Abstract
Background
Hypohidrotic ectodermal dysplasia (HED) is a congenital disorder characterized by sparse hair, oligodontia, and inability to sweat. It is caused by mutations in any of three Eda pathway genes: ectodysplasin (Eda), Eda receptor (Edar), and Edar-associated death domain (Edaradd), which encode ligand, receptor, and intracellular adaptor molecule, respectively. The Eda signaling pathway activates NF-κB, which is central to ectodermal differentiation. Although the causative genes and the molecular pathway affecting HED have been identified, no curative treatment for HED has been established. Previously, we found a rat spontaneous mutation that caused defects in hair follicles and named it sparse-and-wavy (swh). Here, we have established the swh rat as the first rat model of HED and successfully identified the swh mutation.
Results
The swh/swh rat showed sparse hair, abnormal morphology of teeth, and absence of sweat glands. The ectoderm-derived glands, meibomian, preputial, and tongue glands, were absent. We mapped the swh mutation to the most telomeric part of rat Chr 7 and found a Pro153Ser missense mutation in the Edaradd gene. This mutation was located in the death domain of EDARADD, which is crucial for signal transduction and resulted in failure to activate NF-κB.
Conclusions
These findings suggest that swh is a loss-of-function mutation in the rat Edaradd and indicate that the swh/swh rat would be an excellent animal model of HED that could be used to investigate the pathological basis of the disease and the development of new therapies.
Background
Hypohidrotic ectodermal dysplasia (HED) is a genetic disorder characterized by sparse hair, oligodontia, reduced sweating, and defects in a number of other ectodermal organs [1]. A lack of sweat glands can lead to recurrent severe overheating. Thus, children with HED are at substantial risk of sudden death in infancy due to fatal hyperpyrexia [2].
HED is caused by mutations in any of the three Eda pathway genes: ectodysplasin (Eda) [3,4], ED receptor (Edar) [5], and EDAR-associated death domain (Edaradd) [6]. They encode the ligand, receptor, and intracellular signal mediator of a single linear pathway, respectively. The Eda signaling pathway activates transcription factor NF-κB thereby playing an important role in embryonic development, especially in the development of ectodermally derived organs [1].
In humans, there are three types of HED with different inheritance: X-linked HED, autosomal dominant HED, and autosomal recessive HED. X-linked HED is the most common form of HED and is caused by mutations in EDA. Autosomal HED is caused by mutations in EDAR or EDARADD. Currently, over 100 different mutations in the EDA gene are known, while only ~20 and 4 causative mutations have been found in EDAR and EDARADD, respectively [7].
To date, four mouse models of HED are available: Tabby, downless, Sleek, and crinkled. The mutant phenotype of the Tabby mouse is inherited in an X-linked manner and the Tabby mouse carries a mutation in the Eda gene [4]. The recessive downless and dominant Sleek mice carry mutations in the Edar gene [8]. The crinkled mouse carries a mutation in the Edaradd gene [6]. The phenotypes in Eda, Edar, and Edaradd mutant mice are almost identical and include abnormalities in teeth, hair, and sweat glands, the triad of symptoms of HED. Over 20 different glands, including lacrimal, meibomian, salivary, submandibular, and mammary glands, are also affected [9-11]. These mutant mice have been used to study the roles of the Eda pathway in the development and morphogenesis of ectoderm-derived organs and to develop a novel treatment for HED using a recombinant EDA protein [12].
Mutations in some of the genes in the Eda pathway have been identified in various species, such as medaka [13], zebrafish [14], cattle [15-18], and dog [19]. Analyses of these mutations showed critical roles of the Eda pathway in the development of epithelial appendages, as well as in morphological evolution. Thus, the identification of novel mutations in different species emphasized the importance of the Eda pathway, and enabled the phenotypes of the mutated animals to be compared, giving new insights into the functions of the Eda pathway. If such novel mutations can be identified in mammals, then the affected species could be used as a disease model of HED.
