a meaningful confounder.
The abundance of an epigenetic mark cannot be directly correlated with importance. Epigenetic marks may be locally enriched in distinct loci. For example, serine-14 phosphorylation in histone H2B and histone poly(ADP-ribosylation) are detectable only after induction of apoptosis and major DNA damage, respectively, but the role of these epigenetic marks in cell death is clear [4, 31, 32]. Evidence suggests that about one out of three histone H4 molecules might be biotinylated at K12 in telomeric repeats [18]. Importantly, biotinylation of K12 in histone H4 causes chromatin condensation [33], consistent with previous reports that H4K12bio is enriched in repressed loci [10, 11, 15, 17]. Also, please note that a recent Ph.D. thesis in an independent laboratory provides evidence that up to 50% of histone H4 is biotinylated in Candida albicans [34]. These observations suggest that further studies with other model organisms is needed to clarify the biological roles of histone biotinylation.
We agree with Healy et al. that biotinylated histones are difficult to detect by using MS [20], as described in a companion paper (T. Kuroishi et al., in preparation). In about one out of three analytical runs, we observed H4K79bio by using LC/MS/MS, but the signal was barely above background noise and not reproducible in all sample preparations. This observation suggests that histone biotinylation is a modification mark that is at the detection limits of MS analysis. Please note that K79 in histone H4 is a known histone acetylation site [35], consistent with previous observations that biotinylation and acetylation compete for the same residues in histones [7, 14, 15].
Even if biotinylation is a natural histone modification, questions remain as to how such a rare event can cause gene repression and participate in the maintenance genome stability. We devised the following working hypothesis that is consistent with previous reports and ongoing research in our laboratory. In this model, the effects of biotin and HCS in epigenetic pathways of gene regulation are mediated by physical interactions of HCS with other chromatin proteins rather than by biotinylated histones. For example, we have demonstrated that HCS physically interacts with histone H3 [12] and we have generated evidence that HCS interacts with the methylated cytosine binding protein MeCP2 and the histone H3 K9-methyl transferase EHMT-1 by using yeast-two hybrid assays, co-immunoprecipitation, and limited proteolysis assays {[36]; Yong et al., unpublished; Liu et al., unpublished}. We propose that HCS is an integral part of a gene repression complex that may also include histone deacetylases and the nuclear co-repressor N-CoR. Marks and patterns such as DNA methylation, H3K9me2, and histone deacetylation are associated with gene repression [3]. In this model, histone biotinylation marks are created occasionally at HCS docking sites.
Supplementary Material
Radiolabel in [3H]biotin and carboxyl-labeled [14C]biotin can be lost during β-oxidation.
Synthetic peptides H4K8bio and H4K8ac were probed with streptavidin (lanes 1 and 2) and anti-biotin (lanes 3 and 4). Equal loading and transfer of peptides was confirmed using Ponceau stain (lanes 5 and 6). Abbreviations: B = anti-biotin; P = Ponceau stain; SA = streptavidin.
Highlights.
Biotinylation is a natural, albeit rare, modification of histones.
Less than 0.001% of histones H3 and H4 are biotinylated.
The effects of biotin are mediated by HCS/protein interactions.
Acknowledgments
A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945, DK077816, DK082476 and ES015206, USDA CSREES grant 2006-35200-17138, and by NSF grants MCB 0615831 and EPS 0701892.
Abbreviations
- ACC
acetyl-CoA carboxylase
- HCS
holocarboxylase synthetase
- H3K9ac
histone H3, acetylated at lysine-9
- H3K4bio
histone H3, biotinylated at lysine-4
- H3K9bio
biotinylated at lysine-9
- H3K18bio
biotinylated at lysine-18
- H3K4me3
histone H3, trimethylated at lysine-4
- H3K9me2
histone H3, dimethylated at lysine-9
- H3K9me3
histone H3, trimethylated at lysine-9
- H4K12bio
histone H4, biotinylated at lysine-12
- HPLC
high-performance liquid chromatography
- K
lysine
- MCC
3-methylcrotonyl-CoA carboxylase
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- PC
pyruvate carboxylase
- PCC
propionyl-CoA carboxylase
- TAU-PAGE
Triton-Acid-Urea gel electrophoresis
- TPBS
0.05% Tween-20 in PBS
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolffe A. Chromatin. 3. Academic Press; San Diego, CA: 1998. [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2007. pp. 191–209. [Google Scholar]
- 4.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 5.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 6.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 7.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 8.Narang MA, Dumas R, Ayer LM, Gravel RA. Reduced histone biotinylation in multiple carboxylase deficiency patients: a nuclear role for holocarboxylase synthetase. Hum Mol Genet. 2004;13:15–23. doi: 10.1093/hmg/ddh006. [DOI] [PubMed] [Google Scholar]