In a previous study, we described a mutant rat, sparse and wavy hair (swh), which arose spontaneously in a colony of inbred WTC rats in 1998 [20]. The mutant phenotype is characterized by sparse and wavy hair, impaired body weight gain, and hypoplasticity of the mammary gland. The hair follicles in these rats were reduced both in number and size, a characteristic associated with hypoplasia of both the sebaceous glands and the subcutaneous fat tissues. The mammary glands of swh/swh female rats were hypoplastic and differentiation of mammary epithelial and myoepithelial cells was impaired. Thus, it is conceivable that the swh/swh rat will provide a good experimental model to clarify the mechanisms involved in the development of skin appendages, most of which are derived from ectoderm [20].
In our previously reported linkage analysis, swh mapped to the telomeric part of rat Chr 17. At that time, the physical location of the swh locus could not be accurately determined because a SSLP marker, D17Rat140, which defined the distal side of the swh locus was, in the earlier public rat genome linkage map, erroneously assigned to the middle part of Chr 17 and not to the telomeric part of Chr 17. Recently, with the development of more than 20,000 single nucleotide polymorphism (SNP) markers for 167 rat inbred strains and with the haplotype mapping data from the genotyping of these SNPs, the genome linkage map has been improved [21]. In the improved rat genome map, D17Rat140 and its neighboring genes are correctly mapped to the telomeric part of rat Chr 17. Thus, in addition to the 24 candidate genes selected from our previous linkage analysis, we also considered these newly mapped genes to be candidates of swh [20].
In this study, to demonstrate the suitability of the swh rat as an HED model, we investigated the pathology of tissues and organs in which morphological abnormalities in HED are known to occur. Furthermore, we identified the causative mutation of the swh phenotype using a positional cloning approach, and found a missense mutation in the death domain of EDARADD, that might explain the inability of the mutant Edaradd gene to activate NF-κB. Our findings suggest that swh is a loss-of-function mutation of the rat Edaradd and support the swh/swh rat as an excellent animal model of HED that can be used to investigate the pathological basis of the disease and to develop new therapies.
Methods
Animals
ACI/NKyo, WTC/Kyo, and WTC-swh/Kyo rats were provided by the Japanese National BioResource Project for the Rat and kept in our animal facility for all experiments in this study. Animal care and experimental procedures were approved by the Animal Research Committee, Kyoto University, Japan, and were conducted according to the Regulation on Animal Experimentation at Kyoto University.
Histopathology
For light microscopy, the tongue, eyelid, ventral skin, footpad, and preputial gland were harvested from WTC-swh/swh and WTC rats at 8 weeks of age. Tissues were fixed in 10% neutral-buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin (HE).
Sweat tests and whole mount staining of mammary glands
The sweat test was performed as described previously [12]. Briefly, the hind paws of rats anesthetized with sevoflurane were painted with a solution of 3% (wt/vol) iodine in ethanol. Once dry, the paws were painted with a suspension of 40% (wt/vol) starch I mineral oil. Photographs were taken 1 min later and sweat was detected as dark spots. Mammary glands were prepared as a whole mount and stained as described previously [22].
Fine mapping of swh
For fine mapping of swh, F2 animals (n = 769) were produced by intercrossing (ACI/NKyo × WTC-swh) F1 rats. Homozygous swh/swh animals were identified at 3-4 weeks of age based on the appearance of the sparse-and-waved hair phenotype. One hundred and ninety-eight swh/swh homozygotes were used for fine mapping of swh. Genomic DNA was prepared from tail biopsies using the automatic DNA purification system (PI-200; Kurabo, Japan).
RNA extraction, RT-PCR and direct sequencing
Total RNA was extracted from the skin of 2-week-old animals. RNA preparation, RT-PCR and direct sequencing of PCR products were performed as described previously [23]. Rat Edaradd cDNAs were amplified with 6 sets of primers (Table 1). The PCR products overlapped each other and spanned the entire coding sequence of Edaradd.
Table 1.
PCR primers used to amplify rat Edaradd cDNA
| Primer set | Forward (5' > 3') | Reverse (5' > 3') |
|---|---|---|
| Edaradd-1&2 | CTGAGAGAGAGTCGCGCATT | GCCACAGCTGTTCCCATAG |
| Edaradd-3&4 | GCCCAGAAAAGGCAGCTC | GGAAAACCTTTGGAGTTTCTGA |
| Edaradd-5&6 | CGATGAGCCAGCTTTACCTC | GGATAATTGGGTAACTATTCTCAACC |
| Edaradd-7&8 | TCCATCCCAATTTTACCAACA | CGGCAAGCATTTTAATGACC |
| Edaradd-9&10 | CAGTCAGCCCCTTGCACT | GCATGCTCTCATCAACATGG |
| Edaradd-11&12 | TGTCACCAATGTGGTAGAAAAA | CAGGGATAACCACTGCCTGT |
Transient transfection and reporter assays
The NF-κB assay was designed to test for activation of the NF-κB responsive promoter. HEK293T cells grown in poly-L-lysine coated 24-well plates were transfected using SuperFect (Qiagen) with 1.2 μg pNF-κB-Luc (Clontech), 2 μg pRL-TK, and an increasing amount of expression vectors encoding the wild-type EDARADD or the swh-type EDARADD (Pro153Ser). The Luc reporter of the pNF-κB-Luc encodes firefly luciferase. The HSV-TK (herpes simplex virus thymidine kinase) promoter drives renilla luciferase in pRL-TK. Total DNA was adjusted to 2.6 μg by adding pCMV-HA (Clontech) vector as necessary. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) 48 h after transfection, according to the manufacturer's protocol.
Results
Phenotypes of swh/swh rat as hypohidrotic ectodermal dysplasia (HED)
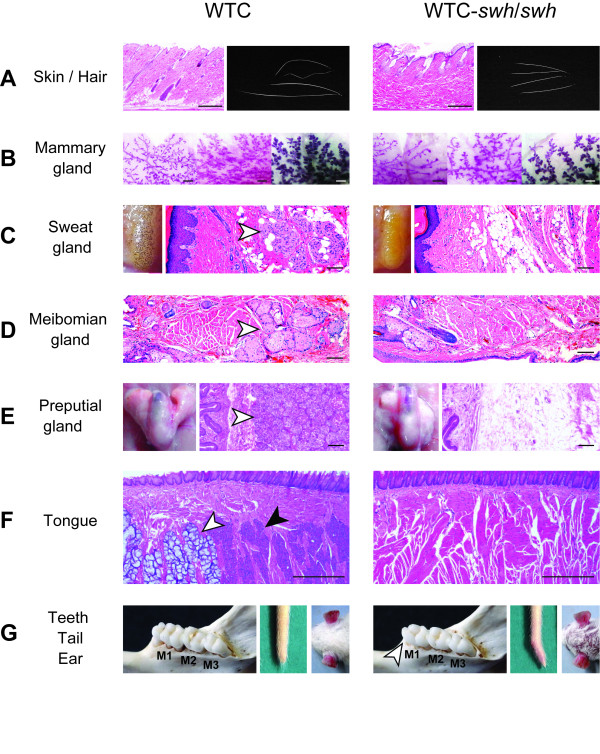
Patients with HED display defective development of hair, teeth, sweat glands, and several exocrine glands, such as sebaceous, salivary, meibomian, and lacrimal [1,24]. To evaluate the relevance of the swh/swh rat as a HED model, we looked for developmental defects in those tissues of swh/swh rats. In addition to defects of the hair, skin, and mammary glands, which have been reported previously [20] (Figure 1A, B), we found defects in the sweat, meibomian, preputial, and tongue glands. In these tissues, the exocrine glands were absent in the swh/swh rats (Figure 1C, D, E, F). In the sweat test, no sweat was detected in swh/swh rats, indicating that the sweat glands were functionally defective (Figure 1C). We also found a reduced number of cusps in the lower first molars in the swh/swh rats (Figure 1G).
Figure 1.