- 9.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–2742. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–408. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, Klinkebiel D, Christman JK, Zempleni J. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J Nutr. 2008;138:2316–2322. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao B, Pestinger V, HYI, Borgstahl GEO, Kolar C, Zempleni J. Holocarboxylase synthetase is a chromatin protein and interacts directly with histone H3 to mediate biotinylation of K9 and K18. J Nutr Biochem. 2011;22:470–475. doi: 10.1016/j.jnutbio.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–315. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–4259. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011;22:328–333. doi: 10.1016/j.jnutbio.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007;18:760–768. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J Nutr Biochem. 2010;21:310–316. doi: 10.1016/j.jnutbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Healy S, Perez-Cadahia B, Jia D, McDonald MK, Davie JR, Gravel RA. Biotin is not a natural histone modification. Biochim Biophys Acta. 2009;1789:719–733. doi: 10.1016/j.bbagrm.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J Nutr. 1995;125:941–946. doi: 10.1093/jn/125.4.941. [DOI] [PubMed] [Google Scholar]
- 22.Zempleni J, Helm RM, Mock DM. In vivo biotin supplementation at a pharmacologic dose decreases proliferation rates of human peripheral blood mononuclear cells and cytokine release. J Nutr. 2001;131:1479–1484. doi: 10.1093/jn/131.5.1479. [DOI] [PubMed] [Google Scholar]
- 23.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–892. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 24.Camporeale G, Zempleni J. Biotin. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. Vol. 1. International Life Sciences Institute; Washington, D.C: 2006. pp. 314–326. [Google Scholar]
- 25.Brady RN, Ruis H, McCormick DB, Wright LD. Bacterial degradation of biotin. Catabolism of 14C-biotin and its sulfoxides. J Biol Chem. 1966;241:4715–4721. [PubMed] [Google Scholar]
- 26.Green NM. Avidin, Adv Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. [DOI] [PubMed] [Google Scholar]
- 27.Healy S, Heightman TD, Hohmann L, Schriemer D, Gravel RA. Nonenzymatic biotinylation of histone H2A. Protein Sci. 2009;18:314–328. doi: 10.1002/pro.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takechi R, Taniguchi A, Ebara S, Fukui T, Watanabe T. Biotin deficiency affects the proliferation of human embryonic palatal mesenchymal cells in culture. J Nutr. 2008;138:680–684. doi: 10.1093/jn/138.4.680. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Melendez R, Cano S, Mendez ST, Velazquez A. Biotin regulates the genetic expression of holocarboxylase synthetase and mitochondrial carboxylases in rats. J Nutr. 2001;131:1909–1913. doi: 10.1093/jn/131.7.1909. [DOI] [PubMed] [Google Scholar]
- 30.Zempleni J, Mock DM. Uptake and metabolism of biotin by human peripheral blood mononuclear cells. Am J Physiol Cell Physiol. 1998;275:C382–C388. doi: 10.1152/ajpcell.1998.275.2.C382. [DOI] [PubMed] [Google Scholar]
- 31.Boulikas T. At least 60 ADP-ribosylated variant histones are present in nuclei from dimethylsulfate-treated and untreated cells. EMBO J. 1988;7:57–67. doi: 10.1002/j.1460-2075.1988.tb02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boulikas T. DNA strand breaks alter histone ADP-ribosylation. Proc Natl Acad Sci USA. 1989;86:3499–3503. doi: 10.1073/pnas.86.10.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filenko NA, Kolar C, West JT, Hassan YI, Borgstahl GEO, Zempleni J, Lyubchenko YL. The role of histone H4 biotinylation in the structure and dynamics of nucleosomes. PLoS ONE. 2011;6:e16299. doi: 10.1371/journal.pone.0016299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh S. Physiology, regulation, and pathogenesis of nitrogen metabolism in opportunistic fungal pathogen Candida albicans. In: Nickerson K, editor. PhD thesis. School of Biological Sciences, University of Nebraska-Lincoln; Lincoln, NE: 2009. advisor. [Google Scholar]
- 35.Zhang L, Eugeni EE, Parthun MR, Freitas MA. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma. 2003;112:77–86. doi: 10.1007/s00412-003-0244-6. [DOI] [PubMed] [Google Scholar]
- 36.Xue J, Zempleni J. Experimental Biology 2011. Washington, DC: 2011. Epigenetic synergies between methylation of cytosines and biotinylation of histones in gene repression. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Radiolabel in [3H]biotin and carboxyl-labeled [14C]biotin can be lost during β-oxidation.
Synthetic peptides H4K8bio and H4K8ac were probed with streptavidin (lanes 1 and 2) and anti-biotin (lanes 3 and 4). Equal loading and transfer of peptides was confirmed using Ponceau stain (lanes 5 and 6). Abbreviations: B = anti-biotin; P = Ponceau stain; SA = streptavidin.