Phenotypes of the swh/swh rat as hypohidrotic ectodermal dysplasia (HED). A, Sections of the dorsal skin (left) and hair (right). Incomplete hair follicles are evident in swh/swh rat. Scale bar, 0.5 mm. The WTC rat has four hair types; auchene, zigzag, awl and guard, while the swh/swh rat have only the abnormal awl hair. B, Whole mount stained mammary glands; 6-week-old (left), 8-week-old (center), and pregnant day 9 (right). Mammary gland branching is poor in swh/swh rat. Scale bar, 1 mm. C, Sweat test results (left) and section of the footpads. Sweat, detected as dark spots, is not seen in swh/swh rat. Sweat glands (arrowhead) are present in WTC rat and absent in swh/swh rat. Scale bar, 100 μm. D, Sections of the eyelid. The meibomian glands (arrowhead) are present in WTC rat and absent in swh/swh rat. Scale bar, 100 μm. E, An entire view (left) and a section of the preputial gland (right). The preputial gland is atrophied in male swh/swh rat. Acinous glands (arrowhead) are present in WTC rat and absent in swh/swh rat. Scale bar, 100 μm. F, Section of the tongue. Both mucous (open arrowhead) and serous (filled arrowhead) glands are present in WTC rat and neither is seen in swh/swh rat. Scale bar, 0.5 mm. G, Buccal views of lower molars (left), tip of tail (center), and posterior auricular region (right). Cusp number is reduced in the first molar (arrow head) in swh/swh rat. Some swh/swh rats show the kink tail. The bald patch behind the ear was not evident in the swh/swh rat.
In the Eda pathway mutant mice, Tabby, downless, and crinkled, a kinked tail tip, a bald patch behind the ear, and abnormal pelage hair composition are characteristic. Similarly, in swh/swh rat, the pelage hair was composed of only an abnormal awl hair (Figure 1A); however, the tail had hair on it, the frequency of kinked tail was low, and the bald patch behind the ear was not found (Figure 1G).
These findings indicate that the mutant phenotypes of swh/swh rats are similar to developmental defects in HED patients and in the established mouse models; therefore, it is likely that the swh/swh rat will be suitable as a model of HED.
Positional cloning of swh
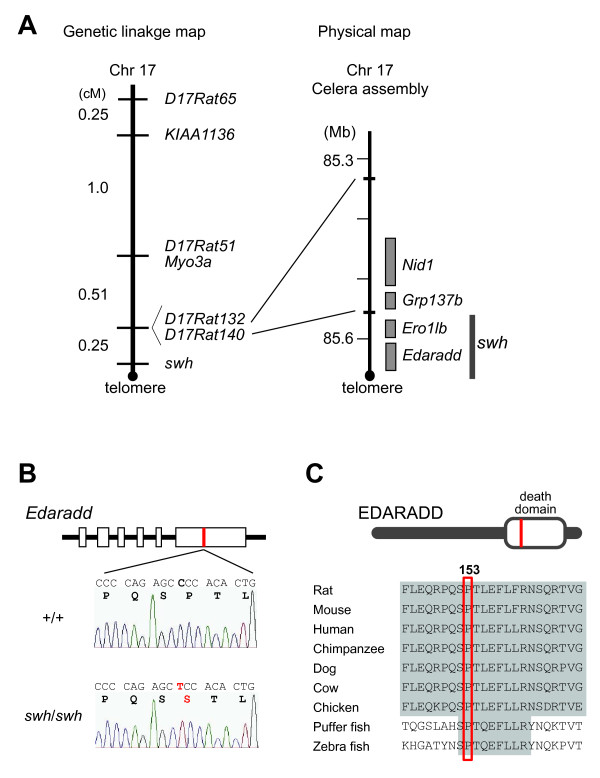
In a previous study, we mapped swh to rat Chr 17 [20]. To more specifically map the position of the swh locus, we genotyped F2 intercross progeny for markers known to be closely linked to swh. There was only one recombinant chromosome between swh and either D17Rat132 or D17Rat140 in 396 meioses (= 198 × 2) and we were able to map swh to the most distal part of Chr17 (Figure 2A). The rat genome map (RGSC v3.4) showed two genes in the swh locus, Ero1lb (ERO1-like beta (S. cerevisiae)) and Edaradd (ectodysplasin-A receptor-associated death domain). The mouse mutant of Edaradd is called crinkled (cr) and mice that carry this mutation show a sparse hair phenotype that is similar to that of the swh rat [25]. Additionally, mutations in the human EDARADD gene have been found in families affected with HED [6,26]. Thus, we considered Edaradd as a good candidate of swh. Although the abnormal expression of Edaradd mRNA was not detected in the skin of swh/swh rats (data not shown), we found a missense mutation (C to T) in exon 6 of the swh/swh Edaradd gene. This mutation was deduced to change proline to serine at the 153rd amino acid (Pro153Ser) of the rat EDARADD protein (Figure 2B). The 153rd amino acid is located in the death domain of EDARADD and is highly conserved in vertebrates (Figure 2C). These findings suggest that the Pro153Ser missense mutation of the Edaradd gene is causative of the phenotypes of swh/swh rats.
Figure 2.
Identification of the rat swh mutation. A, Fine mapping of swh (left) and physical mapping of swh (right). The swh genetically mapped to the most telomeric part of rat Chr17, 0.25-cM distal from D17Rat132 and D17Rat140. In the physical map, the swh locus is localized to a ~0.2-Mb region between D17Rat140 and the telomere. Both Ero1lb and Edaradd have been mapped within the swh locus. B, Sequence analysis of Edaradd gene of wild-type and swh/swh rats. In the genomic DNA of swh/swh rat, a C to T (red) transition is present in exon 6 of rat Edaradd gene. This changes proline to serine at codon 153 of the deduced EDARADD protein. Rat codon 153 corresponds to codon 156 of mouse EDARADD isoform 1 (NP_598398) and codon 153 human EDARADD isoform B (NP_542776). C, Amino-acid sequence alignment of a region of the EDARADD death domain from different species. The 153rd amino acid that is altered in swh/swh rat is highly conserved in the vertebrates.
Reporter assay for the Pro153Ser mutant EDARADD
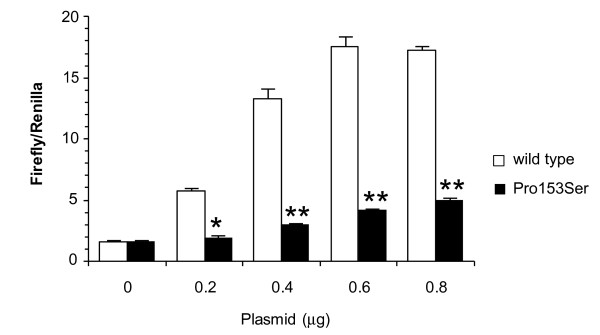
Overexpression of Edaradd in 293T cells activates NF-κB in a dose-dependent manner [25]. To examine whether Pro153Ser Edaradd can activate NF-κB, we carried out a reporter assay. As shown in Figure 3, wild-type Edaradd activated NF-κB in a dose-dependent manner. Meanwhile, Pro153Ser Edaradd showed significantly lower transcriptional activity of NF-κB than the wild type. The expression level of the Pro153Ser EDARADD protein detected by western blotting was not different from that of the wild type (data not shown). These findings indicate that the Pro153Ser missense mutation of the rat Edaradd gene could not activate NF-κB and that the Eda signaling pathway failed to function in swh/swh rats.
Figure 3.
Loss of NF-κB activation by the Pro153Ser Edaradd mutant protein. 293T cells were transfected with 1.2 μg pNF-κB-luciferase reporter gene plasmid, 2 μg pRL-TK, and the amounts of each expression construct were measured in a reporter assay. The expression levels of luciferase were normalized to those of the internal control. Relative NF-κB activity in wild-type transfected cells increased in a dose-dependent manner, while significantly lower relative NF-κB activity was observed in the Pro153Ser Edaradd-transfected cells. *P < 0.01, **P < 0.001.
Discussion
In this study, we demonstrated that the swh/swh rat harbored a Pro153Ser mutation in the Edaradd gene and showed typical symptoms of HED, such as sparse hair, oligodontia, inability to sweat, and developmental defects of the ectoderm-derived glands [27]. Hence, we successfully established the swh/swh rat as a genetically and phenotypically well-characterized disease model of HED.
EDARADD is a 208 amino acid protein consisting of an N-terminal Tnf receptor-associated factor (Traf)-binding consensus sequence and a C-terminal death domain (DD). The Traf-binding consensus sequence of EDARADD is used as a docking site for Traf1, Traf2, and Traf3, thereby recruiting Traf members and leading eventually to the activation of NF-κB [6]. The DD is involved in self-association of EDARADD and its interaction with EDAR [6,25]. Thus, EDARADD is central to Edar signaling. The N-terminal region is responsible for signal transduction and the C-terminal DD is required for receptor engagement.
To date, four EDARADD mutations have been found in a subset of human HED, one leads to autosomal dominant inheritance (Leu112Arg) [26], while the others lead to autosomal recessive inheritance (Glu142Lys, Pro121Ser, and Thr135-Val136del) [6,28,29]. All of these mutations are located in the DD and functional analyses showed that they resulted in the failure of EDARADD to interact with EDAR and to activate NF-κB. In the crinkled mouse, a genomic region of ~66-kb or more which includes exon 6 that encodes the entire DD, is deleted [25]. The crinkled mouse displays developmental defects in hair follicles, teeth, and sweat glands [30,31]. Hence, it is possible that a mutation in the DD of EDARADD is necessary for the HED syndrome to be manifested both in human and mouse.
All members of the DD superfamily form a highly compact structure comprising six antiparallel α-helix that is involved in homotypic and heterotypic protein-protein complex formation [32]. The region spanning the α1 to α4 helices of the DD of MyD88, a member of the death receptor superfamily, is required for its interaction with a downstream kinase [33]. A comparison of the amino acid sequences of the DD superfamily revealed that the Pro153Ser missense mutation found in the present study is located in the α4 helix of the DD of EDARADD. This mutation may cause a profound change in the polarity of a crucial region and eventually diminish NF-κB signaling. It is likely that Pro153Ser affects the structure of the DD thereby interfering in the interaction of EDARADD with EDAR.
Mutations affecting the Eda pathway are known in medaka [13], zebrafish [14], mouse [4,6,8], cattle [15-18], dog [19], and human [3,5,6]. Of them, the mouse mutants have been widely characterized as a model organism of HED. Here we report the swh mutation as the first example of a mutation in the Eda pathway in the rat.
Because the rat is closely related to the mouse, it is important to recognize how the rat Edaradd mutant phenotype matches the mouse Eda pathway mutant phenotypes. Similar to the mouse mutants, the swh/swh rat displayed sparse hair, misshapen teeth, and absence of sweating. Additionally, like the Eda pathway mutant, the swh/swh rat had only abnormal awl hair in the coat. The swh/swh rat showed a lack of the ectoderm-derived glands, meibomian, preputial, and tongue. Interestingly, both serous and mucous glands were absent in the tongue of the swh/swh rat. This is a clear difference from the mouse Eda pathway mutants that lacked mucous glands but had serous glands in the tongue [34]. Moreover, in contrast to the complete absence of tail hair in the Eda pathway mutant mice, the swh/swh rat had hair on its tail. The penetrance of the kink tail phenotype was low in the swh/swh rat, while almost all Eda pathway mutant mice showed the kink tail. Lastly, the bald patch behind the ear was not present in the swh/swh rat, although it was a very characteristic phenotype of the Eda pathway mutant mice.
Why these phenotypes are different between the Eda pathway mutant mice and the swh/swh rats is yet to be explained. However, different types of mutations could possibly explain the differences. The mouse crinkled mutation is a deletion [6], while the swh mutation is missense. Although the Luc-reporter assay strongly suggested that swh is a null mutation, the possibility that swh might be a hypomorphic mutation cannot be eliminated because the activation of NF-κB found in the assay was very low. In the Eda pathway mutant mice, the mammary, salivary and tracheal submucosal glands have been well characterized [9,10]. Further analyses of these glands in swh/swh rats will give further insights into the functions of the Eda pathway genes in the development of these glands.
Conclusions
We successfully established the swh/swh rat as the first rat model of HED and identified swh as a Pro135Ser missense mutation in the Edaradd gene. The Pro135Ser mutant protein failed to activate NF-κB in the Eda signaling pathway. Thus, the swh/swh rat is a good model that can be used to investigate the pathological basis of HED.
Authors' contributions
TK and MY performed the genetic and molecular biological experiments. RH and HH performed the histological examinations. TK wrote the paper and HH and TS revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Takashi Kuramoto, Email: tkuramot@anim.med.kyoto-u.ac.jp.
Mayuko Yokoe, Email: myokoe@anim.med.kyoto-u.ac.jp.
Ryoko Hashimoto, Email: g_remus03@hotmail.com.
Hiroshi Hiai, Email: hiaih@anim.med.kyoto-u.ac.jp.
Tadao Serikawa, Email: serikawa@anim.med.kyoto-u.ac.jp.
Acknowledgements and Funding
The authors are grateful to the National BioResource Project for the Rat for providing the ACI/NKyo, WTC/Kyo, and WTC-swh/Kyo rat strains. This work was supported in part by the Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (21300153 to TK) and by a Grant-in-aid for Cancer Research from the Ministry of Health, Labour and Welfare (to TK).
References
- Mikkola ML, Thesleff I. Ectodysplasin signaling in development. Cytokine Growth Factor Rev. 2003;14(3-4):211–224. doi: 10.1016/S1359-6101(03)00020-0. [DOI] [PubMed] [Google Scholar]
- Salisbury DM, Stothers JK. Hypohidrotic ectodermal dysplasia and sudden infant death. Lancet. 1981;1(8212):153–154. doi: 10.1016/s0140-6736(81)90736-4. [DOI] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, Ferguson B, Munoz F, Morgan D, Clarke A, Baybayan P. et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13(4):409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, Du Y, Ezer S, Jenks T, Shimada T, Pekkanen M, Mikkola ML, Ko MS. et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc Natl Acad Sci USA. 1997;94(24):13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22(4):366–369. doi: 10.1038/11937. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, Tucker AS, Justice MJ, Sharpe PT, Zonana J, Overbeek PA. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414(6866):913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. Molecular aspects of hypohidrotic ectodermal dysplasia. Am J Med Genet A. 2009;149A(9):2031–2036. doi: 10.1002/ajmg.a.32855. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Overbeek PA. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat Genet. 1999;22(4):370–374. doi: 10.1038/11943. [DOI] [PubMed] [Google Scholar]
- Chang SH, Jobling S, Brennan K, Headon DJ. Enhanced Edar signalling has pleiotropic effects on craniofacial and cutaneous glands. PLoS One. 2009;4(10):e7591. doi: 10.1371/journal.pone.0007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick M, Phair RD, Lapidot SA, Jaskoll T. Salivary gland branching morphogenesis: a quantitative systems analysis of the Eda/Edar/NFκB paradigm. BMC Dev Biol. 2009;9:32. doi: 10.1186/1471-213X-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg H. The glandular aspects of the tabby syndrome in the mouse. J Embryol Exp Morphol. 1971;25(1):1–19. [PubMed] [Google Scholar]
- Gaide O, Schneider P. Permanent correction of an inherited ectodermal dysplasia with recombinant EDA. Nat Med. 2003;9(5):614–618. doi: 10.1038/nm861. [DOI] [PubMed] [Google Scholar]
- Kondo S, Kuwahara Y, Kondo M, Naruse K, Mitani H, Wakamatsu Y, Ozato K, Asakawa S, Shimizu N, Shima A. The medaka rs-3 locus required for scale development encodes ectodysplasin-A receptor. Curr Biol. 2001;11(15):1202–1206. doi: 10.1016/S0960-9822(01)00324-4. [DOI] [PubMed] [Google Scholar]
- Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nusslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 2008;4(10):e1000206. doi: 10.1371/journal.pgen.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C, Distl O, Leeb T. Partial deletion of the bovine ED1 gene causes anhidrotic ectodermal dysplasia in cattle. Genome Res. 2001;11(10):1699–1705. doi: 10.1101/gr.182501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemuller C, Peters M, Pohlenz J, Distl O, Leeb T. A single point mutation within the ED1 gene disrupts correct splicing at two different splice sites and leads to anhidrotic ectodermal dysplasia in cattle. J Mol Med (Berl) 2002;80(5):319–323. doi: 10.1007/s00109-002-0320-z. [DOI] [PubMed] [Google Scholar]
- Ogino A, Kohama N, Ishikawa S, Tomita K, Nonaka S, Shimizu K, Tanabe Y, Okawa H, Morita M. A novel mutation of the bovine EDA gene associated with anhidrotic ectodermal dysplasia in Holstein cattle. Hereditas. 2011;148(1):46–49. doi: 10.1111/j.1601-5223.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- Gargani M, Valentini A, Pariset L. A novel point mutation within the EDA gene causes an exon dropping in mature RNA in Holstein Friesian cattle breed affected by X-linked anhidrotic ectodermal dysplasia. BMC Vet Res. 2011;7:35. doi: 10.1186/1746-6148-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal ML, Scheidt JL, Rhodes JL, Henthorn PS, Werner P. Mutation identification in a canine model of X-linked ectodermal dysplasia. Mamm Genome. 2005;16(7):524–531. doi: 10.1007/s00335-004-2463-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto T, Morimura K, Nomoto T, Namiki C, Hamada S, Fukushima S, Sugimura T, Serikawa T, Ushijima T. Sparse and wavy hair: a new model for hypoplasia of hair follicle and mammary glands on rat chromosome 17. J Hered. 2005;96(4):339–345. doi: 10.1093/jhered/esi053. [DOI] [PubMed] [Google Scholar]
- Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P. et al. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet. 2008;40(5):560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild TC, Boylan ES, Calhoon RE, Vonderhaar BK. Transplacental effects of diethylstilbestrol on mammary development and tumorigenesis in female ACI rats. Cancer Res. 1987;47(16):4508–4516. [PubMed] [Google Scholar]
- Kuramoto T, Kuwamura M, Tokuda S, Izawa T, Nakane Y, Kitada K, Akao M, Guenet JL, Serikawa T. A mutation in the gene encoding mitochondrial Mg2+ channel MRS2 results in demyelination in the rat. PLoS Genet. 2011;7(1):e1001262. doi: 10.1371/journal.pgen.1001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed WB, Lopez DA, Landing B. Clinical spectrum of anhidrotic ectodermal dysplasia. Arch Dermatol. 1970;102(2):134–143. doi: 10.1001/archderm.102.2.134. [DOI] [PubMed] [Google Scholar]
- Yan M, Zhang Z, Brady JR, Schilbach S, Fairbrother WJ, Dixit VM. Identification of a novel death domain-containing adaptor molecule for ectodysplasin-A receptor that is mutated in crinkled mice. Curr Biol. 2002;12(5):409–413. doi: 10.1016/S0960-9822(02)00687-5. [DOI] [PubMed] [Google Scholar]
- Bal E, Baala L, Cluzeau C, El Kerch F, Ouldim K, Hadj-Rabia S, Bodemer C, Munnich A, Courtois G, Sefiani A. et al. Autosomal dominant anhidrotic ectodermal dysplasias at the EDARADD locus. Hum Mutat. 2007;28(7):703–709. doi: 10.1002/humu.20500. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262(2):195–205. doi: 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Chassaing N, Cluzeau C, Bal E, Guigue P, Vincent MC, Viot G, Ginisty D, Munnich A, Smahi A, Calvas P. Mutations in EDARADD account for a small proportion of hypohidrotic ectodermal dysplasia cases. Br J Dermatol. 2010;162(5):1044–1048. doi: 10.1111/j.1365-2133.2010.09670.x. [DOI] [PubMed] [Google Scholar]
- Suda N, Bazar A, Bold O, Jigjid B, Garidkhuu A, Ganburged G, Moriyama K. A Mongolian patient with hypohidrotic ectodermal dysplasia with a novel P121S variant in EDARADD. Orthod Craniofac Res. 2010;13(2):114–117. doi: 10.1111/j.1601-6343.2010.01484.x. [DOI] [PubMed] [Google Scholar]
- Kindred B. The expression of the Tabby and crinkled genes in different genetic backgrounds in the mouse. Genetics. 1967;55(1):173–178. doi: 10.1093/genetics/55.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Jaszczak E, Landis SC. Innervation of footpads of normal and mutant mice lacking sweat glands. J Comp Neurol. 1994;346(4):613–625. doi: 10.1002/cne.903460412. [DOI] [PubMed] [Google Scholar]
- Weber CH, Vincenz C. The death domain superfamily: a tale of two interfaces? Trends Biochem Sci. 2001;26(8):475–481. doi: 10.1016/S0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- Loiarro M, Gallo G, Fanto N, De Santis R, Carminati P, Ruggiero V, Sette C. Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J Biol Chem. 2009;284(41):28093–28103. doi: 10.1074/jbc.M109.004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells KL, Mou C, Headon DJ, Tucker AS. Defects and rescue of the minor salivary glands in Eda pathway mutants. Dev Biol. 2011;349(2):137–146. doi: 10.1016/j.ydbio.2010.10.012. [DOI] [PubMed] [Google Scholar]